Abstract
The genomic duplication associated with Potocki-Lupski syndrome (PTLS) maps in close proximity to the duplication associated with Charcot-Marie-Tooth disease type 1A (CMT1A). PTLS is characterized by hypotonia, failure to thrive, reduced body weight, intellectual disability, and autistic features. CMT1A is a common autosomal dominant distal symmetric peripheral polyneuropathy. The key dosage-sensitive genes RAI1 and PMP22 are respectively associated with PTLS and CMT1A. Recurrent duplications accounting for the majority of subjects with these conditions are mediated by nonallelic homologous recombination between distinct low-copy repeat (LCR) substrates. The LCRs flanking a contiguous genomic interval encompassing both RAI1 and PMP22 do not share extensive homology; thus, duplications encompassing both loci are rare and potentially generated by a different mutational mechanism. We characterized genomic rearrangements that simultaneously duplicate PMP22 and RAI1, including nine potential complex genomic rearrangements, in 23 subjects by high-resolution array comparative genomic hybridization and breakpoint junction sequencing. Insertions and microhomologies were found at the breakpoint junctions, suggesting potential replicative mechanisms for rearrangement formation. At the breakpoint junctions of these nonrecurrent rearrangements, enrichment of repetitive DNA sequences was observed, indicating that they might predispose to genomic instability and rearrangement. Clinical evaluation revealed blended PTLS and CMT1A phenotypes with a potential earlier onset of neuropathy. Moreover, additional clinical findings might be observed due to the extra duplicated material included in the rearrangements. Our genomic analysis suggests replicative mechanisms as a predominant mechanism underlying PMP22-RAI1 contiguous gene duplications and provides further evidence supporting the role of complex genomic architecture in genomic instability.
Introduction
Contiguous gene syndromes are caused by chromosome abnormalities that affect multiple contiguous genes and change the gene organization and/or dosage on a chromosome. Such syndromes are mostly rare, sporadic, and heterogeneous with a wide spectrum of clinical phenotypes.1 Contiguous gene syndromes, in contrast to genetic disorders caused by point mutations, result from the rearrangements of large genomic intervals.2, 3 Conditions resulting from such changes are referred to as genomic disorders.4 The rearrangements might disrupt the structural integrity of a gene or alter the copy number of dosage-sensitive genes by generating copy-number variants (CNVs), which are submicroscopic, unbalanced structural variants ranging from kilobases (kb) to megabases (Mb) in size.5 These CNV result in locus specific deviation from the normal diploid state.2
Copy-number gain or loss of a specific genomic interval might result in genomic disorders with distinct phenotypes. For example, duplication of chromosome 17p11.2 results in Potocki-Lupski syndrome (PTLS [MIM 610883]), while its reciprocal deletion leads to Smith-Magenis syndrome (SMS [MIM 182290]).6, 7, 8 Phenotypes resulting from reciprocal CNVs in dosage-sensitive genes can convey mirror image traits as shown for weight/body mass in subjects with PTLS (lean) and SMS (obese), and the corresponding chromosome-engineered mouse models.9, 10
Mechanistically, from a structural variant mutagenesis viewpoint, nonallelic homologous recombination (NAHR) has been demonstrated as the major molecular mechanism for disease-causing recurrent genomic rearrangements.5 NAHR utilizes long homologous repeats, such as low-copy repeats (LCR), as substrates to mediate structural changes of chromosomes. Two major LCR clusters flank the SMS critical region on human chromosome 17p11.2 in direct orientation, termed proximal and distal SMS-REPs.11 Unequal crossover between the proximal and distal SMS-REPs results in the ∼3.6 Mb common recurrent SMS deletion observed in ∼70%–80% of SMS cases, as well as the reciprocal ∼3.6 Mb common recurrent PTLS duplication observed in ∼60%–70% of PTLS cases.7, 12, 13, 14, 15 Besides these NAHR-mediated recurrent CNVs, duplications with a broad size distribution, i.e., nonrecurrent, have been identified in subjects with PTLS phenotypes. These duplications are unique to individual subjects, and arise from mechanisms other than NAHR.16 The smallest region of overlap (SRO) analysis of these duplications narrowed the critical region of PTLS to a 125 kb interval solely encompassing RAI1 (retinoic acid induced 1 [MIM 607642]), a dosage sensitive gene responsible for SMS and likely for PTLS.16, 17, 18, 19
The 17p12 locus, distal to the SMS/PTLS critical region, presents with two other LCR clusters (termed CMT1A-REPs) that can also be substrates for NAHR. NAHR between the two CMT1A REPs results in the common recurrent ∼1.4 Mb duplication associated with Charcot-Marie-Tooth disease type 1A (CMT1A [MIM 118220]).20, 21, 22 The dosage-sensitive gene PMP22 (peripheral myelin protein 22 [MIM 601097]) is included in the CMT1A duplication, and has been shown to be responsible for CMT1A.23 Analogous to the distinct clinical SMS/PTLS phenotypes associated with reciprocal 17p11.2 rearrangements, a reciprocal deletion of CMT1A duplication results in hereditary neuropathy with liability to pressure palsies (HNPP [MIM 162500]), a condition distinct from CMT1A.24
Common recurrent PTLS duplication and CMT1A duplication are independent genomic rearrangement events, because they utilize distinct LCR pairs. RAI1 and PMP22, associated with PTLS and CMT1A, respectively, are in close physical proximity in the human genome—the distance between them is approximately 2.5 Mb. As a result, duplications encompassing both genes are observed.6, 14, 23, 25, 26, 27, 28, 29 We termed such duplications PMP22-RAI1 contiguous gene duplications, or in short, PMP22-RAI1 duplications. However, a systematic analysis and documentation of these duplications with a focus on molecular mechanisms and detailed phenotyping is lacking. In contrast to independent recurrent PTLS and CMT1A duplications determined by LCR substrates, the PMP22-RAI1 duplications are nonrecurrent and not delimited by highly identical LCRs; thus they potentially arise from mechanisms other than NAHR. Replicative mechanisms, such as fork stalling and template switching/microhomology-mediated break-induced replication (FoSTeS/MMBIR), have been proposed to generate nonrecurrent rearrangements independent of LCRs.25, 26 These replication-based mechanisms, as opposed to NAHR, might contribute to the formation of the PMP22-RAI1 duplications. Furthermore, we investigated whether copy-number gains of two distinct dosage-sensitive genes led to a new contiguous gene syndrome that appeared clinically to represent more than the addition of the two resultant genomic disorders.1
The clinical presentation of PTLS includes infantile hypotonia, failure to thrive, intellectual disability, autistic features, sleep apnea, and structural cardiovascular anomalies.7, 14, 27 Phenotypic variability has been observed in subjects harboring different duplication sizes, all of which encompass RAI1.7, 16 CMT1A is the most common inherited peripheral neuropathy in humans and is characterized by distal symmetric polyneuropathy, distal muscle atrophy, foot deformities and a steppage gait, reduced deep tendon reflexes, decreased nerve conduction velocities, and hypertrophic neuropathy on nerve biopsy.22 We hypothesize that duplications encompassing both RAI1 and PMP22 might possibly confer a blended phenotype with potential additive clinical presentations, which might result from the interplay between the phenotypes of infantile hypotonia characteristic of PTLS and peripheral neuropathy, the hallmark of CMT1A.7, 28, 29 This model is consistent with a hypothesis of “mutational load” due to a CNV encompassing two dosage-sensitive genes. It also reflects the feature of a wide spectrum of clinical phenotypes in contiguous gene syndromes, which might be associated with the number of genes involved.1, 30
In the current study, we report 23 subjects identified with nonrecurrent PMP22-RAI1 duplications with sizes ranging from 3.2 Mb to 19.7 Mb. High-resolution array comparative genomic hybridization (array CGH) followed by breakpoint junction sequencing allowed for precise demarcation of the boundaries of the duplications, and revealed mutational signatures that enable us to surmise the potential molecular mechanisms for generating the observed genomic rearrangements. We explore the possibility that perturbation of two well-characterized dosage-sensitive genes might result in a phenotype that is exacerbated in comparison to the individual contribution of each gene. PTLS often presents in infancy, but CMT1A usually presents later in the first or even second decade of life. Thus, the molecular diagnosis in the subjects with PMP22-RAI1 contiguous gene duplication syndrome often precedes the clinical onset of neuropathy and can help facilitate timely intervention and clinical management. Moreover, the concurrence of PTLS and CMT1A suggests an early molecular diagnosis for subjects with PTLS in order to periodically assess for potential neuropathy. In summary, our study, focusing on both genotype-phenotype correlation and the underlying molecular mechanism, provides a systematic investigation of a group of subjects with genomic rearrangements that can lead to overlapping phenotypic outcomes blending clinical aspects of both PTLS and CMT1A.
Experimental Procedures
Human Subjects
A total number of 127 subjects with proximal chromosome 17p duplication encompassing RAI1 were enrolled in a research protocol approved by the Institutional Review Board for Baylor College of Medicine and Affiliated Hospitals. Informed consent was obtained from the subjects and parents or legal guardians of the subjects. Genomic DNA was extracted from whole blood using the Gentra Puregene Blood Extraction Kit per manufacturer’s protocol (QIAGEN). All genomic studies, including array CGH and breakpoint junction sequencing (below), were performed on DNA samples extracted from whole blood.
Twenty-three out of 127 subjects were identified with the PMP22-RAI1 duplication. Molecular data were available for all subjects, while clinical information was available for 17 of the 23 subjects (Table 1 and Table S1). BABs 527,31 1458,32 1861,29 2211,7 2337,33 2362,7 2488,7 and 266128 were reported with both clinical and molecular studies. We revisited their clinical data combining the medical charts and published clinical information. BABs 1229,7 2703,16 2711,25 and 279016 were reported previously without clinical information included in the report. Medical records of these subjects were carefully evaluated. BABs 3813, 6299, 6504, 6635, 6636, 6966, 6967, 6968, 7679, 7873, and 8300 were recently enrolled subjects without any clinical or cytogenetic information reported.
Table 1.
Clinical Information of the Subjects with PMP22-RAI1 Contiguous Gene Duplications
| Identifier | 527 | 1229 | 1458 | 1861 | 2211 | 2337 | 2362 | 2488 | 2661 | 2711 | 3813 | 6504 | 6635 | 6636 | 7679 | 7873 | 8300 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref | 30 | 7 | 31 | 28 | 7 | 32 | 7 | 7 | 27 | 24 | − | − | − | − | − | − | − | |
| Sex | M | M | M | F | M | M | M | M | F | F | M | M | M | M | M | M | F | |
| Age at last exam | 9 y | 4.5 y | 19 m | 7.5 y | 4 y 10 m | 11 m | 11 y 6 m | 9 y | 2 y 9 m | 3 y | 3 y | 3.5 y | 33y | 13 y | 15 m | 22 m | 3 y 11 m | |
| Infancy | Feeding difficulties | + | NR | NR | + | + | NR | + | + | + | + | NR | + | + | + | + | + | − |
| Failure to thrive | + | + | + | − | + | + | − | + | + | − | − | + | + | + | − | + | − | |
| Infantile hypotonia | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Development | Developmental delay | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Age at walking | 6 y | 24 m | NR | 4.5 y | 18 m | NR | 24 m | 15 m | 28 m | 19 m | 24 m | NR | 24 m | 4 y | Not yet at 15 m | Not yet at 22 m | 19 m | |
| Behavioral difficulties | + | + | NR | NR | + | NR | + | + | + | + | + | NR | + | + | NR | − | − | |
| Language delay | + | + | NR | + | + | NR | + | + | + | + | + | + | + | + | + | + | + | |
| Clinical neuropathy (age at onset), exercise intolerance | + (infancy) | + | NR | + | + | NR | + (22 m) | + | + (Before 2 y) | − | + | NR | 15y | + (first decade) | − | − | NR | |
| Sleep disturbance | + | − | NR | NR | + | NR | + | NR | NR | + | − | NR | NR | + | NR | − | + | |
| Physical features | Facial dysmorphisma | + | + | + | + | + | NR | + | + | + | + | + | NR | + | + | + | + | + |
| Foot deformities | Sandle gap, 2-3 syndactyly | − | NR | Congenital talipes equinovarus | hyperpronation of feet | bilateral talipes equinovarus | bilateral pes planus, hindfoot valgus | + | NR | − | − | + | bilateral pes planus, hindfoot valgus | bilateral pes cavus, unilateral equinovarus | − | bilateral pes planus, hindfoot valgus | hindfoot valgus | |
| Unusual gait/ dropped foot | + | + | NR | + | + | NR | Wide-based gait | − | Ataxic gait | + | + | NR | + | Wide-based gait | − | − | + | |
| Distal extremity weakness or atrophy | Hypertonicity lower extremities | + | NR | + | + | NR | + | + | NR | − | + | Generalized hypotonia; increased tone on left arm and leg | + | + | + | hypotonia without muscular weakness | + | |
| Sensory loss | + | − | NR | NR | + | NR | + | − | NR | − | NR | NR | − | + | NR | NR | NR | |
| Reduced or absent ankle DTRs | + | + | NR | + | + | NR | + | + | + | − | + | NR | + | + | + | − | + | |
| Studies and imaging | Brain MRI abnormality | + | − | NR | + | + | NR | − | NR | − | + | NR | + | NR | + | + | − | − |
| Syringomyelia | − | NR | NR | NR | + | NR | NR | NR | + | NR | NR | NR | NR | − | NR | NR | NR | |
| Congenital heart defect | DORV, VSD, ASD, overriding aorta | NR | NR | NR | Dilated aortic root, aortic aneurysm, bicuspid aortic valve, PFO, possible VSD | Bicuspid aortic valve | NR | NR | VSD | − | − | − | Dilated aortic root | − | ASD/PFO, persistent LSVC | Dysplastic aortic valve | Dilated aortic root | |
| Renal abnormality | Small cysts | NR | Alport syndrome | − | Malrotation left kidney | NR | − | NR | NR | − | NR | NR | − | − | hydronephrosis | − | − | |
| Median nerve motor NCV (m/sec) | 20.1 | 19.7 | NR | NR | 15.4 (peroneal) | NR | 23.2 | NR | decreased | NR | NR | NR | NR | 24 | NR | NR | NR | |
| Median nerve distal CMAP (mV) | 6 | 3.4 | NR | NR | 2.4 | NR | 6.8 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | |
Abbreviations are as follows: ASD, atrial septal defect; CMAP, compound muscle action potential; DORV, double outlet right ventricle; LSVC, left superior vena cava; NCV, nerve conduction velocity; NR, not reported; PFO, patent foramen ovale; VSD, ventricular septal defect; y, years; m, months.
Details are included in Table S1.
17p Array CGH
We designed a 4 × 180K format array CGH (AMADID# 032121, Agilent Technologies) with high-density probes tiling the critical region of CMT1A and PTLS in chromosome region 17p11.2p12. The probe density averaged 10 oligonucleotides per kb for 17p11.2p13.1, one oligonucleotide per kb for 17p13.2p13.3, one oligonucleotide per 2 kb for the remainder of chromosome 17, and one oligonucleotide per 100 kb for the rest of the genome. Repetitive sequences and repeats (e.g., LCRs) were not densely interrogated by this design.
We also designed an 8 × 60K format array CGH (AMADID# 070922, Agilent Technologies) tiling the LCR-rich intervals in chromosome regions 17p11.2p12 with an average probe density of 10 oligonucleotides per kb in order to interrogate breakpoint boundaries inside LCRs.
The experimental procedures, including DNA fragmentation and labeling, array hybridization, washing, scanning, and image processing were performed according to the manufacturer’s protocol (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis, Version 7.2, Agilent Technologies) with modifications. 1.2 μg genomic DNA from proband and control (NA15510) were digested with AluI and RsaI (Promega) at 37°C for 2 hr. A 1% agarose gel was run to verify the digestion product. Labeling of the digestion product was performed using BioPrime Array CGH Genomic Labeling Module (Life Technologies) following the manufacturer’s protocol. Cy5-dCTP and Cy3-dCTP from Cyanine Smart Pack dCTP (PerkinElmer) were used for labeling proband and control digestion product, respectively, at 37°C for 2 hr. Amicon Ultra 0.5 Centrifugal Filter (30 kDa, Millipore) was used to purify labeling product. Labeling efficiency was quantitated using NanoDrop ND-1000 UV-VIS spectrophotometer. Labeled proband and control samples were combined in a mass ratio of 1:1 with addition of 5 μg human Cot-1 DNA (Life Technologies), 10X Agilent Blocking Agent (Agilent Technologies), and 2X Hi-RPM Hybridization Buffer (Agilent Technologies). The mixture was incubated at 95°C for 3 min and then at 37°C for 30 min for pre-hybridization, after which the mixture was placed onto the array and into a hybridization chamber (Agilent Technologies) in a rotating oven (Agilent Technologies) for at least 40 hr at 65°C. Washing steps after hybridization were performed following the manufacturer’s protocol with Agilent Oligo CGH Wash Buffer 1 and 2 (Agilent Technologies), acetonitrile (Sigma-Aldrich), and Stabilization and Drying Solution (Agilent Technologies). The dried slides were scanned on Agilent Microarray Scanner with SureScan High-Resolution Technology (model G2565CA, Agilent Technologies) for image acquisition. The scanned images were subsequently processed by Feature Extraction Software version 11.5 (Agilent Technologies) to generate FE files, which were subsequently imported into Agilent Genomic Workbench version 7.0 (Agilent Technologies) for CNV analysis. Copy number was defined by normalized log2 ratio of Cy5/Cy3 fluorescence signal of each probe. The human genome reference sequence GRCh37/hg19 was used to define the genomic coordinates of detected CNVs. Array CGH experiments were performed on DNA samples from trios to identify de novo CNVs.
Breakpoint Junction Sequencing
High-resolution array CGH revealed copy-number transitions. Long-range PCR was performed to amplify the breakpoint junctions. Forward and reverse primers were designed using the sequences from the estimated boundaries of the rearrangements defined by array CGH. TaKaRa LA Taq (Clontech) was used for the PCR amplifications. The thermocycler was programmed at: 98°C × 30 s; 35 cycles of 95°C × 60 s, 65°C × 20 s, 68°C × 20 min; 68°C × 10 min. Sanger sequencing was performed for the PCR products, and the DNA sequences were compared to the reference genome (hg19) using BLAT in order to map the breakpoint junctions.
LCR Density Analysis
A list of LCRs annotated with genomic coordinates was downloaded from the Segmental Dups track in UCSC Genome Browser (GRCh37/hg19). LCR density is estimated by calculating the ratio of the sum of different subsets of genomic intervals annotated with LCR sequences over the entire genomic interval investigated. An interval will be counted multiple times if it is with multiple LCR annotations. Thus the density might be higher than 1. The moving average is plotted by creating a 100 kb sliding window containing the interval to be investigated and sliding each time by 50 kb.
Results
Clinical Profiling of Contiguous Gene PMP22-RAI1 Duplication Syndrome
To date, we have enrolled 127 subjects with proximal chromosome 17p duplication encompassing RAI1 in research protocols, and 84/127 (66%) subjects were identified with the common recurrent PTLS duplication. Of the remaining 43 subjects, 23 (53%) were found to have contiguous gene duplications encompassing both PMP22 and RAI1. Clinical records of 17/23 subjects with such duplications were available for review (Table 1). The clinical and/or molecular findings of 10 of these subjects were included in previous publications (Table 1). Three of the subjects had additional cytogenetic abnormalities on other chromosomes, including a 47,XYY male (Table S1). The most consistently reported clinical features were feeding difficulties, global developmental delay, behavioral difficulties, and language delay. Age at walking ranged from 15 months to 6 years. Nine subjects presented with clinical neuropathy by 10 years of age; six of these were younger than 5 years (BABs 527, 1229, 2211, 2362, 2661, 3813). Data regarding the presence or absence of clinical neuropathy is not available on 4 of the 17 subjects (Table 1). Four subjects had renal involvement, including three subjects with structural renal abnormalities and one previously published subject with Alport syndrome.32 Congenital heart malformations were present in eight subjects, with five of these involving the left ventricular outflow system. Two subjects had syringomyelia. In BAB 2211, the syringomyelia extended from T5 to T12 and was associated with a tethered cord, whereas in BAB 2661 it was a cervical syringomyelia (C4–C7) with an otherwise normal brain and spine MRI.
Nonrecurrent PMP22-RAI1 Duplications
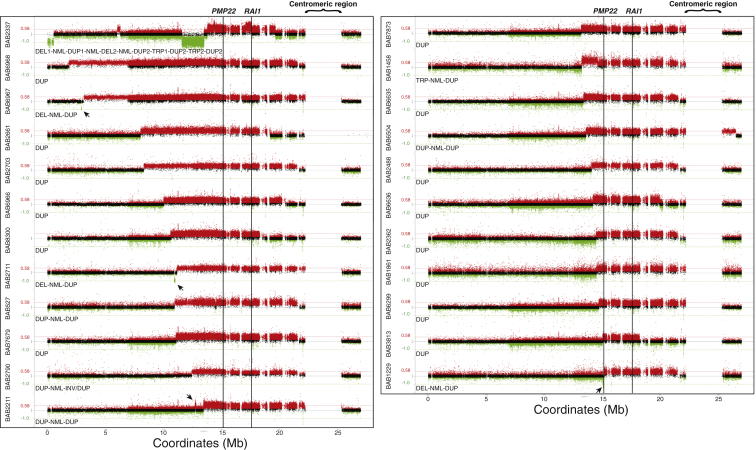
Duplications encompassing both PMP22 and RAI1 were identified in 23 sporadic subjects. Using array CGH targeting chromosome 17p, we obtained a high-resolution delineation of the size, extent, and gene content of these duplications (Figure 1). 14/23 cases (60.87%) had apparently simple duplication rearrangements (Figure 1, Table 2). No additional CNVs other than the PMP22-RAI1 duplications were observed on chromosome 17p in these cases. Interestingly, potential complex genomic rearrangements (CGRs) with higher-level structural complexities were observed in 9/23 cases (39.13%). The CGR pattern in such cases included duplications (DUP), deletions (DEL), and triplications (TRP), often intervened by segments with a normal (NML) copy number (refer to Figure 1 and Table 2 for details). Thus, the PMP22-RAI1 duplications are either simple duplications or involved in CGRs with the SRO encompassing both PMP22 and RAI1 in their entirety. They have variable genomic sizes (from 3.2 Mb in BAB 3813 to 19.7 Mb in BAB 6968) and boundaries of duplicated segments, documenting nonrecurrent duplications.
Figure 1.
Genomic Rearrangements Encompassing PMP22-RAI1 Duplications in 23 Subjects
The genomic rearrangements are shown by array CGH log2 ratio plots. The plot focuses on the short arm of chromosome 17 and a short extension into the long arm, separated by a gap representing the centromeric region. The rearrangement patterns characterized by array CGH are noted underneath each array CGH log2 ratio plot. BABs 1861, 2362, 2488, 2661, 2703, 3813, 6299, 6635, 6636, 6966, 6968, 7679, 7873, and 8300 (14/23, 60.87%) are observed to have simple duplications (DUP). BABs 1229, 2711, and 6967 (3/23, 13.04%) are found to have a distal deletion sequentially followed by a normal copy segment and a proximal duplication (DEL-NML-DUP pattern). BABs 527, 2211, 2790, and 6504 (4/23, 17.39%) are shown to have a distal duplication sequentially followed by a normal copy segment and a proximal duplication (DUP-NML-DUP pattern). Breakpoint sequencing of BAB 2790 further reveals that the distal side of the distal DUP joins the distal side of the proximal DUP in an inverted orientation, representing a DUP-NML-INV/DUP pattern. BAB 1458 (1/23, 4.35%) is found to have a triplication sequentially followed by a normal copy segment and a proximal duplication (TRP-NML-DUP pattern). BAB 2337 (1/23, 4.35%) had a highly complex rearrangement pattern of, from distal to proximal 17p and connected by normal copy segments, a terminal deletion (DEL1), a small duplication (DUP1), a deletion (DEL2) and a large duplication encompassing PMP22 and RAI1 (DUP2) with two small triplicated segments (TRP1 and TRP2) residing within it (DEL1-NML-DUP1-NML-DEL2-NML-DUP2-TRP1-DUP2-TRP2-DUP2 pattern). The plots are arranged according to the starting coordinate of the rearrangements identified in each case. Abbreviations are as follows: DUP, duplication; DEL, deletion; TRP, triplication; NML, normal copy segment. Red (log2 ratio > 0.25), black (−0.25 ≤ log2 ratio ≤ 0.25) and green (log2 ratio < −0.25) dots represent array CGH probes. Red/green horizontal lines mark theoretical log2 ratio of duplication (0.58) / heterozygous deletion (−1.0). Black arrows denote small CNVs. Black vertical lines mark positions of PMP22 and RAI1.
Table 2.
Breakpoint Junction Features of the Rearrangements Identified in 23 Subjects with PMP22-RAI1 Contiguous Gene Duplications
| Individual BAB# | CNV pattern | Inheritance |
Breakpoint junction |
|||||
|---|---|---|---|---|---|---|---|---|
| Segments | Coordinates (Chr17) | Distal boundary | Proximal boundary | Features | Ref | |||
| 527 | DUP-NML-DUP (CGR) |
de novo | DUP | 10954612–14494703 (Min) 10954218–14495430 (Max) |
NA | NA | – | – |
| DUP | 14577325–21557321 (Min) 14576885–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
|||||
| 1229 | DEL-NML-DUP (CGR) |
de novo | DEL | 15075309 a–15113732a | Ua | Ua | 6 bp (ACCTTC) microhomology | 24 |
| DUP | 15117983–21531363 (Min) 15117908–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
|||||
| 1458 | TRP-NML-DUP (CGR) |
de novo | TRP | 13220227–14630812 (Min) 13216466–14630946 (Max) |
NA | NA | – | – |
| DUP | 14634801–22205372 (Min) 14633210–25291809 (Max) |
NA | CEN | |||||
| 1861 | DUP | de novo | DUP | 14541090–22213960 (Min) 14540244–25291809 (Max) |
NA | CEN | – | – |
| 2211 | DUP-NML-DUP (CGR) |
de novo | DUP | 12739434–12805414a (Min) 12738917–12805414a (Max) |
NA | RE (L2a) a |
6 bp (TAATTA) insertion | This study |
| DUP | 13454298a–22171191 (Min) 13454298a–22181378 (Max) |
RE (AluY) a | LCR cluster (PERI-CEN) |
|||||
| 2337 | DEL1-NML-DUP1-NML-DEL2-NML-DUP2-TRP1-DUP2-TRP2-DUP2 (CGR) |
de novo | DEL1 | 0–565389 (Min) 0–566912 (Max) |
NA | NA | Jct1 (AluSp -> T-rich repeats): 18 bp insertionc Jct2 (MER4B-int -> Unique): 33 bp insertionc Jct3 (L1MA4 -> Unique): 257 bp insertionc Jct4 (L1MC4a -> Unique): 551 bp insertionc |
This study |
| DUP1 | 6001081a–6321783a | RE (L1MC4a) a |
RE (AluSp) a |
|||||
| DEL2 | 11587499a–13490898a | Ua | Ua | |||||
| DUP2 | 13797160–19655692a (Min) 13796836–19655692a (Max) |
NA | RE (L1MA4) a |
|||||
| TRP1 | 14362549a–14429100a | RE (T-rich repeats) a |
RE (MER4B-int) a |
|||||
| TRP2 | 17088740a–17596327 (Min) 17088740a–17596376 (Max) |
Ua | NA | |||||
| 2362 | DUP | de novo | DUP | 14507339–21531363 (Min) 14500613–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 2488 | DUP | de novo | DUP | 14062138–21531363 (Min) 14061964–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 2661 | DUP | NA | DUP | 8033957a–19184708a | RE (L1MA7) a |
RE (MER21B) a |
2 bp (AT) microhomology | 24 |
| 2703 | DUP | not maternal | DUP | 8331250–21531363 (Min) 8330470–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 2711 | DEL-NML-DUP (CGR) |
de novo | DEL | 10930335a–11013899a | Ua | RE (MLT1A0) a |
1 bp (C) insertion | 24 |
| DUP | 11164314–22225859 (Min) 11164034–25291809 (Max) |
NA | CEN | |||||
| 2790 | DUP-NML-INV/DUP (CGR) |
NA | DUP | 12434682a–12973003 (Min) 12434682a–12973388 (Max) |
RE (L1MEg) a |
NA | 4 bp (CTTT) insertion | 15 |
| DUP | 12999819a–21531363 (Min) 12999819a–21701186 (Max) |
U A | LCR cluster (17p-PROX) |
|||||
| 3813 | DUP | not maternal | DUP | 15060680a–18212609a | RE (MLT1E2) a |
RE (L1MB5) a |
17 bpb insertion | This study |
| 6299 | DUP | de novo | DUP | 14736306–22234459 (Min) 14735780–25291809 (Max) |
NA | CEN | – | – |
| 6504 | DUP-NML-DUP (CGR) |
de novo | DUP | 13582799–13805140 (Min) 13582620–13805545 (Max) |
NA | NA | – | – |
| DUP | 13825917–26507332 (Min) 13825439–26507855 (Max) |
NA | NA | |||||
| 6635 | DUP | NA | DUP | 13376683–21508916 (Min) 13376614–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 6636 | DUP | not maternal | DUP | 14197653a–19928406a | RE (HAL1b) a |
Ua | 1 bp (A) microhomology | This study |
| 6966 | DUP | NA | DUP | 10016908a–20511549a | Ua | LCR cluster (SMS-REP) a |
4 bp (TCAG) microhomology | This study |
| 6967 | DEL-NML-DUP (CGR) |
NA | DEL | 2930963–2954284 (Min) 2930296–2967871 (Max) |
NA | LCR cluster (distal 17p) |
– | – |
| DUP | 3156474–22234459 (Min) 3143016–25291809 (Max) |
NA | CEN | – | – | |||
| 6968 | DUP | NA | DUP | 1859192–21531363 (Min) 1858544–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 7679 | DUP | de novo | DUP | 11065839–21531640 (Min) 11064378–21701186 (Max) |
NA | LCR cluster (17p-PROX) |
– | – |
| 7873 | DUP | de novo | DUP | 13188763–22234459 (Min) 13187737–25291809 (Max) |
NA | CEN | – | – |
| 8300 | DUP | de novo | DUP | 10608712a–18506553a | Ua | LCR cluster (SMS-REP) a |
3 bp (CCT) microhomology | This study |
Abbreviations are as follows: The CGR patterns of DUP-NML-DUP, DEL-NML-DUP, TRP-NML-DUP and DEL1-NML-DUP1-NML-DEL2-NML-DUP2-TRP1-DUP2-TRP2-DUP2 are described in Figure 1; DUP-NML-INV/DUP, a CGR pattern of DUP-NML-DUP with the distal side of the distal DUP being connected to the distal side of the proximal DUP in an inverted orientation (INV/DUP). DUP, duplication; DEL, deletion; TRP, triplication; NML, normal-copy segment; INV/DUP, inverted duplication; CGR, complex genomic rearrangement; U, Unique sequences; RE, Repetitive elements; CEN, centromere; PERI-CEN, pericentromeric; NA, not available; Min, boundaries of CNVs defined by the two flanking aberration probes; Max, boundaries of CNVs defined by the two flanking normal probes.
Precise position of the boundaries for each CNV by breakpoint sequencing. For the boundaries of CNVs without the breakpoint junction sequenced, we determined the CNV boundaries using the probes flanking the region showing the copy-number transition.
GCTGTGATTCTGTAAGC.
Detailed information is in Figure S3.
We performed array CGH analysis on 14 trios (subjects and their unaffected parents) to investigate the inheritance pattern of the rearrangements observed in the subjects. The results suggested that the rearrangements arose de novo in all the 14 subjects (Table 2). Among these, 7/14 (50%) were potential de novo CGRs with higher-level structural complexity (Table 2). Trio analysis was not performed for the remaining nine families, as no or only one parental sample was available for each affected subject. Thus the inheritance pattern of the rearrangements identified in the subject was not available (Table 2).
Replicative Mutational Mechanism Underlying PMP22-RAI1 Duplications
We attempted long-range PCR for the breakpoint junctions, boundaries of which were estimated by high-resolution array CGH. Sanger sequencing was performed for the breakpoint junctions that achieved successful PCR amplification. Potential molecular mechanisms underlying the genomic rearrangements could be surmised from the “mutational signatures” uncovered at the breakpoint junctions.
Among the 23 nonrecurrent rearrangements identified in 23 subjects, 14 were characterized as simple duplications by array CGH. In these subjects, we expect 14 putative breakpoint junctions, five of which were successfully amplified and sequence determined (Figure S1). Microhomologies were identified in BABs 2661 (AT),25 6636 (A), 6966 (TCAG), and 8300 (CCT) at the breakpoint junction, which are consistent with a potential FoSTeS/MMBIR mechanism.25, 26, 34 Other mechanisms, including microhomology-mediated end-joining (MMEJ),35 might provide alternative explanations for the mechanism underlying these simple duplications observed on 17p. The microhomology-mediated template-switch event might occur once, at this resolution level, for generating each of the observed duplications in BABs 2661, 6636, 6966, and 8300 (Figure S1A). In contrast, in the fifth sample BAB 3813, a 17 bp inserted sequence (GCTGTGATTCTGTAAGC) was identified at the breakpoint junction. This inserted genomic segment could be partially derived from a nearby region (Chr17: 15060672–15060686, Figure S1B). This breakpoint junction feature might be explained by FoSTeS/MMBIR via iterative template switches copying genomic sequences from multiple nearby regions of the breakpoint junction (Figure S1B).
Breakpoint junction sequences were not obtained in the remaining nine cases, because they map at the pericentromeric or centromeric regions within extensive repeats and repetitive sequences, such as LCR or satellite DNA sequences (Figure 2). These sequences limit the experimental approaches to sequencing breakpoint junctions not involving long stretch of homologous sequences. Array CGH targeting LCR sequences on 17p were designed to narrow down those breakpoint junctions within LCRs. However, it was not always successful due to the limited dynamic range and capability of array CGH to reveal transitions between higher order copy number (e.g. quadruplication versus triplication).
Figure 2.
Summary Diagram of the Genomic Rearrangements Identified in 17p
The genomic rearrangements identified in 23 subjects are shown together with common recurrent PTLS duplication.
(A) Ideogram showing the entire short arm of chromosome 17.
(B) Detailed information of the genomic rearrangements in 23 subjects. Different types of CNV identified by array CGH were presented in colored segments. Segments in green, heterozygous deletion; red, duplication; blue, triplication; white, normal copy number; maroon, LCRs or repetitive sequences (e.g., satellite DNA sequences) at the breakpoint junctions. The plots are arranged according to the starting coordinate of the rearrangements identified in each case. Colored vertical lines represent genomic location of critical genes or LCR clusters. Vertical lines in light blue, CMT1A-REPs; magenta, SMS-REPs; maroon, 17p-PROX; black, genes PMP22 and RAI1. Centromeric region is also noted.
(C) Count of incidences of each genomic rearrangement. CR, common recurrent PTLS duplication. The incidence is one for the rearrangements labeled with BAB numbers.
PMP22-RAI1 Duplications Can Be Part of CGRs on 17p
Several PMP22-RAI1 duplications were identified previously using conventional cytogenetic diagnostic methods, such as fluorescence in situ hybridization (FISH) or chromosome G-banding analyses.28, 29, 32 However, these methods are not able to detect submicroscopic structural complexities. High-resolution array CGH can reveal more extensive structural complexities with high-density probes interrogating the region of interest. We utilized high-resolution 17p array CGH and found evidence for other CNVs in addition to the PMP22-RAI1 duplications, constituting potential CGRs.
Three cases were identified with a CGR pattern of “DEL-NML-DUP” (Figure 1). The CGR in BABs 1229 and 2711 were not identified in either parent, and likely arose de novo in the subject (Table 2). Breakpoint junction sequencing revealed 1 bp insertion (C) and 6 bp (ACCTTC) microhomology at the deletion breakpoint in BABs 2711 and 1229 respectively.25 The duplication breakpoints were not sequenced as they map in pericentromic and centromeric regions (Figure 2). The inheritance pattern of the rearrangement observed in BAB 6967 is unknown as the parental samples were not available.
Four cases were identified with a CGR pattern of “DUP-NML-DUP” (Figure 1). The CGR in BABs 527, 2211, and 6504 were de novo according to the array CGH analysis in trio studies (Table 2). The partially mapped breakpoint junction sequence in BAB 2211 revealed that the proximal side of the distal DUP was connected to the distal side of the proximal DUP in a direct orientation (Figure S2A). A 6 bp inserted palindrome sequence (TAATTA) was observed at the breakpoint. The inheritance pattern of the CGR in BAB 2790 was unknown as parental samples were unavailable. Breakpoint junction sequencing revealed that the distal side of the distal DUP was connected to the distal side of the proximal DUP in an inverted orientation (INV/DUP), consistent with a potential DUP-NML-INV/DUP rearrangement (Figure S2B). A 4 bp inserted sequence (CTTT) at the breakpoint was identified previously, which possibly originated from a nearby sequence (Chr17: 12999823–12999830), with two 2 bp microhomologies (TT and CA) flanking the 4 bp insertion (Figure S2B).16 The mutational signatures identified in the CGRs of BABs 2211 (insertion) and 2790 (insertion and microhomology) are indicative of potential replication-based mechanisms such as FoSTeS/MMBIR.
De novo CGR with a pattern of “TRP-NML-DUP” was identified by high-resolution array CGH in BAB 1458 (Table 2). Breakpoint junction sequencing was attempted but not successfully obtained. The lack of amplification might be due to additional structural complexities not resolved by array CGH (such as inversions) in addition to the apparent CNVs.
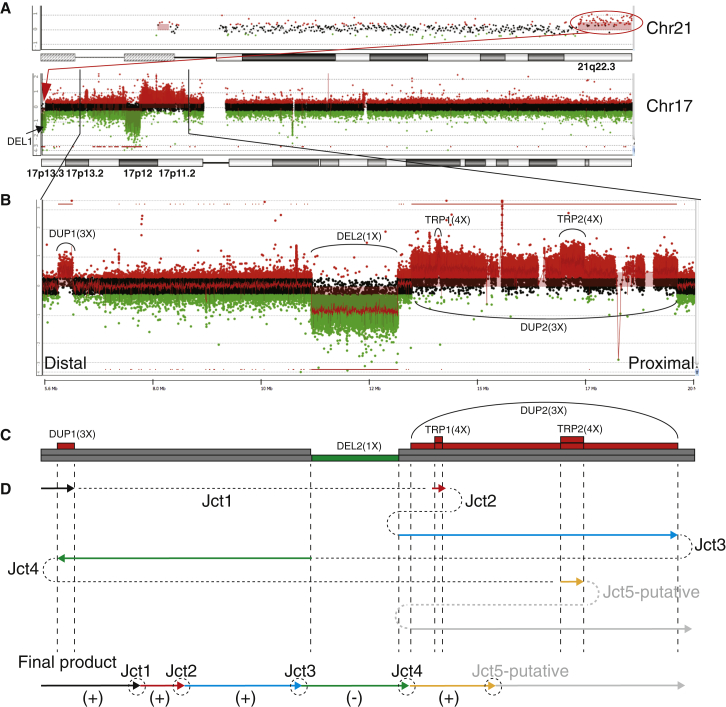
PMP22-RAI1 Duplication Can Be Part of a Highly Complex Chromothripsis/Chromoanasynthesis Event
The CGRs involving the PMP22-RAI1 duplication can be exceedingly complex. Cytogenetic and molecular analysis, including FISH, chromosome analysis and low-resolution BAC array CGH, were used to investigate the genome of BAB 2337, and a complex rearrangement was revealed. The rearrangements included a terminal deletion of 17p13.3 (DEL1), a deletion of 17p12 (DEL2), and a duplication of 17p11.2p12 (DUP2: a PMP22-RAI1 duplication)33 (Figures 3A and 3B). FISH experiments also revealed a balanced insertional translocation event, including a segment of 17p13.3 (Miller-Dieker lissencephaly syndrome [MDLS, MIM 247200] region) being inserted in an inverted orientation amidst the middle SMS-REP/LCR17pB block.33 Additionally, FISH revealed unbalanced translocation of a duplicated segment of 21q22.3qter to 17p13.3pter (Figure 3A).33
Figure 3.
Complex Genomic Rearrangement in BAB 2337
(A) An overview of genomic rearrangements identified in the entire genome of BAB 2337. Top panel shows duplicated segment in 21q22.3, which is translocated to 17pter shown by FISH.33 Bottom panel shows rearrangements occur in chromosome 17. The unbalanced translocation between 17p and 21q is shown by the red oval and arrow.
(B) Zoom-in view of genomic rearrangements in 17p. It excludes the terminal deletion in 17p13.3.
(C) Diagram showing the dosage changes based on array CGH analysis. Colored segments represent the dosage changes. Number of segments stacking at the same position indicates absolute copy number. Segments in red, copy-number gain; green; copy-number loss; gray, normal copy number. Abbreviations are as follows: DEL, deletion; DUP, duplication; TRP, triplication. “X,” copy number.
(D) Schematic diagram showing the potential rearrangement-generating mechanism. Colored arrows indicate the replication direction. Dashed lines between arrows represent breakpoint junctions (Jct). Arrows in black, red, blue, and yellow indicate replications on the (+) strand in a same direction. The arrows in green indicate a replication on the (−) strand. The arrow and dashed line in gray represents a potential process to generate the unmapped, putative Jct5. The potential final product of rearrangement is shown at the bottom.
High-resolution array CGH targeting 17p uncovered additional structural abnormalities. An excess number of CNVs were identified on the short arm of chromosome 17. In addition to the CNVs identified by FISH, a small duplication (∼300 kb in size) distal to the PMP22-RAI1 duplication (DUP1) and two triplications (TRP1 and TRP2, approximately 70 kb and 510 kb in size, respectively) located within the PMP22-RAI1 duplication region were revealed (Figures 3B and 3C). The overall pattern of “DEL1-NML-DUP1-NML-DEL2-NML-DUP2-TRP1-DUP2-TRP2-DUP2” was reminiscent of chromothripsis or chromoanasynthesis, a phenomenon consisting of excessive rearrangements apparently concentrated on one chromosome or one arm of a chromosome.36, 37, 38 Breakpoint sequencing revealed four junction sequences (Jct1, Jct2, Jct3, and Jct4, Figure 3D and Table 2). Jct1 connected the proximal end of DUP1 and the distal end of TRP1 with an 18 bp insertion observed at the breakpoint junction (Figure 3D and Figure S3A). This insertion could not be found in the adjacent genomic regions (40 kb) surrounding each breakpoint, indicating a DNA sequence potentially synthesized from a region beyond the nearby region of the breakpoints. Jct2 connected the proximal end of TRP1 and the proximal end of DEL2 (Figure 3D). A 33 bp insertion was observed at the breakpoint junction Jct2 and could be split into two fragments, including a 2 bp fragment (CT) and a 31 bp fragment potentially derived from a nearby region (Chr17: 8607208–8607238) with a 4 bp microhomology (GACA) at the junction (Figure S3A). Jct3 connected the proximal end of DUP2 and the distal end of DEL2 in an inverted orientation (Figure 3D). A 257 bp insertion was observed at Jct3 and could be divided into four fragments, including a 4 bp fragment (ACTT), a 128 bp fragment (ins1, derived from Chr17: 17575856–17575983), an 85 bp fragment (ins2, derived from Chr17: 17576136–17576220) and a 32 bp fragment (ins3, derived from Chr17: 17568995–17569026) (Figure S3A). Microhomologies (TGC, CA and GCA, respectively) were observed at each transition point (Figure S3A). Jct4, connecting the distal end of DUP1 and the distal end of TRP2 in an inverted orientation, was observed with a 551 bp insertion that might be partially derived from sequences in chromosomes 21 (Chr21: 43658009–43658301) and 17 (Chr17: 17597062–17597133) (Figure 3D and Figure S3A). At least two template switches are required to generate one intact inserted sequence; thus at least (N+1) template switches are potentially required to generate N number of intact insertions (Figures S3B and S3C). DNA repair and replication after double-strand break is error-prone and might undergo iterative template switches due to the usage of a low-processivity DNA polymerase.26, 34, 39 Multiple insertion fragments were revealed by the breakpoint junction sequences of BAB 2337, indicating iterative template switches before the establishment of a processive replicative fork. In addition, microhomologies were observed to catenate DNA fragments from different origin. The cumulative evidence suggests FoSTeS/MMBIR as the likely mechanism for forming the CGR in BAB 2337. The breakpoint junction sequence connecting the proximal end of TRP2 and the distal end of DUP2 was not determined. It indicated that potential cryptic complexity undetectable by array CGH might underlie the rearrangement. A balanced insertional translocation event of a segment from the 17p13.3 MDLS region being invertedly inserted in the middle SMS-REP/LCR17pB block has been reported previously33—this rearrangement is apparently copy-number neutral and thus cannot be detected by array CGH technique. Such type of complexity might evade detection and breakpoint junction determination by currently used methodologies.40
Discussion
PTLS has an estimated incidence of 1:50,000, while CMT (includes all of its molecular genetic forms, of which CMT1A is the most commonly observed) has a total prevalence rate of 1:2,500. Thus the expected prevalence of cases with concurrence of PTLS and CMT1A can be estimated at 1:125,000,000 based on the hypothesis that PTLS and CMT1A are independent events. However, because the two loci are in relatively close physical proximity (mapping ∼2.5 Mb apart, Figure 4B), they can theoretically be involved in a single mutational event with both dosage sensitive genes mapping within a duplicated genomic segment. In our study, 23 subjects have been identified with PMP22-RAI duplication within a cohort of 127 subjects (23/127, 18.11%) with duplications encompassing RAI1. Specifically, PMP22-RAI1 duplications account for 23/43 (53%) of subjects with uncommon or noncurrent rearrangements, which emphasizes the importance of pursuing a molecular diagnosis in subjects with PTLS (Figure S4). Moreover, prognostic information provided for PTLS subjects with PMP22-RAI1 duplications is different from that for typical PTLS resulting from common recurrent duplication. Furthermore, neuropathy can be monitored for in the relevant cases guided by molecular diagnosis.
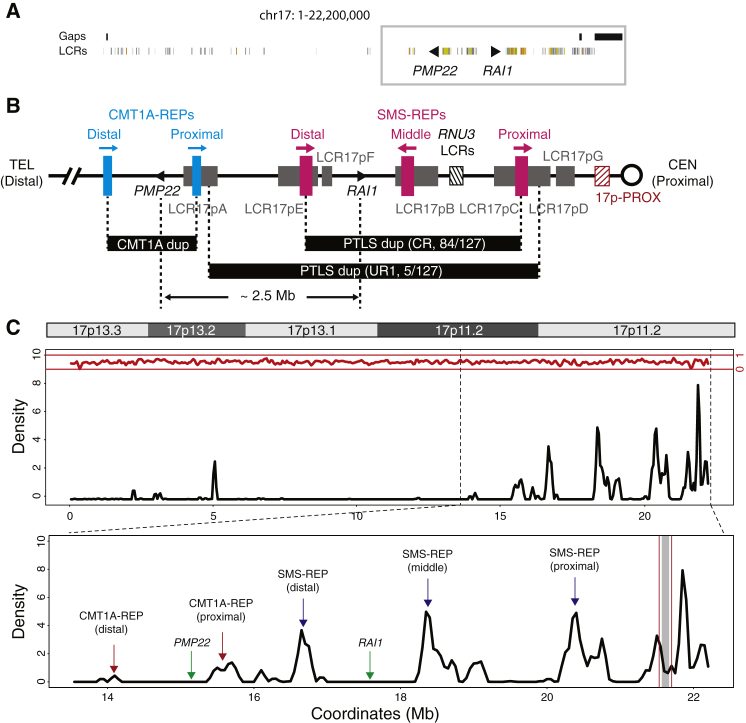
Figure 4.
Genomic Architecture in the Chromosome 17p Region
(A) Distribution of LCRs in chromosome 17p shown by the Segmental Dups track in the UCSC genome browser (hg19). Colored segments represent LCRs. Black triangles represent PMP22 and RAI1. Regions with gaps are shown as black segments on top of the LCR track. The gray box indicates proximal 17p region.
(B) LCR-rich genomic structure in the proximal 17p region. LCR clusters are shown by colored segments. Segments in blue, CMT1A-REPs; magenta, SMS-REPs; gray, LCR17pA-G; black hatched, RNU3 LCRs responsible for idic(17q); maroon hatched, 17p-PROX. The common recurrent CMT1A duplication, common recurrent (CR) and uncommon recurrent (UR1) PTLS duplications are shown as black segments between vertical dashed lines. Distance between PMP22 and RAI1 is approximately 2.5 Mb.
(C) LCR density plot in chromosome 17p region. The plot is based on the annotation in the Segmental Dups track from the UCSC Genome Browser. Top panel shows the ideogram of chromosome 17p. Middle panel is the LCR density plot showing the moving-average of LCR density along the chromosome 17p region. LCR density might be higher than 1 if multiple overlapping LCRs cluster in a given window. Density of other repetitive sequences from RepeatMasker is shown at the top in red. The bottom panel is zoom-in view of the middle panel focusing on the proximal 17p region. Major LCR clusters and genes are annotated. Region between vertical maroon lines indicate the 17p-PROX, and the region with gray shade represents the gap region in hg19, which is recently filled by satellite DNA sequences in hg38.
Infantile hypotonia, failure to thrive, and developmental delay are common features of PTLS,7, 28, 41 whereas the clinical manifestations of CMT1A do not present during infancy, yet develop gradually during the first 2 decades. Delayed walking is seen in the vast majority of persons with PTLS, while individuals with CMT1A typically attain early developmental milestones without difficulty.42 In the current cohort, delayed walking (beyond age 15 months) was observed in 13 of 14 subjects; and clinical neuropathy developed in the first decade in 9 of 10 subjects for which this information is available (Table 1). Although it is possible that the molecular diagnosis provided an ascertainment bias and clinicians were more likely to diagnose the peripheral neuropathy at an early age, it is plausible to suggest that both the motor delay and the neuropathy were exacerbated by the combination of central and peripheral nervous system abnormalities in subjects with PMP22-RAI1 duplications.
Talipes equinovarus has been reported in association with PTLS, and was suggested to be a result of perturbed regulation of PMP22 in a subject with the common recurrent PTLS duplication.10, 15 A case (261273) with a PMP22-RAI1 duplication has been reported in DECIPHER with indications including intellectual disability, micrognathia, muscular hypotonia, prominent nasal bridge, and talipes equinovarus. Interestingly, 2 of 15 subjects in our cohort had congenital talipes equinovarus, possibly related to their peripheral neuropathy. Syringomyelia, a fluid-filled cyst within the spinal cord, was found in two of the four subjects who had a spinal MRI (Table 1, Table S1). This malformation is not typically associated with CMT1A or PTLS and might suggest that a spine MRI be considered in subjects with the PMP22-RAI1 duplication.
While structural renal abnormalities have been reported in 19%–35% of subjects with Smith-Magenis syndrome,43, 44 they are considered relatively infrequent (<10%) in subjects with PTLS.16 Goh et al. highlighted FLCN (MIM 607273) as a possible candidate gene contributing to renal abnormalities in subjects with PTLS-associated duplications.45 The association between gain-of-function mutations (e.g., duplications) of FLCN and renal abnormalities warrants further investigation. The slightly higher incidence (3/11 or 27%) of structural renal abnormalities in our cohort might suggest the consideration of an additional dosage sensitive gene involved in kidney development, possibly located in between the map positions of the recurrent CMT1A and PTLS duplications.
Boundaries of the PMP22-RAI1 duplication are not confined by the LCR-rich architecture on 17p. LCR-rich architecture in the 17p11.2p12 region determines recurrent, disease-causing CNVs. The genomic architecture of major LCR clusters, including CMT1A-REPs, SMS-REPs, RNU3 LCRs, and LCR17pA/B/C/D/E/F/G clustering with the SMS-REPs, has been delineated in several studies using BAC clone sequencing, FISH, Southern blot, and pulsed-field gel electrophoresis (Figure 4B).11, 21, 46, 47, 48 These LCR clusters are involved in the formation of uncommon recurrent 17p11.2 deletion/duplication CNV (UR1 and UR2), translocations and marker chromosomes identified in subjects with SMS/PTLS phenotypes, as well as somatic isochromosome 17q leading to malignancies.16, 46, 49, 50, 51, 52, 53 Although LCR clusters surround both the PMP22 and RAI1 loci, the CMT1A-REPs (flank PMP22 and mediate CMT1A duplication) and SMS-REPs (flank RAI1 and mediate PTLS duplication) do not share extensive homology. In fact, no extensive directly-oriented homology distal to PMP22 has been observed to partner the LCRs proximal to RAI1; thus a pairing LCR defined genomic interval encompassing both PMP22 and RAI1 is lacking (Figure 4C). As a result and consistent with our experimental observations, a recurrent rearrangement for PMP22-RAI1 duplication is unlikely.
Other than NAHR mediated by homologous LCRs, structural rearrangements might be generated by replicative mechanisms, which result from DNA replication error and repair. FoSTeS/MMBIR has been proposed to generate rearrangements with microhomologies at the breakpoint junction.25, 26 Single-end, double-strand breaks can result from mitotic replication fork collapse, which allows the single-strand 3′ end to undergo template switches to a nearby replication fork primed by Watson-Crick base pairing via microhomologies to continue replication.23, 24, 34 The position of re-establishment of a processive replication fork determines the pattern (duplication, deletion, inversion, translocation, etc.) and size of rearrangements. The process of template switching with subsequent replication can be homology-independent, or mediated by limited homology, termed microhomology; chromosomal structural changes resulting from such replicative processes are attributed to the FoSTeS/MMBIR model.26, 34, 40 The observation of microhomology at the breakpoint junction of several rearrangements encompassing PMP22-RAI1 duplications is consistent with the FoSTeS/MMBIR model (Table 2). The small-scale complexities in the form of templated insertions from an adjacent region in close proximity inserted at the breakpoint junctions are also observed in a number of cases, which are characteristic of the low-processivity, low-fidelity replication process in MMBIR.38, 39 FoSTeS/MMBIR requires microhomology or no homology, thus it is independent of the long homology provided by LCRs concentrated in the chromosome 17p11.2p12 region.
The distribution of copy-number variants is not random based on experimental observation.26 This might be affected by multiple factors, such as DNA sequence features. CNVs are more frequently observed at the centromeric or pericentromeric regions, which are also enriched with LCR content.54, 55 LCRs have been proposed to catalyze genomic rearrangement; nonrecurrent and complex structural changes tend to occur close to LCRs, and a breakage/recombination-stimulating role has been proposed for LCRs.38, 46 Secondary structure may form within LCRs, and thereby cause replication fork stalling followed by template switches during DNA replication.40 Chromosomal region 17p11.2p12 is enriched with LCRs, which are also termed segmental duplications based on computational characterization.56 According to our analysis, LCRs constitute approximately 36.52% of genomic sequences of 17p11.2, 23.06% of 17p11.2p12 and 12.86% of the entire short arm of chromosome 17. These regions thus have approximately seven-, four-, and two-fold greater segmental duplications respectively as compared to the genome average of 5.13%.
In a total number of 36 putative breakpoint junctions characterized by array CGH on 17p, we observe involvement of LCRs at 13 junctions (termed LCR-associated junctions), 12 of which are located in the 17p11.2 region (Table 2), constituting 12/36 (33.33%) of total breakpoint junctions. The fraction of LCR-associated events observed in the 17p11.2 region is similar to the LCR concentration (36.52%) in the same region, thus the enrichment of LCRs at breakpoint junctions cannot be implicated. It might be explained by: (1) small sample size, and (2) using a selected cohort with a specific phenotype.
Interestingly, nine LCR-associated junctions in nine samples (Figure 2, Table 2) are located at an LCR cluster in the proximal 17p region (termed 17p-PROX), constituting 75% (9/12) of the entire observed LCR-associated junctions. Among the entire LCR sequences in 17p11.2, 17p-PROX is ∼160 kb (7.07%) in contrast to ∼2,104 kb (92.93%) remaining LCR sequences in 17p11.2. The observation of 9 out of 12 LCR-associated junctions demonstrates an enrichment of junctions in the 17p-PROX (chi-square test, p < 0.0001). Remarkably, the enrichment of breakpoint in LCR has also been demonstrated at other loci, such as MECP2 locus, by high-resolution array CGH and breakpoint junction sequencing.57
There exists a gap in the middle of 17p-PROX in hg19 surrounded by highly enriched LCR sequences (Figure 4C). This gap was recently resolved with additional LCR sequences in hg38 (GRCh38/hg38) together with, interestingly, an ∼141 kb stretch of microsatellite DNA sequences (AT content 61.36%). Satellite DNA sequences are hypermutable and present in pericentromeric regions or centromeres and are involved in the formation of Robertsonian translocations, pericentric inversions, and translocations involving 17p11.46, 58, 59, 60, 61 Furthermore, it is noteworthy that there are six rearrangements identified in this study to have at least one breakpoint that resides in the centromeric 17p11q11 region (Figure 2, Table 2); karyotyping and FISH analysis for two of these six subjects, BABs 1458 and 1861, have demonstrated that the duplicated segment mapped on chromosome 17 in both cases and created an isodicentric 17 in BAB 1861.29, 32 These findings suggest that satellite DNA sequences might affect genomic instability and result in rearrangements. Mechanisms including homologous recombination and DNA breakage followed by erroneous DNA repair have been proposed for satellite DNA sequence-mediated translocations.59 For instance, satellite III, composed of short monomers (∼5 bp), might be involved in forming nonrecurrent translocations.59 Thus, the long stretch of satellite DNA sequences residing in the 17p-PROX and 17p11 region might be the responsible genome architectural feature for the nonrecurrent duplications observed in our study. These might also act in concert with the LCR-rich genomic architecture in the pericentromeric region to incite genomic instability and facilitate genomic rearrangement formation (Figure 4C). However, due to the highly complex and repetitive nature of the satellite DNA sequences, they are recalcitrant to precise breakpoint mapping and sequencing. Thus, single-molecule long-read sequencing—perhaps single-molecule real-time (SMRT) sequencing62—might be required to resolve breakpoint junctions of the rearrangements that occur at pericentromeric or centromeric regions.63, 64 The single-molecule long-read sequencing technologies might also be beneficial to the sequencing of breakpoint junctions of rearrangements involving other repeats (e.g., LCRs) or repetitive sequences (e.g., Alus) in this study.
In conclusion, we characterized the genomic rearrangements in subjects with PMP22-RAI1 duplications by high-resolution array CGH. Subjects with such duplication might have a blended CMT1A and PTLS phenotypes. The motor delay and neuropathy might be exaggerated by the combined abnormalities of the central (PTLS) and peripheral (CMT1A) nervous system with a potential earlier onset. Additional clinical presentations, including structural renal abnormalities, might be observed in subjects with PMP22-RAI1 duplications due to the extra duplicated genomic material with a potential gene contributing to renal development. Breakpoint junction sequencing revealed likely replicative mechanisms, perhaps FoSTeS/MMBIR, contribute to the rearrangements involving PMP22-RAI1 duplications. Such mechanism might be independent of pairing LCR substrate architecture, and result in larger CNVs leading to a contiguous gene syndrome with multiple disease loci affected; i.e., the PMP22-RAI1 contiguous gene duplication syndrome. Breakpoint junctions with potential repetitive sequence origin were significantly enriched in the cohort studied. It indicates that, at least in chromosome 17p, repetitive sequences, especially satellite DNA sequences, might predispose to genomic rearrangement resulting in the PMP22-RAI1 duplication.
Conflict of Interest
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen Inc., is a member of the Scientific Advisory Board of Baylor Miraca Genetics Laboratories, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory.
Acknowledgments
We thank the patients and their families for their participation. We also thank Dr. Michael Wright for kindly providing both a patient DNA specimen and clinical profile of subject BAB 6635. J.R.L received grant support from the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) Grant No. HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG); the National Institute of Neurological Disorders and Stroke (NINDS) NS058529, the National Institute of General Medical Sciences (NIGMS) GM106373, the National Institute of Child Health and Development (NICHD) HD024064 Intellectual and Developmental Disabilities Research Center (IDDRC), the National Eye Institute (NEI) EY021163 and EY019861, and the Smith-Magenis Syndrome Research Foundation (SMSRF). T.H. was supported by the Medical Genetics Research Fellowship Program NIH T32 GM007526. C.R.B. is an HHMI Fellow of the Damon Runyon Cancer Research Foundation (DRG 2155-13).
Published: November 5, 2015
Footnotes
Supplemental Data include four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.10.003.
Web Resources
The URLs for data presented herein are as follows:
DECIPHER, http://decipher.sanger.ac.uk/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Schmickel R.D. Contiguous gene syndromes: a component of recognizable syndromes. J. Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. [DOI] [PubMed] [Google Scholar]
- 2.Lupski J.R. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ. Mol. Mutagen. 2015;56:419–436. doi: 10.1002/em.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 4.Lupski J.R. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg F., Guzzetta V., Montes de Oca-Luna R., Magenis R.E., Smith A.C., Richter S.F., Kondo I., Dobyns W.B., Patel P.I., Lupski J.R. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2) Am. J. Hum. Genet. 1991;49:1207–1218. [PMC free article] [PubMed] [Google Scholar]
- 7.Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neira-Fresneda J., Potocki L. Neurodevelopmental Disorders Associated with Abnormal Gene Dosage: Smith–Magenis and Potocki–Lupski Syndromes. J. Pediatr. Genet. 2015 doi: 10.1055/s-0035-1564443. Published online September 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacaria M., Saha P., Potocki L., Bi W., Yan J., Girirajan S., Burns B., Elsea S., Walz K., Chan L. A duplication CNV that conveys traits reciprocal to metabolic syndrome and protects against diet-induced obesity in mice and men. PLoS Genet. 2012;8:e1002713. doi: 10.1371/journal.pgen.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricard G., Molina J., Chrast J., Gu W., Gheldof N., Pradervand S., Schutz F., Young J.I., Lupski J.R., Reymond A. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S.S., Stankiewicz P., Bi W., Shaw C., Lehoczky J., Dewar K., Birren B., Lupski J.R. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K.S., Manian P., Koeuth T., Potocki L., Zhao Q., Chinault A.C., Lee C.C., Lupski J.R. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 13.Bi W., Park S.S., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potocki L., Chen K.S., Park S.S., Osterholm D.E., Withers M.A., Kimonis V., Summers A.M., Meschino W.S., Anyane-Yeboa K., Kashork C.D. Molecular mechanism for duplication 17p11.2- the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 15.Magoulas P.L., Liu P., Gelowani V., Soler-Alfonso C., Kivuva E.C., Lupski J.R., Potocki L. Inherited dup(17)(p11.2p11.2): expanding the phenotype of the Potocki-Lupski syndrome. Am. J. Med. Genet. A. 2014;164A:500–504. doi: 10.1002/ajmg.a.36287. [DOI] [PubMed] [Google Scholar]
- 16.Zhang F., Potocki L., Sampson J.B., Liu P., Sanchez-Valle A., Robbins-Furman P., Navarro A.D., Wheeler P.G., Spence J.E., Brasington C.K. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am. J. Hum. Genet. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi W., Yan J., Stankiewicz P., Park S.S., Walz K., Boerkoel C.F., Potocki L., Shaffer L.G., Devriendt K., Nowaczyk M.J. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 2002;12:713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slager R.E., Newton T.L., Vlangos C.N., Finucane B., Elsea S.H. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 19.Walz K., Paylor R., Yan J., Bi W., Lupski J.R. Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2) J. Clin. Invest. 2006;116:3035–3041. doi: 10.1172/JCI28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pentao L., Wise C.A., Chinault A.C., Patel P.I., Lupski J.R. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat. Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- 21.Inoue K., Dewar K., Katsanis N., Reiter L.T., Lander E.S., Devon K.L., Wyman D.W., Lupski J.R., Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 23.Patel P.I., Roa B.B., Welcher A.A., Schoener-Scott R., Trask B.J., Pentao L., Snipes G.J., Garcia C.A., Francke U., Shooter E.M. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- 24.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treadwell-Deering D.E., Powell M.P., Potocki L. Cognitive and behavioral characterization of the Potocki-Lupski syndrome (duplication 17p11.2) J. Dev. Behav. Pediatr. 2010;31:137–143. doi: 10.1097/DBP.0b013e3181cda67e. [DOI] [PubMed] [Google Scholar]
- 28.Doco-Fenzy M., Holder-Espinasse M., Bieth E., Magdelaine C., Vincent M.C., Khoury M., Andrieux J., Zhang F., Lupski J.R., Klink R. The clinical spectrum associated with a chromosome 17 short arm proximal duplication (dup 17p11.2) in three patients. Am. J. Med. Genet. A. 2008;146A:917–924. doi: 10.1002/ajmg.a.32195. [DOI] [PubMed] [Google Scholar]
- 29.Shaw C.J., Stankiewicz P., Christodoulou J., Smith E., Jones K., Lupski J.R. A girl with duplication 17p10-p12 associated with a dicentric chromosome. Am. J. Med. Genet. A. 2004;124A:173–178. doi: 10.1002/ajmg.a.20355. [DOI] [PubMed] [Google Scholar]
- 30.Campbell I.M., Yatsenko S.A., Hixson P., Reimschisel T., Thomas M., Wilson W., Dayal U., Wheless J.W., Crunk A., Curry C. Novel 9q34.11 gene deletions encompassing combinations of four Mendelian disease genes: STXBP1, SPTAN1, ENG, and TOR1A. Genet. Med. 2012;14:868–876. doi: 10.1038/gim.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupski J.R., Wise C.A., Kuwano A., Pentao L., Parke J.T., Glaze D.G., Ledbetter D.H., Greenberg F., Patel P.I. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1:29–33. doi: 10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- 32.Balarin M.A., da Silva Lopes V.L., Varella-Garcia M. A dup(17)(p11.2p11.2) detected by fluorescence in situ hybridization in a boy with Alport syndrome. Am. J. Med. Genet. 1999;82:183–186. [PubMed] [Google Scholar]
- 33.Vissers L.E., Stankiewicz P., Yatsenko S.A., Crawford E., Creswick H., Proud V.K., de Vries B.B., Pfundt R., Marcelis C.L., Zackowski J. Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies. Hum. Genet. 2007;121:697–709. doi: 10.1007/s00439-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McVey M., Lee S.E. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maher C.A., Wilson R.K. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P., Erez A., Nagamani S.C., Dhar S.U., Kolodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho C.M., Pehlivan D., Ramocki M.B., Fang P., Alleva B., Franco L.M., Belmont J.W., Hastings P.J., Lupski J.R. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 41.Soler-Alfonso C., Motil K.J., Turk C.L., Robbins-Furman P., Friedman E.M., Zhang F., Lupski J.R., Fraley J.K., Potocki L. Potocki-Lupski syndrome: a microduplication syndrome associated with oropharyngeal dysphagia and failure to thrive. J. Pediatr. 2011;158:655–659. doi: 10.1016/j.jpeds.2010.09.062. e652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brennan K.M., Bai Y., Shy M.E. Demyelinating CMT-what’s known, what’s new and what’s in store? Neurosci. Lett. 2015;596:14–26. doi: 10.1016/j.neulet.2015.01.059. [DOI] [PubMed] [Google Scholar]
- 43.Potocki L., Shaw C.J., Stankiewicz P., Lupski J.R. Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome. Genet. Med. 2003;5:430–434. doi: 10.1097/01.gim.0000095625.14160.ab. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg F., Lewis R.A., Potocki L., Glaze D., Parke J., Killian J., Murphy M.A., Williamson D., Brown F., Dutton R. Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2) Am. J. Med. Genet. 1996;62:247–254. doi: 10.1002/(SICI)1096-8628(19960329)62:3<247::AID-AJMG9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Goh E.S., Perez I.C., Canales C.P., Ruiz P., Agatep R., Yoon G., Chitayat D., Dror Y., Shago M., Goobie S. Definition of a critical genetic interval related to kidney abnormalities in the Potocki-Lupski syndrome. Am. J. Med. Genet. A. 2012;158A:1579–1588. doi: 10.1002/ajmg.a.35399. [DOI] [PubMed] [Google Scholar]
- 46.Stankiewicz P., Shaw C.J., Dapper J.D., Wakui K., Shaffer L.G., Withers M., Elizondo L., Park S.S., Lupski J.R. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am. J. Hum. Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stankiewicz P., Shaw C.J., Withers M., Inoue K., Lupski J.R. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004;14:2209–2220. doi: 10.1101/gr.2746604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stankiewicz P., Park S.S., Inoue K., Lupski J.R. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 2001;11:1205–1210. doi: 10.1101/gr.181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stankiewicz P., Park S.S., Holder S.E., Waters C.S., Palmer R.W., Berend S.A., Shaffer L.G., Potocki L., Lupski J.R. Trisomy 17p10-p12 resulting from a supernumerary marker chromosome derived from chromosome 17: molecular analysis and delineation of the phenotype. Clin. Genet. 2001;60:336–344. doi: 10.1034/j.1399-0004.2001.600503.x. [DOI] [PubMed] [Google Scholar]
- 50.Shaw C.J., Stankiewicz P., Bien-Willner G., Bello S.C., Shaw C.A., Carrera M., Perez Jurado L., Estivill X., Lupski J.R. Small marker chromosomes in two patients with segmental aneusomy for proximal 17p. Hum. Genet. 2004;115:1–7. doi: 10.1007/s00439-004-1119-5. [DOI] [PubMed] [Google Scholar]
- 51.Yatsenko S.A., Treadwell-Deering D., Krull K., Lewis R.A., Glaze D., Stankiewicz P., Lupski J.R., Potocki L. Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications. Am. J. Med. Genet. A. 2005;138A:175–180. doi: 10.1002/ajmg.a.30948. [DOI] [PubMed] [Google Scholar]
- 52.Barbouti A., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004;74:1–10. doi: 10.1086/380648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carvalho C.M., Lupski J.R. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18:1724–1732. doi: 10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emanuel B.S., Shaikh T.H. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 55.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 56.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 57.Carvalho C.M., Zhang F., Liu P., Patel A., Sahoo T., Bacino C.A., Shaw C., Peacock S., Pursley A., Tavyev Y.J. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Earle E., Shaffer L.G., Kalitsis P., McQuillan C., Dale S., Choo K.H. Identification of DNA sequences flanking the breakpoint of human t(14q21q) Robertsonian translocations. Am. J. Hum. Genet. 1992;50:717–724. [PMC free article] [PubMed] [Google Scholar]
- 59.Page S.L., Shin J.C., Han J.Y., Choo K.H., Shaffer L.G. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum. Mol. Genet. 1996;5:1279–1288. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- 60.Gravholt C.H., Friedrich U., Caprani M., Jorgensen A.L. Breakpoints in Robertsonian translocations are localized to satellite III DNA by fluorescence in situ hybridization. Genomics. 1992;14:924–930. doi: 10.1016/s0888-7543(05)80113-2. [DOI] [PubMed] [Google Scholar]
- 61.Ramesh K.H., Verma R.S. Breakpoints in alpha, beta, and satellite III DNA sequences of chromosome 9 result in a variety of pericentric inversions. J. Med. Genet. 1996;33:395–398. doi: 10.1136/jmg.33.5.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 63.English A.C., Salerno W.J., Hampton O.A., Gonzaga-Jauregui C., Ambreth S., Ritter D.I., Beck C.R., Davis C.F., Dahdouli M., Ma S. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics. 2015;16:286. doi: 10.1186/s12864-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M., Beck C.R., English A.C., Meng Q., Buhay C., Han Y., Doddapaneni H.V., Yu F., Boerwinkle E., Lupski J.R. PacBiol.-LITS: a large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations. BMC Genomics. 2015;16:214. doi: 10.1186/s12864-015-1370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.