Abstract
Stuttering is a common, highly heritable neurodevelopmental disorder characterized by deficits in the volitional control of speech. Whole-exome sequencing identified two heterozygous AP4E1 coding variants, c.1549G>A (p.Val517Ile) and c.2401G>A (p.Glu801Lys), that co-segregate with persistent developmental stuttering in a large Cameroonian family, and we observed the same two variants in unrelated Cameroonians with persistent stuttering. We found 23 other rare variants, including predicted loss-of-function variants, in AP4E1 in unrelated stuttering individuals in Cameroon, Pakistan, and North America. The rate of rare variants in AP4E1 was significantly higher in unrelated Pakistani and Cameroonian stuttering individuals than in population-matched control individuals, and coding variants in this gene are exceptionally rare in the general sub-Saharan West African, South Asian, and North American populations. Clinical examination of the Cameroonian family members failed to identify any symptoms previously reported in rare individuals carrying homozygous loss-of-function mutations in this gene. AP4E1 encodes the ε subunit of the heterotetrameric (ε-β4-μ4-σ4) AP-4 complex, involved in protein sorting at the trans-Golgi network. We found that the μ4 subunit of AP-4 interacts with NAGPA, an enzyme involved in the synthesis of the mannose 6-phosphate signal that targets acid hydrolases to the lysosome and the product of a gene previously associated with stuttering. These findings implicate deficits in intracellular trafficking in persistent stuttering.
Introduction
Stuttering is a common neurodevelopmental speech disorder characterized by repetitions, prolongations, and interruptions in the flow of speech.1 Although twin and adoption studies have demonstrated high heritability of this disorder,2, 3, 4, 5 Mendelian segregation does not typically occur,6 which has rendered mutation identification difficult. We have previously used consanguineous families to identify associated loci,7, 8, 9 and at one of these loci on chromosome 12, we have identified rare coding variants in GNPTAB (MIM: 607840), as well as in the functionally related genes GNPTG (MIM: 607838) and NAGPA (MIM: 607985), which are associated with stuttering in populations from North America, England, Brazil, Pakistan, and Cameroon.10 Although rare coding variants in these genes together might account for 8%–16% of cases of familial persistent stuttering,11 a large fraction of the heritable causes of stuttering remain unidentified.
We have previously reported a large polygamous kindred from Cameroon, West Africa, in which many members are affected by persistent developmental stuttering. A linkage study indicated that multiple associated genes located on chromosomes 2, 3, 14, and 15 are acting in different branches of this family.12 We performed whole-exome sequencing, which revealed that two rare coding variants in adaptor-related protein complex 4, epsilon 1 subunit (AP4E1 [MIM: 607244]) co-segregate with stuttering in 11 members of sub-pedigree E of this family, which had previously demonstrated linkage to this region of chromosome 15. Additional sequencing in unrelated stuttering individuals from three different continental populations revealed additional rare variants in this gene and motivated cell biological and clinical studies of the effects of these variants.
Material and Methods
Individuals with non-syndromic persistent developmental stuttering were enrolled with written informed consent under NIH protocol 97-DC-0057, as previously described.10, 12 Documented neurologically normal control DNA samples (NDPT; n = 368) were obtained from the National Institute of Neurological Disorders and Stroke (NINDS) panels NDPT006, NDPT020, NDPT023, NDPT079, NDPT082, and NDPT093 from the Coriell Cell Repository. Unrelated affected individuals and population-matched control individuals included Pakistani affected (PKST + PKSTR; n = 132) and control (PKNR; n = 96) individuals, Cameroonian affected (STCR + CAMST01; n = 93) and control (RC; n = 94) individuals, and North American affected individuals, including those from our NIH group (NA + NIH = 711). Stuttering was diagnosed according to the Stuttering Severity Index 3 (SSI-3) and as previously described.13, 14 Individuals were classified as affected if they displayed stuttering dysfluencies at a rate of ≥4% of syllables or words. Affected individuals were classified as unrelated by self-report, and no genotypic evidence for relatedness among these subjects was observed.
Whole-exome sequencing was performed with the Agilent SureSelect Human All Exon V4+UTRs (71 MB) exome capture kit and subsequent analysis on the AB5500 SOLiD sequencer. Dideoxy sequencing of exons and their 50-bp flanking regions was performed with an AB 3730xl instrument, and sequence traces were analyzed with DNASTAR SeqMan Pro version 9.1.0. The sequence data of random individuals were obtained from the final phase of the 1000 Genomes Project (3,068 individuals) and the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (∼6,400 individuals) and by dideoxy sequencing of population-specific matched control individuals (the 558 subjects described above). The chromosomal phase of the c.1549G>A (p.Val517Ile) and c.2401G>A (p.Glu801Lys) variants in AP4E1 was inferred from familial segregation and was measured directly by cloning of RT-PCR products covering exons 13–21 of AP4E1 mRNA from Cameroonian unrelated individuals and by subsequent sequencing of individual clones.
Clinical examinations were performed at the NIH Clinical Center and included assessment of medical history, physical examination, neurological evaluation, audiological examination, X-rays of lower limbs, and brain MRI and fMRI.
Construction of AP-4 ε subunit expression vectors involved the pGADT7-human AP-4 ε subunit construct,15 which was subjected to site-directed mutagenesis (QuickChange, Agilent) for removal of the internal XhoI site. The human ε cDNA without the internal XhoI site was subsequently amplified by PCR with primers including KpnI and XhoI sites at the 5′ and 3′ ends, respectively. The resulting PCR product was subsequently ligated into the KpnI and XhoI sites of a modified pcDNA 3.1 vector containing Strep and FLAG epitopes. This ligation generated pcDNA 3.1-TSF-ε, a construct that is based on the pcDNA 3.1/Myc-His A MCS (Invitrogen) backbone and directs the expression of human ε tagged at its N terminus with TSF (two Strep and one FLAG) epitopes. The pcDNA 3.1-TSF-ε was subjected to site-directed mutagenesis for the generation of the ε variants analyzed in this study. All engineered variants were confirmed by sequencing in both directions.
A cDNA encoding the cytosolic tail (residues 475–515) of the human N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) was amplified by PCR from the pCMV6-Entry-h NAGPA construct (Origene Technologies) with primers containing EcoRI and XhoI sites. The PCR fragment was subsequently subcloned into the corresponding sites of the Gal4-binding domain (BD) yeast two-hybrid (Y2H) vector pGBKT7 (Clontech). The p.Tyr486Ala, p.Tyr488Ala, and p.Leu491Ala substitutions were introduced by site-directed mutagenesis of the pGBKT7-NAGPA tail construct.
HEK293T cells were cultured in DMEM with high glucose supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin on 100-mm dishes at 37°C under 95:5 air:CO2. Cells were transfected with pcDNA 3.1-TSF-ε constructs at ∼30%–50% confluency at a ratio of 7 μg of DNA per 100-mm dish with the X-tremeGENE9 reagent (Roche) and Opti-MEM I Reduced Serum Medium (Life Technologies). Approximately 24 hr after transfection, cells were gently washed twice with PBS at room temperature and lysed for 30 min at 4°C in 800 μl of 50 mM Tris/HCl (pH 7.4), 75 mM NaCl, and 0.8% (v/v) Triton X-100 supplemented with protease inhibitors (EDTA-free Complete, Roche). Cell lysates were spun for 10 min at 21,000 × g and 4°C, and the supernatants were kept at −80°C.
AP-4 complex assembly was assayed with thawed lysates of HEK293T cells transfected with TSF-ε constructs that were centrifuged for 2 min at 16,000 × g and 4°C. The supernatants were incubated overnight at 4°C with 60 μl of StrepTactin Sepharose beads (IBA) that had been previously washed twice by resuspension in PBS containing 0.01% (v:v) Triton X-100 and centrifugation at 4,000 × g. Beads with immobilized complexes were washed three times by resuspension in 50 mM Tris/HCl (pH 7.4), 75 mM NaCl, and 0.1% (v/v) Triton X-100 and microfuge centrifugation (2 min at 4,000 × g and 4°C) and washed twice by resuspension in PBS followed by centrifugation. Washed beads were eluted by incubation with 50 μl of 1× d-desthiobiotin-containing elution buffer (IBA) in PBS for 20 min at 25°C and subsequently centrifuged for 2 min at 16,000 × g and room temperature. Supernatants with eluted proteins were subjected to SDS-PAGE and immunoblotting with rabbit antiserum against the C-terminal region of the β4 subunit of AP-4 (anti-β4c)16 and with mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich). Samples of the original cell lysates were also subjected to SDS-PAGE and immunoblotted with anti-FLAG antibody for analysis of the differences in the expression levels of the different recombinant ε constructs. Immunoblots were developed with horseradish-coupled secondary antibodies and Western Lightning Plus-ECL reagent (PerkinElmer). Films were analyzed by densitometry with Image J software (version 1.48v).
The constructs encoding the μ subunits of the adaptor complexes AP-1A, AP-1B, AP-2, AP-3A, AP-3B, and AP-4 subcloned into the Gal4-activation domain Y2H vector pACT2 (Clontech) have been described.17 Y2H assays were performed as previously described18 with the AH109 reporter yeast strain. Positive and negative controls for the assays are described in the Figure 3 legend.
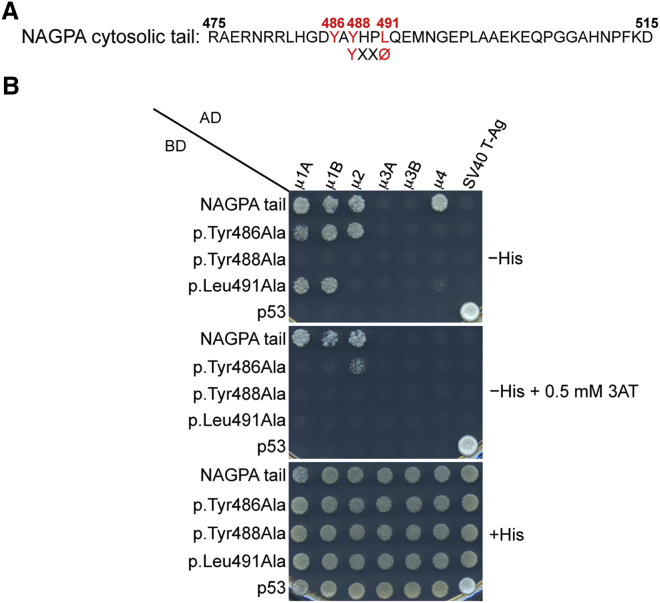
Figure 3.
The NAGPA Cytosolic Tail Interacts with μ Subunits of AP Complexes
(A) Amino acid sequence of the human NAGPA cytosolic tail shows a YXXØ motif characteristic of endocytic and lysosome-targeting signals (X is any amino acid, and Ø is a bulky hydrophobic residue). Numbering corresponds to the isoform comprising 515 residues (UniProt: Q9UK23).
(B) Y2H analysis showed that the NAGPA cytosolic tail interacts with the μ subunits of AP-1 (both the μ1A and μ1B isoforms), AP-2, and AP-4. Interactions depend on residues Tyr488 and Leu491 in the YXXØ-based sequence of the NAGPA tail and on the Tyr486 residue located immediately upstream. The NAGPA cytosolic tail was subcloned in a Y2H Gal4 BD vector, whereas the AP μ subunits were subcloned in a Gal4 AD vector. Growth on the −His plates is indicative of interactions, whereas growth on the +His plate is a control for viability and loading of all double transformants. Co-transformation of the BD-NAGPA tail constructs with AD-SV40 T-Ag and of AD-μ subunits with the BD-p53 construct provided negative controls for the assay. Co-transformation of BD-p53 with AD-SV40 T-Ag provided a positive control for interactions. Plating on −His plates containing 0.5 mM 3-amino-1,2,4 triazole (3AT) provided an assay with increased stringency conditions (3AT is a competitive inhibitor of HIS3).
Results
Stuttering in Family CAMSTO1
Cameroonian family CAMST01 has been previously described12 and contains a large fraction of individuals who have persistent stuttering with an onset at age 2–5 years. The family members evaluated in this study speak multiple languages (English, French, and/or Lamnso) and stutter in all of the languages they speak. They report that their stuttering is exacerbated when they are under stress or use the telephone and that some improvement in fluency has occurred with age. No non-stuttering speech pathologies were observed in these family members during evaluation at the NIH Clinical Center. Overall, the speech history and symptoms in all eight subjects are consistent with typical persistent developmental stuttering.
Identification of Variants in AP4E1
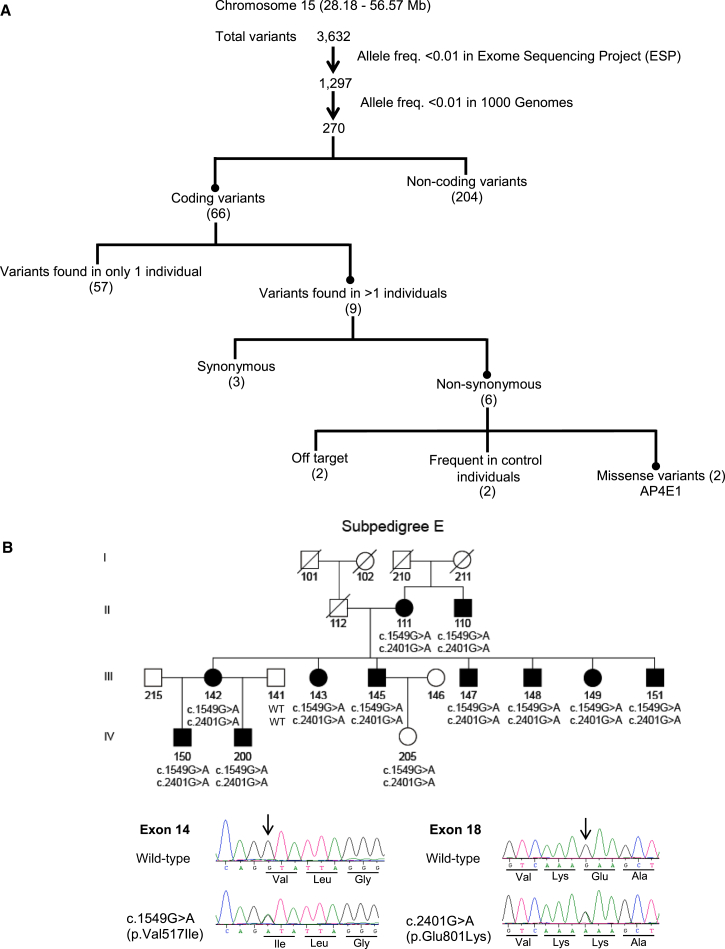
Family CAMST01 displays significant evidence of genetic linkage between persistent stuttering and multiple loci, including a discontinuous region in 15q.12 Whole-exome sequencing was performed on affected individuals II-111, III-142, III-145, III-147, III-148, and IV-200 in sub-pedigree 1E, as shown in Figure 1B, and on affected individuals III-125, IV-175, and IV-184 in sub-pedigree B, as shown in Figure S3. These sub-pedigrees displayed linkage to two closely linked but non-overlapping loci in chromosomal region 15q12 (Figure 1A). Within this region, three positional candidate genes (GANC [MIM: 104180], GATM [MIM: 602360], and GALK2 [MIM: 137028]) were also fully dideoxy sequenced. The whole-exome single-ended sequencing with 70 bp reads produced a mean coverage depth of 25.6× (range 19.0×–37.9×). Analysis using Lifescope identified a mean of 20,655 variants (range 19,604–21,708 variants) per exome. These variants were subjected to bioinformatics filtering analysis, summarized in Figure 1A. In the first step, variants outside the linkage intervals in sub-pedigrees 1E and 1B were eliminated, leaving a total of 3,632 variants, all residing within the previously demonstrated linkage regions and the short region between them12 in 15q. Many of these variants occurred commonly in the 1000 Genomes phase 3 dataset (which contains 405 individuals with sub-Saharan West African ancestry similar to that of Cameroonians) or in the NHLBI ESP6400 dataset. These were eliminated from consideration, and rare variants, defined as those with a frequency of 0.01 or lower in 1000 Genomes, were considered further. Of the remaining 270 variants, we focused on the 66 non-synonymous coding variants on the basis of the hypothesis that the causative allele in sub-pedigree 1B and/or 1E of CAMST01 is a coding allele of large effect. Nine of these 66 variants occur in more than one affected individual, and six of these nine are predicted to cause non-synonymous changes. Of these six, two occur frequently in our normal Cameroonian control individuals, and two reside in the area between the two linkage regions in 15q and are considered off target. The remaining two are in AP4E1 (GenBank: NM_007347.4): c.1549G>A (p.Val517Ile), which occurs at the beginning of exon 14 and is predicted to cause a valine-to-isoleucine substitution, and c.2401G>A (p. Glu801Lys), which occurs in exon 18 and is predicted to cause the substitution of a lysine for the normal glutamic acid (Figure 1B). These two variants co-segregate with the disorder in 11 members of sub-pedigree 1E, and all of the affected individuals in this sub-pedigree carry one chromosome with these variants, which does not occur in any other branches of this family. The wild-type valine and glutamic acid at these positions are highly conserved across mammals (Figure S1) and completely conserved in normal Cameroonian control individuals, normal Pakistani control individuals, neurologically normal North American control individuals, 1000 Genomes individuals, and NHLBI ESP6400 individuals, who together represent >19,000 chromosomes.
Figure 1.
Identification of Rare Variations in AP4E1
(A) Bioinformatic-analysis pipeline applied to whole-exome sequence data from chromosomal region 15q.
(B) Co-segregation of the chromosome containing the c.1549G>A and c.2401G>A variations of AP4E1 in Cameroonian family CAMST01, sub-pedigree E. Dideoxy sequencing traces of representative heterozygous c.1549G>A (p.Val517Ile) and c.2401G>A (p.Glu801Lys) variations are shown.
Sequencing 96 unrelated Cameroonians who stutter identified the same two rare AP4E1 coding variants in two additional individuals. A survey of 12 SNP variants extending across 47 kb of AP4E1 genomic sequence containing these two variants identified a SNP haplotype shared by these two individuals and the affected members of Cameroonian family CAMST01. This haplotype was observed in 1/42 (2.4%) Cameroonian chromosomes not containing these variants. This suggests that the c.1549G>A and c.2401G>A variants share a common origin on a haplotype that is uncommon in the Cameroonian population.
Sequencing 96 normally fluent Cameroonian control individuals identified a single missense variant (c.632G>A [p.Arg211Gln]) in one individual. Sequencing AP4E1 in unrelated affected individuals from Cameroon (n = 93), Pakistan (n = 132), and North America (n = 711) revealed 23 other rare variants in this gene, including small (1- to 3-bp) deletions, insertions, duplications, frameshifts, and stop codons (Table 1). No germline small deletion, insertion, frameshift, or stop-codon variants of AP4E1 occur in any of our Cameroonian (n = 94), Pakistani (n = 96), or North American (n = 368) control individuals (total n = 558).
Table 1.
Summary of Rare AP4E1 Variants in Population-Matched Stuttering Individuals
| cDNA Change | Amino Acid Change | SNP ID |
Stuttering Individuals and Families (n = 936)a |
Population-Matched Control Individuals (n = 558) |
No. of Individuals in Population Databases |
PolyPhen-2 Score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAMST01 (1 Family) | STCR ( n = 92) | PKST (60 Families) | PKSTR (n = 72) | NA + NIH (n = 711) | 1000 Genomes | NHLBI ESP6400 | |||||
| c.−2_1−18(1_3) | non-coding | unknown | − | − | PKST9 [3] | − | NA276, NA363 | − | − | − | − |
| c.−4G>A | non-coding | unknown | − | STCR132, STCR201 | − | − | − | − | − | 2 | − |
| c.−7G>A | non-coding | unknown | − | − | − | − | Na0672 | − | − | − | − |
| c.286T>G | p.Phe96Val | unknown | − | − | PKST27 [1] | − | − | − | − | − | 1.000 |
| c.542+3A>G | non-coding | unknown | − | − | − | − | Na0689 | − | − | − | − |
| c.613C>A | p.His205Asn | rs148499164 | − | − | − | − | NA182 | − | − | 5 | 0.924 |
| c.833delT | p.Leu278∗ | unknown | − | − | − | − | NA272 | − | − | − | − |
| c.741dup | p.Val248Cysfs∗41 | unknown | − | − | − | − | NA275 | − | − | − | − |
| c.932A>G | p.Asn311Ser | rs536656846 | − | − | − | − | Na0166 | − | − | − | 0.910 |
| c.977C>T | p.Ser326Phe | rs372479885 | − | − | − | − | Na0507 | − | − | 1 | 0.897 |
| c.1151A>G | p.His384Arg | unknown | − | − | − | − | NIH25 | − | − | − | 1.000 |
| c.1276A>C | p.Ile426Leu | rs148817957 | − | − | − | − | NA22 | ND05461 | 3 | 11 | 0.924 |
| c.1424C>T | p.Ala475Val | rs200678853 | − | − | PKST95 [2] | − | − | − | − | − | 1.000 |
| c.1549G>A | p.Val517Ile | unknown | 1E [12] | STCR102, STCR199 | − | − | − | − | − | − | 1.000 |
| c.1624A>G | p.Met542Val | unknown | − | − | PKST26 [1], PKST81 [3] | − | NA136 | − | − | − | 0.000 |
| c.1851+1_4dupGTAA | non-coding | unknown | − | − | − | PKSTR25 | − | − | − | − | − |
| c.1867T>C | p.Ser623Pro | unknown | − | − | − | − | NA41 | ND08851 | – | – | 1.000 |
| c.2383delA | p.Arg795Glyfs∗19 | unknown | − | − | − | − | NA173 | − | − | − | − |
| c.2401G>A | p.Glu801Lys | unknown | 1E [12] | STCR102, STCR199 | − | − | − | − | − | − | 0.006 |
| c.2570C>G | p.Ser857∗ | unknown | − | − | PKST44 [2] | − | − | − | − | − | − |
| c.2713T>C | p.Ser905Pro | unknown | − | − | − | − | NA217 | − | − | − | 0.013 |
| c.2932C>T | p.Pro978Ser | rs141278078 | − | − | PKST87 [2] | − | − | − | − | 6 | 0.001 |
| c.3238A>G | p.Ile1080Val | unknown | − | − | PKST69 [1] | − | − | − | − | − | 0.000 |
| c.3266T>G | p.Leu1089Arg | unknown | − | − | PKST32 [1] | PKSTR59 | − | − | − | − | 0.990 |
| c.3314G>A | p.Arg1105Gln | rs139640763 | 1E [1] | − | − | − | − | − | 2 | 1 | 1.000 |
Abbreviations are as follows: CAMST01, Cameroonian family; STCR, Cameroonian unrelated stuttering individuals; PKST, Pakistani families with stuttering individuals; PKSTR, Pakistani unrelated stuttering individuals; NA, unrelated North American stuttering individuals; Na, single unrelated stuttering individuals from North American families; −, variant has zero allele frequency.
Brackets indicate the number of affected individuals within the family.
To gain a better idea of the rate of such loss-of-function variants in the general population, we investigated the 1000 Genomes and NHLBI ESP (ESP6400) databases, which represent individuals who are unphenotyped for speech fluency. We found a total of three such variants—one stop-gain and two splice-site variants—in individuals of East-Asian origin (the CDX [Chinese Dai in Xishuangbanna, China] population, consisting of 93 individuals). Our finding of three loss-of-function variants in the ∼19,000 chromosomes in these databases—which contain individuals of similar sub-Saharan West African (the YRI [Yoruba in Ibadan, Nigeria], GWD [Gambian in Western Divisions in the Gambia], MSL [Mende in Sierra Leone], and ESN [Esan in Nigeria] populations; n = 405), South Asian (the ITU [Indian Telugu from the UK], STU [Sri Lankan Tamil from the UK], BEB [Bengali from Bangladesh], PJL [Punjabi from Lahore, Pakistan], and GIH [Gujarati Indian from Houston, Texas] populations; n = 489), and North American ancestries as our stuttering and control individuals—suggests that germline AP4E1 loss-of-function variants are very rare in the general population.
In addition to loss-of-function variants, rare missense variants in AP4E1 were also observed (Tables 1 and 2). We compared the rate of such variants in affected and control individuals within sub-populations. AP4E1 variants were found at a higher frequency in Pakistani affected individuals (11/132) than in Pakistani control individuals (1/96; Chi-square p = 0.0149) and at a higher frequency in Cameroonian affected individuals (9/93) than in Cameroonian control individuals (1/94; Chi-square p = 0.0088). Variants occurred at a frequency of 2.1% (15/711) in North American affected individuals and a frequency of 1.9% (7/368) in North American control individuals (Chi-square p = 0.81). Thus, AP4E1 variants were significantly more frequent in affected individuals than in control individuals in two of the three populations in the study.
Table 2.
Summary of Rare AP4E1 Variants in Population-Matched Control Individuals
| cDNA Change | Amino Acid Change | SNP ID |
Population-Matched Control Individuals (n = 558) |
Stuttering Individuals (n = 936) |
No. of Individuals in Population Databases |
PolyPhen-2 |
|||
|---|---|---|---|---|---|---|---|---|---|
| RC (n = 94) | PKNR (n = 96) | NDPT (n = 368) | 1000 Genomes | NHLBI ESP6400 | Score | ||||
| c.254T>C | p.Ile85Thr | rs147005786 | − | − | ND05681 | − | − | 2 | 0.890 |
| c.434C>G | p.Thr145Ser | rs200034177 | − | − | ND07655 | − | − | 1 | 0.966 |
| c.632G>A | p.Arg211Gln | unknown | RC25 | − | − | − | − | − | 0.507 |
| c.791A>G | p.Asn264Ser | rs145541719 | − | PKNR87 | − | − | 7 | 1 | 0.002 |
| c.1276A>C | p.Ile426Leu | rs148817957 | − | − | ND05461 | NA22 | 3 | 11 | 0.924 |
| c.1852G>A | p.Val618Ile | rs142215198 | − | − | ND05770 | − | − | 4 | 0.548 |
| c.1867T>C | p.Ser623Pro | unknown | − | − | ND08851 | NA41 | − | − | 1.000 |
| c.2117T>A | p.Ile706Lys | unknown | − | − | ND05125 | − | − | − | 0.211 |
| c.2437A>G | p.Met813Val | unknown | − | − | ND07113 | − | − | − | 0.000 |
Abbreviations are as follows: RC, random Cameroonian control individuals; PKNR, random Pakistani population control individuals; NDPT, neurologically normal control individuals; NA, unrelated North American stuttering individuals; −, variant has zero allele frequency.
We sought to estimate the effects of the rare missense variants found in affected and control individuals by using PolyPhen-2.19 These results are shown in Tables 1 (affected individuals) and 2 (control individuals). The mean PolyPhen-2 score of the missense variants found in affected individuals was 0.67, whereas the mean score of those found in control individuals was 0.56.
A total of 30 germline rare AP4E1 variants of all types (missense and loss of function) exist in the 1000 Genomes phase 3 dataset, consisting of 6,508 chromosomes that include individuals of sub-Saharan West African, South Asian, and European populations, indicating that such variants are rare worldwide. Although individuals in databases have unknown speech-fluency phenotypes, the rate of rare AP4E1 variants in our stuttering individuals is higher than the rate of such variants in similar populations within the 1000 Genomes phase 3 dataset. For example, we observed a higher rate of variants in our Cameroonian affected individuals (9/93) than in the unphenotyped West African individuals (5/405; Chi-square = 31.81, p = 8.9 × 10−6) and in Pakistani affected individuals (11/132) than in unphenotyped South Asians (8/489; Chi-square = 15.72, p = 7.35 × 10−5). Our North American stuttering individuals showed a trend toward a higher rate of rare AP4E1 variants (15/711) than did ethnically similar unphenotyped Europeans in the 1000 Genomes database (4/503; Chi-square = 3.3, p = 0.069).
The variants observed in our unrelated stuttering individuals were all rare, but two were observed in unrelated affected individuals from different populations (Table 1). We also evaluated the hypothesis that rare variants in genes encoding the three other subunits of the AP-4 complex (AP4M1 [MIM: 602296], AP4S1 [MIM: 607243], and AP4B1 [MIM: 607245]) are more frequent in stuttering individuals than in control individuals. We observed rare non-synonymous coding variants in these three genes at similar rates in affected and control individuals (Tables S1A–S1C).
Clinical Studies
Homozygous AP4E1 deficiency (MIM: 607244) has been described in five individuals to date,20, 21, 22 although the effects of heterozygosity for such variants have not been carefully evaluated. We sought to determine whether our stuttering subjects, all of whom are heterozygous for an AP4E1 variant, display any symptoms previously associated with homozygous AP4E1 deficiency. In family CAMST01 sub-pedigree 1E, eight members (IV-150, III-142, III-143, III-145, III-147, III-148, III-149, and III-151 in Figure 1B), all of whom carry the AP4E1 c.1549G>A and c.2401G>A variants, underwent clinical examinations at the NIH Clinical Center. Particular attention was paid to potential symptoms previously observed in the five individuals carrying homozygous loss-of-function mutations in AP4E1.20, 21, 22 These symptoms included intellectual disability and pseudobulbar symptoms such as drooling, stereotypic laughter, and oral motor spasticity. These individuals also demonstrated spasticity and hyperreflexia in the extremities, microcephaly, severe ambulation problems, seizures, incontinence, conductive hearing loss, severe speech impairments, and mycobacterial disease.
Given these findings, our evaluation procedures included examination of general cognitive function, speech, and language, a comprehensive neurological evaluation including H-reflex and limb-dexterity tests, audiological evaluation, and electroencephalography (EEG). In addition, individuals also underwent electromyography (EMG) and nerve conduction velocity (NCV) tests, electrocardiography, MRI, X-rays of long bones, complete blood count (CBC), and a comprehensive blood-chemistry panel, as well as assessment of general medical history and physical examination.
No consistent notable clinical findings were observed in our eight subjects. There was no evidence of developmental delay or mycobacterial disease. Cognitive and language assessments revealed no evidence of general intellectual or cognitive dysfunction or of deficits in speech and language comprehension or production other than stuttering. Head circumference was within normal limits in all subjects. Neurological evaluation revealed no evidence of cranial-nerve abnormalities or pseudobulbar symptoms. Motor examination was normal in that no weakness or hypertonia was noted. Sensory examination was normal. Deep tendon reflexes were normal, pathological reflexes were absent, and cerebellar exam was intact. H-reflexes and manual dexterity were normal in the subset of individuals tested. Audiometry and auditory brainstem responses were normal, except for in one individual (age 63 years) who showed hearing changes consistent with age-related hearing loss. EEG showed no evidence of epileptiform activity. EMG and NCV tests were normal. X-rays revealed no evidence of long-bone deformities. CBC and blood chemistries were essentially within normal limits.
The heterozygous carrier parents of the severely affected AP4E1-deficient individuals have been previously reported to be medically normal. Our stuttering subjects with rare variants in AP4E1 are all heterozygotes carrying a single copy of the variants, consistent with a lack of the severe symptoms reported in homozygotes.
Biochemical Effects of AP4E1 Variants Found in Stuttering
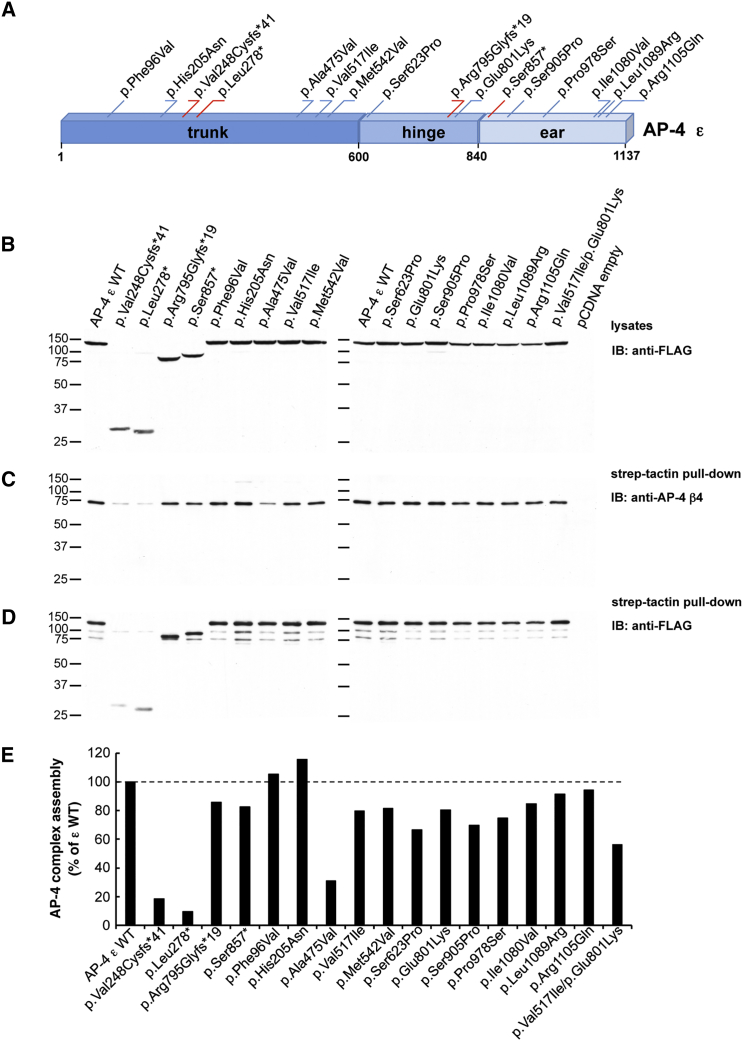
AP4E1 encodes the ε subunit of AP-4, a heterotetrameric (ε-β4-μ4-σ4) complex involved in the sorting of transmembrane proteins at the trans-Golgi network (TGN).16, 23, 24, 25 The rare variants identified in the stuttering subjects mapped to all three domains of the ε subunit: trunk, hinge, and ear (Figure 2A). To better understand the effects of these variants, we constructed vectors that express wild-type and variant AP-4 ε proteins tagged at the N terminus with TSF epitopes. Immunoblot analysis of HEK293T cells transfected with an antibody to the FLAG epitope showed that all the variant genes were expressed and that their protein products had the predicted apparent molecular masses (Figure 2B). The truncating variants c.741dup (p.Val248Cysfs∗41), c.833delT (p.Leu278∗), c.2383delA (p.Arg795Glyfs∗19), and c.2570C>G (p.Ser857∗) produced ε proteins of reduced apparent molecular mass, as expected (Figure 2B). The effects of AP4E1 variants on the assembly of the AP4 complex were measured with a well-characterized antibody to the β4 subunit of this complex. Transgenic-protein pull-down using StrepTactin beads and subsequent immunoblotting for the endogenous β4 subunit of AP-4 showed reduced co-isolation of β4 with the products of the p.Val248Cysfs∗41 and p.Leu278∗ truncating variants and the p.Ala475Val missense variant in AP4E1 (Figures 2C–2E), indicating that these rare coding variants reduce the assembly of the AP-4 complex. Other variants had no appreciable effect on AP-4 assembly (Figures 2C–2E), although many are predicted to be deleterious by PolyPhen-2, and the mean effect prediction score was slightly higher for missense variants found in our stuttering individuals (0.67) than for missense variants in our matched control individuals (0.56) (Tables 1 and 2). We also tested the effects of the rare AP4E1 missense variants found in our control populations. None displayed a consistent effect on the assembly of the AP-4 complex within the limits of this assay (Figure S2).
Figure 2.
Assembly of AP-4 Complexes by Variant ε Subunits
(A) Schematic representation of the AP-4 ε subunit. Single amino acid substitutions are indicated in blue, whereas truncating variants are depicted in red. The numbering corresponds to the human AP-4 ε isoform, comprising 1,137 residues (GenBank: NP_031373.2); assignment of domains is as in Boehm and Bonifacino.26
(B) Expression of wild-type (WT) and variant human AP-4 ε constructs tagged at their N termini with TSF-tagged ε constructs in HEK293T cells. Transfected cell lysates were subjected to SDS-PAGE followed by immunoblotting with anti-FLAG antibody.
(C) Assembly of WT and variant TSF-tagged ε constructs into AP-4 complexes. Lysates of transfected HEK293T cells were incubated with StrepTactin Sepharose beads. Pulled-down complexes were subsequently eluted with d-desthiobiotin and subjected to SDS-PAGE and immunoblotting with anti-AP-4 β4 antiserum.16
(D) Immunoblot membranes with samples of the StrepTactin pull-down assays shown in (C) were stripped and subjected to additional immunoblotting with anti-FLAG antibody.
(E) The assembly of AP-4 complexes by WT and variant ε constructs (pulled-down complexes detected by anti-β4; blots in C) was calculated in relation to the total expression of the cognate ε constructs in transfected HEK293T cells (blots in B). Blots were subjected to densitometric analysis with Image J software. Results are expressed as the percentage of assembly measured for WT ε (percentage of WT ε).
Other stuttering individuals were previously found to have mutations in NAGPA,10 encoding the enzyme NAGPA, which is involved in the synthesis of the mannose 6-phosphate signal for sorting of acid hydrolases to lysosomes.10 Interestingly, this enzyme localizes at least in part to the TGN,27 where AP-4 is also located.16, 23 Moreover, the cytosolic tail of NAGPA contains the sequence YAYHPLQE, which includes a tyrosine-based signal (YHPL) fitting the YxxØ (X is any amino acid, and Ø is a bulky hydrophobic residue) consensus motif for binding to the μ subunits of AP complexes, including AP-4.23, 24, 28 Residues in the YAYHPLQE sequence have been shown to mediate sorting at different compartments, including the TGN, endosomes, and plasma membrane.29 We used a Y2H system to test for interaction between the NAGPA cytosolic tail (amino acids 475–515) and the μ subunits from the AP-1, AP-2, AP-3, and AP-4 complexes. We tested both the wild-type cytosolic tail of NAGPA and three variants (p.Tyr486Ala, p.Tyr488Ala, and p.Leu491Ala), each containing single amino acid substitutions within the YAYHPLQE sequence (Figure 3A). Interactions were observed between the NAGPA tail and the μ4 subunit of AP-4, as well as the μ1 and μ2 subunits of AP-1 and AP-2, respectively (Figure 3B). In contrast, no interactions between the NAGPA tail and the μ3 subunits of AP-3A and AP-3B were observed. Importantly, the interaction with μ4 was abolished when alanine substitutions were introduced in the Tyr486, Tyr488, and Leu491 residues, believed to be essential components of the YAYHPLQE recognition motif. These observations thus demonstrate a physical interaction between the NAGPA cytosolic tail and the μ4 subunit of AP-4.
Discussion
Genetic contributions to stuttering have long been documented, and recent progress has been made in the identification of the specific genes that underlie this disorder. Previously, we identified rare GNPTAB, GNPTG, and NAGPA coding variants that together could account for 8%–16% of cases of persistent developmental stuttering.11 These genes encode the components of the pathway that generates the mannose 6-phosphate signal that targets a diverse group of hydrolytic enzymes to their ultimate location in the lysosome. Rare NAGPA coding variants found in stuttering subjects have been shown to impair the biochemical function of the enzyme encoded by this gene.30 However, it is not yet clear how such deficits in lysosomal enzyme targeting give rise to non-syndromic persistent stuttering, and identification of additional genes that contribute to this disorder could aid in the understanding of the pathophysiology involved. Our results now implicate rare coding variants in AP4E1, encoding the ε subunit of AP-4, in the genesis of additional cases of stuttering.
A number of variants, including loss-of-function variants in AP4E1, have been described in dbSNP. However, these variants were identified in tumors rather than in constitutional DNA and are not germline variants, such as those we have identified in stuttering individuals. We estimate that rare variants in AP4E1 underlie 2.1%–3.7% of unrelated cases of persistent stuttering. The higher end of this range is the rate at which we found rare variants in our combined stuttering individuals. The lower range of this estimate is the rate in our combined affected individuals (35/936 [3.7%]) minus the rate of such variants in combined population-matched control individuals (9/558 [1.6%]). These estimates are subject to several possible sources of error, including the unknown penetrance of these variants in stuttering (given that less-than-complete penetrance would reduce their contribution to the disorder) and potential differential contributions to the disorder from the different populations that make up our study.
In the three other genes that encode components of the AP4 complex (AP4B1, AP4M1, and AP4S1), we found similar rates of rare non-synonymous coding variants in affected and control individuals. This indicates that the population sample and the analytic methods we used do not show a higher rate of rare variants in stuttering individuals in these genes, all of which reside at loci with no evidence of linkage to stuttering.7, 8, 9, 31, 32 In addition, our dideoxy sequencing of GANC, GATM, and GALK2, which all lie within the sub-pedigree 1E linkage interval in chromosomal region 15q, revealed no difference in the rate of rare variants in affected and matched control individuals. Together, these results support the view that rare variants in other genes, either functionally related or positionally close to AP4E1, do not show an association with stuttering.
Adaptor protein complexes are ubiquitous eukaryotic cell components that control trafficking in the endomembrane system. Our results support a direct interaction between the AP-4 complex and NAGPA, which are known to reside at the TGN. Thus, our findings of rare variants in AP4E1 not only account for additional cases of persistent stuttering but also establish a direct link between this gene and the products of the previously identified stuttering-associated genes. Although our knowledge of the full spectrum of genetic causes of stuttering remains incomplete, our results to date suggest that deficits in intracellular trafficking, particularly that of the endosomal transport system, contribute to persistent developmental stuttering.
This intracellular trafficking system is ubiquitous in most eukaryotic cells, so it is not surprising that homozygous mutations in AP4E1 (along with mutations in GNPTAB and GNPTG) lead to rare syndromes with severe effects in many different organs and tissues.20, 21, 22, 33 Because our stuttering individuals all carry only a single copy of a rare coding variant in AP4E1, they were not expected to display any of the symptoms previously observed in rare individuals carrying homozygous loss-of-function mutations in this gene. Our clinical evaluations identified a scattering of abnormal findings in this group of eight Cameroonian subjects. However, no spastic paraplegia, developmental disability, microcephaly, foot abnormalities, short stature, or mycobacterial disease was identified in these subjects. Although deficits in speech have been reported in AP4E1-deficient homozygotes, stuttering per se has not. These homozygous individuals have multiple developmental disabilities, and their speech deficits might be secondary to their general neurologic deficits. Our subjects did not display speech deficits other than stuttering. Thus, we could find no evidence supporting the view that non-syndromic persistent developmental stuttering is a mild presentation of the syndrome seen in homozygous AP4E1 deficiency.
Our current results establish a possible link between the AP-4 complex and the product of NAGPA, which has previously been associated with stuttering.10, 11 These proteins participate in the control of endosomal and/or lysosomal trafficking. Defects in this process are an increasingly recognized cause of neurologic disorders, ranging from rare Mendelian disorders such as Charcot-Marie-Tooth disease type 1C (MIM: 601098), Niemann-Pick disease type C (MIM: 257220), and Perry syndrome (MIM: 168605)34, 35, 36 to more common genetically complex disorders such as Alzheimer and Parkinson diseases.37 Further studies are needed to address the question of how AP-4 deficits could lead to a specific deficit in speech and to better elucidate the contribution of such deficits to the total number of stuttering cases. However, our results suggest a cellular mechanism for the neuronal pathology underlying stuttering and place this poorly understood disorder in the mainstream of neurological disease.
Acknowledgments
This work was supported by NIH National Institute of Child Health and Human Development (NICHD) intramural grants Z1A-DC000046-15 (to D.D.), Z1A DC 000039-15 (to T.B. Friedman), and Z1A HD001607 (to J.B.). We thank the Stuttering Foundation of America, the Hollins Communications Research Institute, and the National Stuttering Association for assistance and M. Hallett, P. Friedman, S. Solomon, S. Inati, T. Lehky, L. Hathway, P. Penyakaew, S. Hove, J. Greenfield, S. Lischynsky, and H. Chow for assistance in clinical evaluations.
Published: November 5, 2015
Footnotes
Supplemental Data include three figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.10.007.
Accession Numbers
The AP4E1 SNPs reported in this article were submitted to dbSNP. The online article will be updated with the accession numbers when they are available.
Web Resources
The URLS for data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org
1000 Genomes AP4E1, http://browser.1000genomes.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000081014;r=15:51200869-51298097
Coriell Cell Repository, http://www.ccr.coriell.org/Sections/Collections/NINDS/DNAPanels.aspx?PgId=195&coll=ND
Image J, http://www.imagej.nih.gov
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS
OMIM, http://www.omim.org/
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Bloodstein O., Ratner N.B. Sixth Edition. Cengage Learning; 2008. A handbook on stuttering. [Google Scholar]
- 2.Dworzynski K., Remington A., Rijsdijk F., Howell P., Plomin R. Genetic etiology in cases of recovered and persistent stuttering in an unselected, longitudinal sample of young twins. Am. J. Speech Lang. Pathol. 2007;16:169–178. doi: 10.1044/1058-0360(2007/021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fagnani C., Fibiger S., Skytthe A., Hjelmborg J.V. Heritability and environmental effects for self-reported periods with stuttering: a twin study from Denmark. Logoped. Phoniatr. Vocol. 2011;36:114–120. doi: 10.3109/14015439.2010.534503. [DOI] [PubMed] [Google Scholar]
- 4.Felsenfeld S., Plomin R. Epidemiological and offspring analyses of developmental speech disorders using data from the Colorado Adoption Project. J. Speech Lang. Hear. Res. 1997;40:778–791. doi: 10.1044/jslhr.4004.778. [DOI] [PubMed] [Google Scholar]
- 5.Rautakoski P., Hannus T., Simberg S., Sandnabba N.K., Santtila P. Genetic and environmental effects on stuttering: a twin study from Finland. J. Fluency Disord. 2012;37:202–210. doi: 10.1016/j.jfludis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Kidd K.K., Heimbuch R.C., Records M.A. Vertical transmission of susceptibility to stuttering with sex-modified expression. Proc. Natl. Acad. Sci. USA. 1981;78:606–610. doi: 10.1073/pnas.78.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raza M.H., Amjad R., Riazuddin S., Drayna D. Studies in a consanguineous family reveal a novel locus for stuttering on chromosome 16q. Hum. Genet. 2012;131:311–313. doi: 10.1007/s00439-011-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raza M.H., Riazuddin S., Drayna D. Identification of an autosomal recessive stuttering locus on chromosome 3q13.2-3q13.33. Hum. Genet. 2010;128:461–463. doi: 10.1007/s00439-010-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riaz N., Steinberg S., Ahmad J., Pluzhnikov A., Riazuddin S., Cox N.J., Drayna D. Genomewide significant linkage to stuttering on chromosome 12. Am. J. Hum. Genet. 2005;76:647–651. doi: 10.1086/429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang C., Riazuddin S., Mundorff J., Krasnewich D., Friedman P., Mullikin J.C., Drayna D. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N. Engl. J. Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raza M.H., Domingues C.E., Webster R., Sainz E., Paris E., Rahn R., Gutierrez J., Chow H.M., Mundorff J., Kang C.S. Mucolipidosis types II and III and non-syndromic stuttering are associated with different variants in the same genes. Eur. J. Hum. Genet. 2015 doi: 10.1038/ejhg.2015.154. Published online July 1, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raza M.H., Gertz E.M., Mundorff J., Lukong J., Kuster J., Schäffer A.A., Drayna D. Linkage analysis of a large African family segregating stuttering suggests polygenic inheritance. Hum. Genet. 2013;132:385–396. doi: 10.1007/s00439-012-1252-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley, G.D. (1994). SSI-3: Stuttering Severity Instrument for Children and Adults, Third Edition (PRO-ED). [DOI] [PubMed]
- 14.Webster R.L. Evolution of a Target-Based Behavioral-Therapy for Stuttering. J. Fluency Disord. 1980;5:303–320. [Google Scholar]
- 15.Mattera R., Boehm M., Chaudhuri R., Prabhu Y., Bonifacino J.S. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J. Biol. Chem. 2011;286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dell’Angelica E.C., Mullins C., Bonifacino J.S. AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem. 1999;274:7278–7285. doi: 10.1074/jbc.274.11.7278. [DOI] [PubMed] [Google Scholar]
- 17.Guo X., Mattera R., Ren X., Chen Y., Retamal C., González A., Bonifacino J.S. The adaptor protein-1 μ1B subunit expands the repertoire of basolateral sorting signal recognition in epithelial cells. Dev. Cell. 2013;27:353–366. doi: 10.1016/j.devcel.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattera R., Arighi C.N., Lodge R., Zerial M., Bonifacino J.S. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 2003;22:78–88. doi: 10.1093/emboj/cdg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abou Jamra R., Philippe O., Raas-Rothschild A., Eck S.H., Graf E., Buchert R., Borck G., Ekici A., Brockschmidt F.F., Nöthen M.M. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am. J. Hum. Genet. 2011;88:788–795. doi: 10.1016/j.ajhg.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X.F., Bousfiha A., Rouissi A., Itan Y., Abhyankar A., Bryant V., Okada S., Ailal F., Bustamante J., Casanova J.L. A novel homozygous p.R1105X mutation of the AP4E1 gene in twins with hereditary spastic paraplegia and mycobacterial disease. PLoS ONE. 2013;8:e58286. doi: 10.1371/journal.pone.0058286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-De-Luca A., Helmers S.L., Mao H., Burns T.G., Melton A.M., Schmidt K.R., Fernhoff P.M., Ledbetter D.H., Martin C.L. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J. Med. Genet. 2011;48:141–144. doi: 10.1136/jmg.2010.082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirst J., Bright N.A., Rous B., Robinson M.S. Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell. 1999;10:2787–2802. doi: 10.1091/mbc.10.8.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgos P.V., Mardones G.A., Rojas A.L., daSilva L.L., Prabhu Y., Hurley J.H., Bonifacino J.S. Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell. 2010;18:425–436. doi: 10.1016/j.devcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda S., Miura E., Matsuda K., Kakegawa W., Kohda K., Watanabe M., Yuzaki M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Boehm M., Bonifacino J.S. Adaptins: the final recount. Mol. Biol. Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohrer J., Kornfeld R. Lysosomal hydrolase mannose 6-phosphate uncovering enzyme resides in the trans-Golgi network. Mol. Biol. Cell. 2001;12:1623–1631. doi: 10.1091/mbc.12.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilar R.C., Boehm M., Gorshkova I., Crouch R.J., Tomita K., Saito T., Ohno H., Bonifacino J.S. Signal-binding specificity of the mu4 subunit of the adaptor protein complex AP-4. J. Biol. Chem. 2001;276:13145–13152. doi: 10.1074/jbc.M010591200. [DOI] [PubMed] [Google Scholar]
- 29.Lee W.S., Rohrer J., Kornfeld R., Kornfeld S. Multiple signals regulate trafficking of the mannose 6-phosphate-uncovering enzyme. J. Biol. Chem. 2002;277:3544–3551. doi: 10.1074/jbc.M108531200. [DOI] [PubMed] [Google Scholar]
- 30.Lee W.S., Kang C., Drayna D., Kornfeld S. Analysis of mannose 6-phosphate uncovering enzyme mutations associated with persistent stuttering. J. Biol. Chem. 2011;286:39786–39793. doi: 10.1074/jbc.M111.295899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suresh R., Ambrose N., Roe C., Pluzhnikov A., Wittke-Thompson J.K., Ng M.C., Wu X., Cook E.H., Lundstrom C., Garsten M. New complexities in the genetics of stuttering: significant sex-specific linkage signals. Am. J. Hum. Genet. 2006;78:554–563. doi: 10.1086/501370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittke-Thompson J.K., Ambrose N., Yairi E., Roe C., Cook E.H., Ober C., Cox N.J. Genetic studies of stuttering in a founder population. J. Fluency Disord. 2007;32:33–50. doi: 10.1016/j.jfludis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornfeld S., Sly W. The Metabolic Bases of Inherited Disease. Eight Edition. McGraw-Hill; 2001. I-Cell Disease and Pseudo-Hurler Polydystrophy: Disorders of Lysosomal Enzyme Phosphorylation and Localization. [Google Scholar]
- 34.Lee S.M., Chin L.S., Li L. Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J. Cell. Biol. 2012;199:799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrer M.J., Hulihan M.M., Kachergus J.M., Dächsel J.C., Stoessl A.J., Grantier L.L., Calne S., Calne D.B., Lechevalier B., Chapon F. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanier M.T. Niemann-Pick disease type C. Orphanet J. Rare Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neefjes J., van der Kant R. Stuck in traffic: an emerging theme in diseases of the nervous system. Trends Neurosci. 2014;37:66–76. doi: 10.1016/j.tins.2013.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.