Abstract
S-adenosylmethionine (SAM) is the predominant methyl group donor and has a large spectrum of target substrates. As such, it is essential for nearly all biological methylation reactions. SAM is synthesized by methionine adenosyltransferase from methionine and ATP in the cytoplasm and subsequently distributed throughout the different cellular compartments, including mitochondria, where methylation is mostly required for nucleic-acid modifications and respiratory-chain function. We report a syndrome in three families affected by reduced intra-mitochondrial methylation caused by recessive mutations in the gene encoding the only known mitochondrial SAM transporter, SLC25A26. Clinical findings ranged from neonatal mortality resulting from respiratory insufficiency and hydrops to childhood acute episodes of cardiopulmonary failure and slowly progressive muscle weakness. We show that SLC25A26 mutations cause various mitochondrial defects, including those affecting RNA stability, protein modification, mitochondrial translation, and the biosynthesis of CoQ10 and lipoic acid.
Keywords: mitochondria, mitochondrial dysfunction, methylation, SAM, SLC25A26, S-adenosylmethionine
Main Text
Altered S-adenosylmethionine (SAM) concentrations in the cytoplasm have been suggested to be involved in the pathophysiology of disease and in the natural aging process.1, 2 Highly specialized methyltransferases, encoding approximately 1%–2% of eukaryotic genomes,3 use SAM as a methyl group donor to methylate their targets. The human mitochondrial SAM carrier (SAMC), encoded by SLC25A26 (MIM: 611037), is expressed in all human tissues examined and is believed to be the only route of SAM entry into mitochondria.4 However, regulatory mechanisms of intra-mitochondrial SAM (mtSAM) concentrations or other pathways modulating mtSAM levels are unknown, and so far the pathophysiological consequences of reduced mitochondrial SAM import are unclear.
We identified three families with different ethnic origins and a complex biochemical phenotype caused by mutations in SLC25A26. Individual 1 (P1, individual II:2 from family 1 in Figure 1A) was born to consanguineous parents from Iraq and presented at 4 weeks with acute circulatory collapse and pulmonary hypertension, requiring extra-corporeal membrane oxygenation for 5 days. He had severe lactic acidosis around 20 mmol/l (reference: 0.5–2.3). Sodium dichloroacetic acid had good effect, and the boy slowly normalized. At 3.5 years, he had a second episode of pulmonary hypertension, which also normalized. At 6 years 3 months, the boy had increasing muscle weakness, fatigue, recurrent abdominal pain, lack of appetite, and slightly delayed development. Investigation of mitochondrial function from a muscle biopsy revealed reduced activities of complexes I and IV and a reduced ATP production rate, in particular when pyruvate was used as a substrate (Figures S1A and S1B). Histology showed the presence of COX-negative muscle fibers (Figure S1C). Additionally, Blue-native PAGE (BN-PAGE) revealed reduced levels of assembled complexes I and IV (Figure S1D). Individual 2 (P2, II:1 from family 2 in Figure 1A), born to Japanese parents, developed severe lactic acidosis up to 42 mmol/l (reference: <1.8), an elevated pyruvate level (0.65 mmol/l; reference: <0.1), and respiratory failure 11 hr after birth, prompting mechanical ventilation and dichloroacetic acid treatment. The child improved, and gross development was normal until 2 years of age, when she experienced an additional episode of severe lactic acidosis (36 mmol/l) followed by cardiopulmonary arrest and hypoxic brain damage. After this episode, the individual has remained severely handicapped. Activities of respiratory-chain enzymes were normal in fibroblasts but showed decreased activities of complexes I, III, and IV in skeletal muscle (Figure S1E). Muscle histology was normal at day 6 but revealed both ragged red fibers and COX-negative fibers when individual 2 was 3 years of age (Figure S1F). Individual 3 (P3, individual II:4 from family 3 in Figure 1A), born to consanguineous parents of Moroccan decent, was delivered by caesarean section at 30 weeks 5 days after reduced fetal movements, polyhydramnios, fetal hydrops, and poor cardiotocography (CTG) readings were noted from 27 weeks of gestational age. She had normal antropometric parameters (birth weight 1,300 g, length 38 cm, and head circumference 27.5 cm) but presented with a poor Apgar score (3-5-6) due to bradycardia, hypotonia, and respiratory insufficiency, necessitating assisted ventilation with high-frequency oscillation. Urine lactate and pyruvate levels were 18 mmol/mmol creatinine (reference: 1–285 μmol/mmol creatinine) and 1.2 mmol/mmol creatinine (reference: 1–130 μmol/mmol creatinine), respectively. Brain ultrasound demonstrated cystic necrosis of the germinal matrix (extensive symmetrical caudothalamic germinolysis) and mild striatal arteriopathy. The child died of respiratory and multiple organ failure at 5 days of age. Measurement of respiratory-chain activity in fibroblasts demonstrated decreased complex IV activity. Additional clinical descriptions and experimental details are provided in the Supplemental Note.
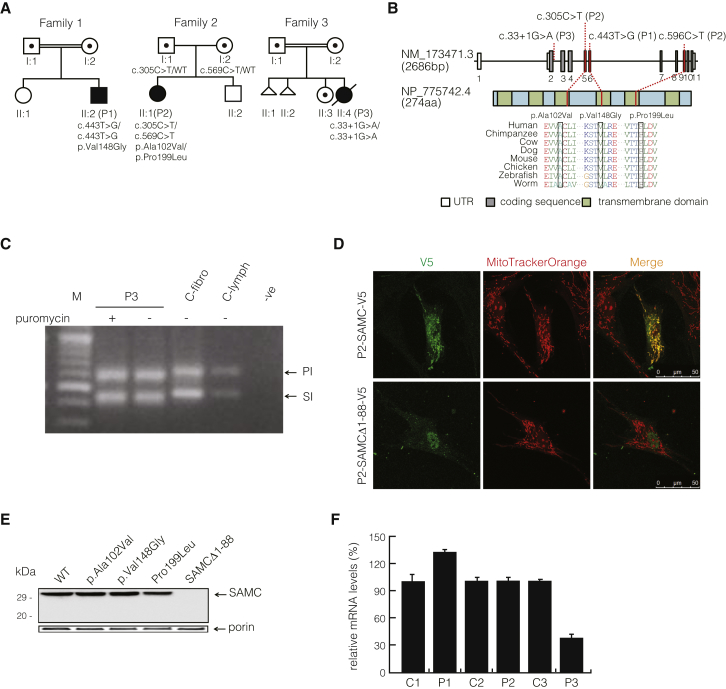
Figure 1.
Identification of Mutations in SLC25A26
(A) Pedigrees of individuals P1–P3 indicate the inheritance patterns in the individuals’ families. P1 was born to consanguineous parents from Iraq after a normal pregnancy and neonatal period. P2 was born full term to unrelated parents from Japan with an Apgar score of 9-10. P3 was born to consanguineous parents of Moroccan descent. Symbols and colors are defined as follows: square, male; circle, female; triangle, miscarriage with unknown gender; white, unaffected; dot, unaffected carrier; black, affected. WT indicates wild-type.
(B) Diagram representing the relative positions of SLC25A26 mutations (NM_173471.3) and SLC25A26 alterations (GenBank: NP_775742.4). Amino acid alignments of eight species show the regions of each mutation.
(C) SLC25A26 mutation c.33+1G>A causes an RNA-splicing defect: the top band in lanes 2–5 indicates the amplification of the principal isoform (PI; Ensembl: ENST00000354883), and the lower band in lanes 2–5 indicates the amplification of the shorter isoform (SI; Ensembl: ENST00000336733). As a result of the mutation, PCR products from the individual, treated both with and without puromycin, were observed to be shorter in length (top band: PI around 572 bp; lower band: SI around 415 bp) than those of the control fibroblasts and lymphocytes (top band: PI 617 bp; lower band: SI 450 bp). No difference was observed between the puromycin-treated and non-puromycin-treated P3 samples. Lane contents are as follows: lanes 1 and 7, 100 bp DNA ladder (Fermentas); lane 2, PCR products amplified from cDNA extracted from P3 fibroblasts treated with puromycin; lane 3, PCR products amplified from cDNA extracted from P3 fibroblasts cultured without puromycin; lane 4, PCR products amplified from cDNA extracted from control fibroblasts; lane 5, PCR products amplified from cDNA extracted from control lymphocytes; and lane 6, PCR reaction blank.
(D) Subcellular localization of C-terminal V5-tagged SAMC (p2-SAMC-V5) and the shortened SAMCΔ1–88 (p2-SAMCΔ-V5) in P2 fibroblasts stained with MitoTrackerOrange.
(E) Amounts of wild-type (WT) SAMC, p.Ala102Val SAMC, p.Val148Gly SAMC, p.Pro199Leu SAMC, SAMCΔ1–88, and endogenous porin in mitochondria from SAM5Δ yeast transformed with WT SAMC-pYES2 (SAMC), p.Ala102Val SAMC-pYES2 (p.Ala102Val), p.Val148Gly SAMC-pYES2 (p.Val148Gly), p.Pro199Leu SAMC-pYES2 (p.Pro199Leu), and short SAMC-pYES2 (SAMCΔ1–88). Equal amounts of mitochondrial lysates (30 μg protein) were separated by SDS-PAGE, transferred to nitrocellulose, and immunodecorated with the anti-hemagglutinin or the anti-porin antibody.
(F) Relative SLC25A26 mRNA steady-state levels in fibroblasts as determined by qRT-PCR. Values are normalized to 18S rRNA levels. Error bars show the SEM.
Written informed consent was obtained from the parents, and investigations were performed according to the regional ethics committees at the Karolinska Institutet (Sweden), the Saitama Medical University (Japan), and Antwerp University Hospital (Belgium).
Homozygosity mapping, exome sequencing,5, 6, 7, 8, 9, 10, 11 and Sanger confirmation (Figures 1A and 1B and Figure S2A) revealed SLC25A26 mutations (GenBank: NM_173471.3) in all affected individuals and their parents. We identified conserved missense mutations in P1, homozygous for a c.443T>C (p.Val148Gly) substitution, and P2, compound heterozygous for c.305C>T (p.Ala102Val) and c.596C>T (p.Pro199Leu). P3 was homozygous for a splice mutation (c.33+1G>A) (Figure 1C), which results in either a frameshift mutation in SLC25A26, when an alternative splice site in exon 2 is used, or a shorter polypeptide lacking the first 88 amino acids (SAMCΔ1–88), as a result of an alternative translation initiation site in exon 4 (Figure S2B). Cloning and sequencing of cDNA from P3 fibroblasts of this region confirmed the presence of exclusively alternative splice variants (Figure S2C). The shortened transcript lacks the first two transmembrane helices (Figure S3) and failed to co-localize (Figure 1D) or be detected in mitochondria by western blot analysis (Figure 1E), indicating that it does not encode a functional mitochondrial carrier protein. Additionally, the splice mutation resulted in reduced SLC25A26 mRNA transcript levels in fibroblasts from P3, whereas P1 and P2 samples were unaffected (Figure 1F).
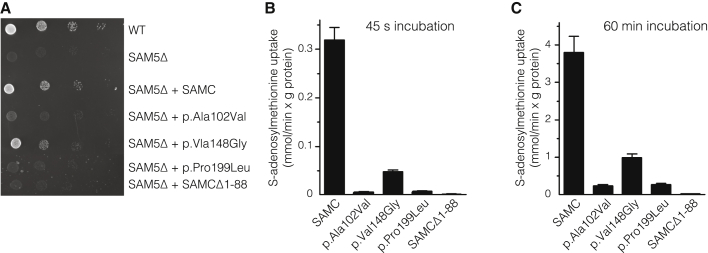
The conservation of all three missense mutations among 87 species (Ala102 [84%], Val148 [100%, including Leu and Ile], and Pro199 [100%]) suggests that their replacement might disrupt protein function. We also considered the transversal scores of the altered SAMC residues (these scores are a measure of the strength of the evolutionary selection acting on the residues) from a study of the rate of single-nucleotide evolution.12 These values (4.52 for Ala102, 3.68 for Val148, and 5.15 for Pro199) are all close to or greater than 3.7, previously shown to represent sites of functional importance in mitochondrial carriers.12 Furthermore, the position of all three SLC25A26 missense mutations in the structural homology model of SAMC also suggested a pathogenic effect of the mutations (Figure S4).13 We confirmed pathogenicity by complementation studies in an S. cerevisiae SAMC-null strain (SAM5Δ)14 by revealing that the growth phenotype of SAM5Δ cells on non-fermentable carbon sources could not be restored by complementation of the knockout strain with the p.Ala102Val, p.Pro199Leu, or SAMCΔ1–88 variant. Only the p.Val148Gly altered SAMC partially rescued the growth defect of SAM5Δ cells (Figure 2A). Additionally, we measured SAM transport capacity in reconstituted liposomes as previously described15, 16, 17, 18, 19 and demonstrated a severe abrogation of SAM transport capacity for all altered proteins (Figures 2B and 2C and Figure S5). SAMCΔ1–88 was completely inactive, whereas p.Ala102Val and p.Pro199Leu variants exhibited negligible activity, and p.Val148Gly strongly inhibited SAMC activity (15% of wild-type SAMC). All together, conservation scores, yeast complementation, and in vitro reconstitution studies confirm the deleterious consequences of the SLC25A26 mutations on SAMC function. Also supporting this is that the various degrees of residual SAM-import capacity correlated well with the severity of the clinical presentation and biochemical phenotype in the affected individuals.
Figure 2.
In Vivo and In Vitro Pathology of the SLC25A26 Mutations
(A) 4-fold serial dilution of wild-type (WT) yeast cells, SAM5Δ cells, and SAM5Δ cells transformed with WT SAMC-pYES2 (SAMC), p.Ala102Val SAMC-pYES2 (p.Ala102Val), p.Val148Gly SAMC-pYES2 (p.Val148Gly), p.Pro199Leu SAMC-pYES2 (p.Pro199Leu), and short SAMC-pYES2 (SAMCΔ1–88) were plated on YP medium supplemented with 3% glycerol and 0.05% galactose for 72 hr at 30°C.
(B and C) Liposomes reconstituted with WT or the indicated SAMC variants were preloaded with 10 mM S-adenosylmethionine at 25°C. Transport was started with 1 mM [3H]S-adenosylmethionine and terminated after (B) 45 s or (C) 60 min. The values are means ± SD of at least four independent experiments.
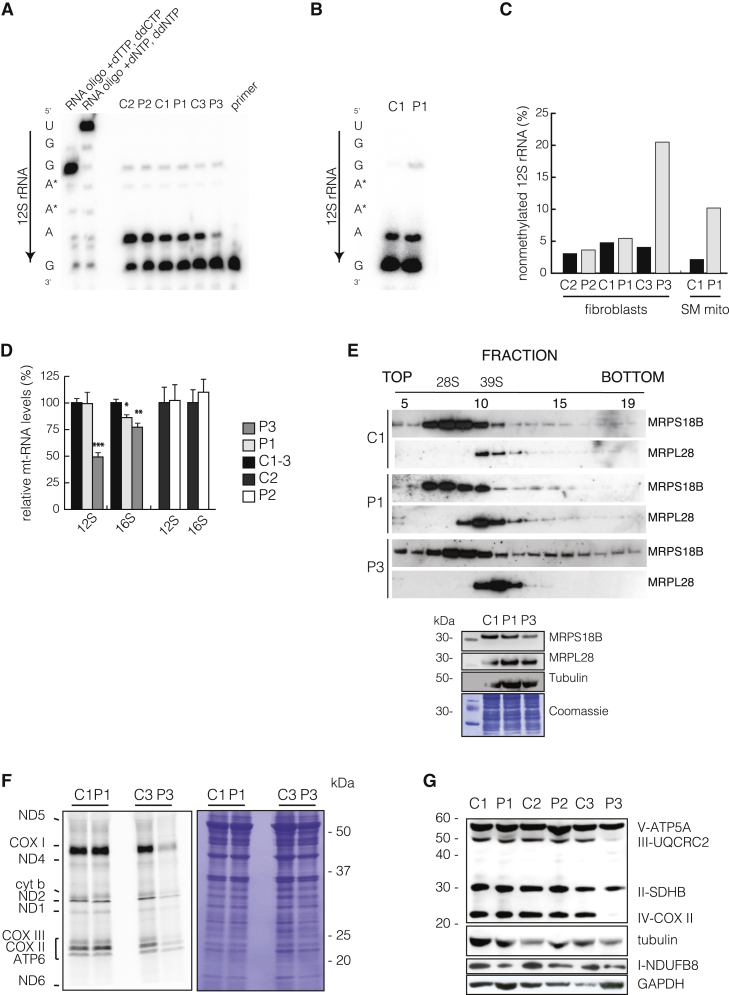
Methylation is required for a multitude of mitochondrial processes, including RNA and protein modifications, and we therefore investigated the status of adenine dimethylation in the hairpin loop at the 3′ end of the mitochondrial 12S rRNA by poisoned primer extension,20 known to be methylated via mtSAM.21, 22, 23 In control samples, the majority of 12S rRNA molecules were dimethylated at adenines 936–937, whereas fibroblasts from P3 (Figure 3A) and skeletal-muscle mitochondria from P1 (Figure 3B) revealed a substantial shift from methylated to non-methylated ribosomal transcripts (Figure 3C). Surprisingly, not only did we fail to observe a methylation defect in fibroblast samples from individuals P1 and P2, but there was also substantial termination of primer extension in P3 fibroblasts, suggesting some methylation of 12S rRNA despite the complete lack of SAMC activity. 12S rRNA steady-state levels are dependent on adenine dimethylation,23 and in agreement with this, 12S rRNA steady-state levels in fibroblasts from P3 were decreased (Figure 3D), whereas all other transcripts tested had only mild changes (Figures S6A and S6B). Additionally, mitochondrial ribosomal assembly was only moderately affected in P3, who showed reduced amounts of the small and possible stabilization of the large mitochondrial ribosome subunits (Figure 3E). Despite the mild effect on mitochondrial ribosome assembly, de novo mitochondrial translation25 was severely affected in P3 fibroblasts (Figure 3F), possibly because methylation is required for tRNA maturation. This defect is also reflected by the reduced steady-state level of COXII (Figure 3G), a subunit of complex IV, and most likely contributes to the mitochondrial dysfunction in P1 skeletal muscle, which showed reduced levels of complexes I and IV (Figure S1).
Figure 3.
Affected Mitochondrial Translation
(A and B) Poisoned primer extension on total RNA from (A) fibroblasts or (B) skeletal-muscle mitochondria and subsequent size separation by denaturing PAGE. [32P] end-labeled oligo complement to the 3′ terminus of 12S rRNA was annealed to RNA extracts and elongated in the presence of dTTP and ddCTP by M-MLV reverse transcriptase. In the case of adenine dimethylation, reverse transcription will terminate upstream of the dimethylation, whereas in its absence, termination will occur immediately downstream of the first guanidine residue because of ddCTP.
(C) Quantification of termination and read-through of (A) and (B).
(D) qRT-PCR of the steady-state levels of 12S and 16S rRNA in fibroblasts. The mean value of two independent experiments performed in triplicate is shown.
(E) Ribosomal gradients (top panel) from fibroblast mitochondria of P1 and P3. Ribosomes were separated in 10%–30% sucrose gradient by centrifugation and then fractionated as previously described,24 with slight modifications. Western blot analysis against subunits of the small ribosomal subunit (28S; MRPS18B) or large subunit (39S; MRPL28) revealed their individual migration and ribosomal monosome (55S) formation. Loading onto the gradient was controlled by input western blot analysis (bottom panel) against mtSSU (MRPS18B), mt-LSU (MRPL28), and tubulin. Additionally, a Coomassie stain is shown.
(F) For determining de novo translation,25 fibroblasts were cultured for 45 min in the presence of [35S] methionine and cysteine; then, protein extracts were separated by SDS-PAGE, and the gel was exposed. The low-molecular-weight subunits of ND3, ATP8, and ND4L are not shown.
(G) Western blot analysis of fibroblasts used antibodies against nuclear-encoded subunits of complexes I–V.
Several mitochondrial proteins are known to be methylated by S-adenosylmethionine-dependent methyltransferases.26, 27 We studied the methylation status of three known mitochondrial SAM targets, ADP/ATP translocators ANT1 and ANT2, and the electron-transferring flavoprotein ETFB. Western blot analysis against di- and tri-methyl lysine (DTML) revealed decreased methylation levels in all fibroblast samples from affected individuals, and P3 was the most severely affected (Figure 4A). Transfection of cell lines from affected individuals with exogenous ANT1 and ANT2 further confirmed the methylation deficiency (Figure 4B). Loss of protein methylation was further rescued by wild-type SAMC in fibroblasts from P2 and P3 (Figure 4C).
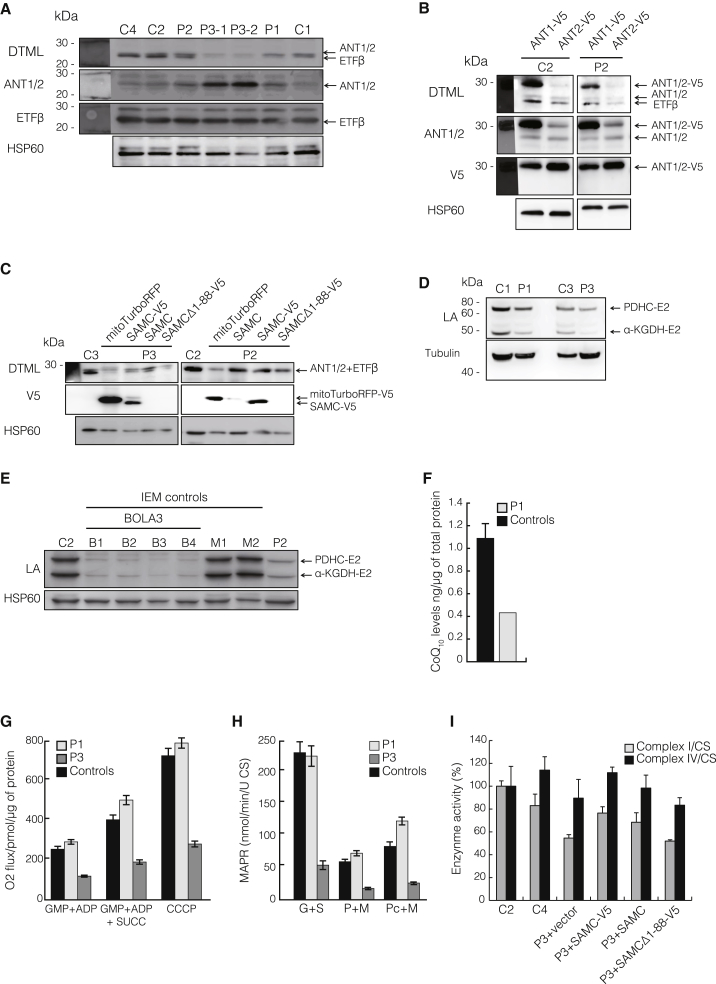
Figure 4.
Effects of Reduced Mitochondrial Methylation
(A) Steady-state levels of ANT1, ANT2, and ETFβ (middle panels) in individuals P1–P3 and control cells (C1, C2, and C4), as well as DTML levels (upper panel) normalized to HSP60.
(B) Control (C2) or P2 fibroblasts were transfected with V5-tagged isoforms of ANT (ANT1-V5 and ANT2-V5) for determining DTML methylation of ANT1-V5 and ANT2-V5.
(C) Western blot analysis of DTML levels in samples from control (C2 and C3) and P2 and P3 fibroblasts transfected with empty vector (mitoTurboRFP), wild-type SAMC (SAMC), V5-tagged SAMC (SAMC-V5), or the N-terminal-truncated SAMC (SAMCΔ-V5).
(D and E) Western blot analysis of the lipoic acid (LA) subunits pyruvate dehydrogenase complex E2 (PDHC-E2) and alpha-ketoglutarate dehydrogenase E2 (α-KGDH-E2) in (D) control (C1 and C3) and P1 and P3 samples or in (E) control (C2) or affected (B1–B4, M1 and M2, and P2) samples. Control samples were obtained from individuals with non-related inborn errors of metabolism (IEMs) and either mutations in BOLA3 (BolA family member 3) (B1–B4) or unrelated mitochondrial diseases (M1 and M2).
(F) CoQ10 levels in mitochondrial extracts from skeletal muscle were determined by ultra-performance liquid chromatography tandem mass spectrometry7, 28 in four control samples (black) and muscles from affected individuals (gray). Control values are the mean ± SD of four control samples.
(G) Mitochondrial oxygen consumption of control (black) or P1 and P3 (gray) fibroblasts. Measurements were performed on an Oroboros oxygraph in the presence of (left) complex I substrates glutamate, malate, pyruvate (GMP), and ADP; (middle) complex I and II substrates GMP, succinate, and ADP; or (right) complex I and II substrates GMP, ADP, succinate, and the mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Error bars indicate the SEM of three independent experiments.
(H) Mitochondrial ATP production rate (MAPR)29 in control (C1–C3; black) and P1 and P3 (gray) fibroblasts was determined by a firefly-luciferase-based method using glutamate and succinate (G+S), pyruvate and malate (P+M), or palmitoyl-L-carnitine and malate (Pc+M) as a substrate at 25°C. Results are presented as the ATP synthesis rate (units) per unit of citrate synthase (CS) activity. Values are the mean ± SEM of three independent experiments.
(I) Isolated enzyme activities29, 30 of complexes I (gray) and IV (black) are normalized to citrate synthase (CS) activities from control (C2 and C4) and P3 fibroblast cell lines after transfection with empty vector (mitoTurboRFP; P3), V5-tagged SAMC (SAMC-V5), SAMC, or V5-tagged SAMCΔ1–88.
Lipoic acid (LA) metabolism depends heavily on SAM-dependent methylation within mitochondria.31 Individual P1 presented with high plasma glycine and low ATP production in muscle when pyruvate was used as a substrate, consistent with deficiencies of the glycine cleavage system and the pyruvate dehydrogenase complex, both of which require LA. These measurements were not performed for individual P2 or P3. Fibroblasts from individuals P1–P3 showed reduced levels of the LA subunits pyruvate dehydrogenase complex E2 (PDHC-E2) and alpha-ketoglutarate dehydrogenase E2 (α-KGDH-E2) (Figures 4D and 4E), and P3 was the most severely affected. This decrease was not secondary to the mitochondrial dysfunction observed, given that two independent samples from individuals with unrelated mitochondrial diseases showed normal levels of LA (M1 and M2 in Figure 4E), whereas samples from individuals with mutations affecting LA biosynthesis were severely reduced (B1–B4 in Figure 4E).
The final steps of coenzyme Q10 (CoQ10) biosynthesis, including several methylation steps of the benzoquinone ring, are performed within the mitochondrial network.32 We therefore measured CoQ10 levels in isolated skeletal-muscle mitochondria from P1 as previously described7, 28 and observed that they were severely decreased, presumably as a result of impaired CoQ10 biosynthesis (Figure 4F). In order to investigate the bioenergetic consequences of reduced mtSAM import, we measured both oxygen consumption (Figure 4G) and mitochondrial ATP production rates (Figure 4H) in fibroblasts carrying the mildest (P1) or null (P3) mutations. Fibroblasts from P3 showed reduced oxygen consumption (Figure 4G) and reduced mitochondrial ATP production rates (Figure 4H), whereas P1 fibroblasts, in contrast to muscle samples (Figure S1A), showed no defect. Finally, the biochemical defects of P3 fibroblasts in the activity of complexes I and IV was rescued by transiently expressing wild-type and tagged wild-type SAMC, but not SAMCΔ1–88 (Figure 4I).
In summary, we have presented three individuals affected by a primary defect in the mitochondrial methylome. Our results show that impaired SAM transport into mitochondria causes a complex syndrome causing multiple primary defects, including those affecting RNA stability, protein modification, mitochondrial translation, and the biosynthesis of CoQ10 and LA. We identified three individuals who originate from different ethnic groups and share striking similarities both biochemically and clinically, consistent with the degree of residual SAM-import capacity. Surprisingly, even though we studied SAMC-null samples, we detected some degree of intra-mitochondrial methylation, suggesting that other forms of methylation or recycling of methyl groups originating from imported methylated proteins might occur within mitochondria.
Acknowledgments
We thank the families who participated in this study; Y. Mogami, K. Tominaga, Y. Tokuzawa, H. Nyuzuki, Y. Yatsuka, S. Tamaru, C. Shimizu, and S. Suzuki for technical assistance; and H. Miyoshi of Keio University and RIKEN BioResource Center for the CS-CA-MCS plasmid. This work was supported by the Japanese Ministry of Education, Culture, Sports, Science, and Technology (Innovative Cell Biology by Innovative Technology and Strategic Research Centers at private universities) and the Takeda Science Foundation to Y.O.; Japan Society for the Promotion of Science KAKENHI 20634398 to Y.K.; Research on Intractable Diseases (Mitochondrial Disorder) from the Ministry of Health, Labor, and Welfare of Japan to A.O.; and Grants-in-Aid for the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development to K.M. Support was also given by the Swedish Research Council (A. Wedell [VR12198]; A. Wredenberg [VR521-2012-2571]); Karolinska Institutet (A. Wredenberg [2013fobi38557]; C.F. [2013fobi37932]); Åke Wiberg Foundation (A. Wredenberg [738762088]; C.F. [367990950]); Stockholm County Council (A. Wedell [20140053]; A. Wredenberg [K0176-2012]); Swedish Foundation for Strategic Research (A. Wredenberg [ICA 12-0017]); and Knut & Alice Wallenberg Foundation (A. Wedell and A. Wredenberg [KAW 20130026]). A. Wredenberg is a Ragnar Söderberg fellow (M77/13). F.P. received grants from the Comitato Telethon Fondazione Onlus (GGP11139) and Italian Human ProteomeNet (RBRNO7BMCT-009). B.L.L. is a senior clinical investigator of the Fund for Scientific Research, Flanders (FWO, Belgium), holds a European Research Council (ERC) starting grant, and received additional support from the FWO (G.0221.12) and ERC.
Published: October 29, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Data include a Supplemental Note and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.09.013.
Contributor Information
Ferdinando Palmieri, Email: ferdinando.palmieri@uniba.it.
Anna Wredenberg, Email: anna.wredenberg@ki.se.
Web Resources
The URLs for data presented herein are as follows:
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Carrasco M., Rabaneda L.G., Murillo-Carretero M., Ortega-Martínez S., Martínez-Chantar M.L., Woodhoo A., Luka Z., Wagner C., Lu S.C., Mato J.M. Glycine N-methyltransferase expression in the hippocampus and its role in neurogenesis and cognitive performance. Hippocampus. 2014;24:840–852. doi: 10.1002/hipo.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Infantino V., Castegna A., Iacobazzi F., Spera I., Scala I., Andria G., Iacobazzi V. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione levels in Down’s syndrome. Mol. Genet. Metab. 2011;102:378–382. doi: 10.1016/j.ymgme.2010.11.166. [DOI] [PubMed] [Google Scholar]
- 3.Clarke S.G. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 2013;38:243–252. doi: 10.1016/j.tibs.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrimi G., Di Noia M.A., Marobbio C.M.T., Fiermonte G., Lasorsa F.M., Palmieri F. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 2004;379:183–190. doi: 10.1042/BJ20031664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranneheim, H. (2014). Mutation Identification Pipeline (MIP), https://github.com/henrikstranneheim/MIP.
- 6.Stranneheim H., Engvall M., Naess K., Lesko N., Larsson P., Dahlberg M., Andeer R., Wredenberg A., Freyer C., Barbaro M. Rapid pulsed whole genome sequencing for comprehensive acute diagnostics of inborn errors of metabolism. BMC Genomics. 2014;15:1090. doi: 10.1186/1471-2164-15-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freyer C., Stranneheim H., Naess K., Mourier A., Felser A., Maffezzini C., Lesko N., Bruhn H., Engvall M., Wibom R. Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J. Med. Genet. 2015 doi: 10.1136/jmedgenet-2015-102986. Published online June 17, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohtake A., Murayama K., Mori M., Harashima H., Yamazaki T., Tamaru S., Yamashita Y., Kishita Y., Nakachi Y., Kohda M. Diagnosis and molecular basis of mitochondrial respiratory chain disorders: exome sequencing for disease gene identification. Biochim. Biophys. Acta. 2014;1840:1355–1359. doi: 10.1016/j.bbagen.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Haack T.B., Jackson C.B., Murayama K., Kremer L.S., Schaller A., Kotzaeridou U., de Vries M.C., Schottmann G., Santra S., Büchner B. Deficiency of ECHS1 causes mitochondrial encephalopathy with cardiac involvement. Ann. Clin. Transl. Neurol. 2015;2:492–509. doi: 10.1002/acn3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharpf R.B., Parmigiani G., Pevsner J., Ruczinski I. Hidden Markov models for the assessment of chromosomal alterations using high-throughput SNP arrays. Ann. Appl. Stat. 2008;2:687–713. doi: 10.1214/07-AOAS155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandeweyer G., Reyniers E., Wuyts W., Rooms L., Kooy R.F. CNV-WebStore: online CNV analysis, storage and interpretation. BMC Bioinformatics. 2011;12:4. doi: 10.1186/1471-2105-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierri C.L., Palmieri F., De Grassi A. Single-nucleotide evolution quantifies the importance of each site along the structure of mitochondrial carriers. Cell. Mol. Life Sci. 2014;71:349–364. doi: 10.1007/s00018-013-1389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 2013;34:465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Marobbio C.M.T., Agrimi G., Lasorsa F.M., Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003;22:5975–5982. doi: 10.1093/emboj/cdg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmieri F., Indiveri C., Bisaccia F., Iacobazzi V. Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Methods Enzymol. 1995;260:349–369. doi: 10.1016/0076-6879(95)60150-3. [DOI] [PubMed] [Google Scholar]
- 16.Capobianco L., Bisaccia F., Mazzeo M., Palmieri F. The mitochondrial oxoglutarate carrier: sulfhydryl reagents bind to cysteine-184, and this interaction is enhanced by substrate binding. Biochemistry. 1996;35:8974–8980. doi: 10.1021/bi960258v. [DOI] [PubMed] [Google Scholar]
- 17.Palmieri L., De Marco V., Iacobazzi V., Palmieri F., Runswick M.J., Walker J.E. Identification of the yeast ARG-11 gene as a mitochondrial ornithine carrier involved in arginine biosynthesis. FEBS Lett. 1997;410:447–451. doi: 10.1016/s0014-5793(97)00630-3. [DOI] [PubMed] [Google Scholar]
- 18.Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M.I., Studart-Guimaraes C., Fernie A.R., Palmieri F. Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 2006;142:855–865. doi: 10.1104/pp.106.086975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmieri F., Klingenberg M. Direct methods for measuring metabolite transport and distribution in mitochondria. Methods Enzymol. 1979;56:279–301. doi: 10.1016/0076-6879(79)56029-7. [DOI] [PubMed] [Google Scholar]
- 20.Rio D.C., Ares M., Hannon G.J., Nilsen T.W. Cold Spring Harbor Laboratory Press; 2011. RNA: A Laboratory Manual. [Google Scholar]
- 21.Helser T.L., Davies J.E., Dahlberg J.E. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat. New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 22.Poldermans B., Van Buul C.P., Van Knippenberg P.H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J. Biol. Chem. 1979;254:9090–9093. [PubMed] [Google Scholar]
- 23.Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.-G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Rorbach J., Boesch P., Gammage P.A., Nicholls T.J.J., Pearce S.F., Patel D., Hauser A., Perocchi F., Minczuk M. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell. 2014;25:2542–2555. doi: 10.1091/mbc.E14-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leary S.C., Sasarman F. Oxidative phosphorylation: synthesis of mitochondrially encoded proteins and assembly of individual structural subunits into functional holoenzyme complexes. Methods Mol. Biol. 2009;554:143–162. doi: 10.1007/978-1-59745-521-3_10. [DOI] [PubMed] [Google Scholar]
- 26.Rhein V.F., Carroll J., Ding S., Fearnley I.M., Walker J.E. NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I. J. Biol. Chem. 2013;288:33016–33026. doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhein V.F., Carroll J., He J., Ding S., Fearnley I.M., Walker J.E. Human METTL20 methylates lysine residues adjacent to the recognition loop of the electron transfer flavoprotein in mitochondria. J. Biol. Chem. 2014;289:24640–24651. doi: 10.1074/jbc.M114.580464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C., Larsson N.G. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 2015;208:429–442. doi: 10.1083/jcb.201411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wibom R., Hagenfeldt L., von Döbeln U. Measurement of ATP production and respiratory chain enzyme activities in mitochondria isolated from small muscle biopsy samples. Anal. Biochem. 2002;311:139–151. doi: 10.1016/s0003-2697(02)00424-4. [DOI] [PubMed] [Google Scholar]
- 30.Kirby D.M., Thorburn D.R., Turnbull D.M., Taylor R.W. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- 31.Booker S.J., Cicchillo R.M., Grove T.L. Self-sacrifice in radical S-adenosylmethionine proteins. Curr. Opin. Chem. Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laredj L.N., Licitra F., Puccio H.M. The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie. 2014;100:78–87. doi: 10.1016/j.biochi.2013.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.