Abstract
Cnidarians, in general, are long-lived organisms and hence may repeatedly encounter common pathogens during their lifespans. It remains unknown whether these early diverging animals possess some type of immunological reaction that strengthens the defense response upon repeated infections, such as that described in more evolutionary derived organisms. Here we show results that sea anemones that had previously encountered a pathogen under sub-lethal conditions had a higher survivorship during a subsequently lethal challenge than naïve anemones that encountered the pathogen for the first time. Anemones subjected to the lethal challenge two and four weeks after the sub-lethal exposure presented seven- and five-fold increases in survival, respectively, compared to the naïve anemones. However, anemones challenged six weeks after the sub-lethal exposure showed no increase in survivorship. We argue that this short-lasting priming of the defense response could be ecologically relevant if pathogen encounters are restricted to short seasons characterized by high stress. Furthermore, we discovered significant changes in proteomic profiles between naïve sea anemones and those primed after pathogen exposure suggesting a clear molecular signature associated with immunological priming in cnidarians. Our findings reveal that immunological priming may have evolved much earlier in the tree of life than previously thought.
The ability of the immune system to respond more rapidly and effectively to a pathogen that has been encountered previously is a trait well-characterized in vertebrates and mechanistically explained by the functional uniqueness of the adaptive immune system1. This trait has profound implications for a wide array of epidemiological and evolutionary phenomena. In contrast, it has long been assumed that invertebrates have an immune response that differs considerably from the acquired immune response found in vertebrates1.Invertebrates possess only an innate immune system, which is characterized by invariable pattern recognition receptors (PRRs) that target general pathogens-associated molecular patterns (PAMPs). The current consensus is that invertebrates lack the components of the adaptive immune system, such as those well characterized in vertebrates including highly variable major histocompatibility complex (MHC) receptors, immunoglobulins, and B and T cells that undergo clonal expansion and long term cell survival following antigen induced activation1,2,3,4. These elements of the adaptive immune system underlie the mechanism of the immunological memory phenomenon demonstrated in vertebrates. However, increasing evidence over the past years suggests that invertebrate immunity is much more complex than was generally believed. Immunological priming, the stimulation of the immune system with long-lasting effects that accelerate subsequent exposures to infectious pathogens, has been documented for a few groups of invertebrates, such as insects and crustaceans5,6,7,8,9,10. The first studies to demonstrate that initial pathogen exposure confers lasting specific protection were for the crustacean copepod Macrocyclops albidus6 and for the social insect Bombus terrestris9. Nevertheless, evidence of immunological priming in many other invertebrates remains absent, particularly for early diverging animals.
Cnidarians, including corals and sea anemones, are evolutionarily early-diverged metazoans and of great interest since some of these invertebrates can live for hundreds of years, suggesting they are potentially exposed to the same pathogens on many occasions during their lifespans. This has led to the mystery of how these long-lived organisms have done so well with only an innate immune system as the protective mechanism against infectious agents. As of yet, it is unknown if the defense response of these organisms strengthens upon repeated infections. Pioneering studies on coral skin grafts conducted in the late 1970s demonstrated that corals were able to reject skin grafts from genetically distinct donors more rapidly the second time the grafts were applied, suggesting a capacity for non-self recognition11,12,13. Self- and allogeneic recognition has also been described in sea anemones14 and soft corals15. While this phenomenon is not directly comparable with a defense against a pathogen, it might indicate that these basal metazoans have the capacity to remember foreign biological interactions. Additionally, understanding this aspect of immunology in corals and other cnidarians is imperative in light of the global concern of increasing epizootic disease outbreaks currently affecting the health of corals16,17,18,19 and the persistence of these fragile coral reef ecosystems20,21,22,23.
In the present study, we investigated whether priming is existent in cnidarians in response to pathogenic infections. If the protective response and survivorship of the host to pathogen challenges improve as a result of repetitive encounters with the infectious agent as compared to single encounters, it would suggest the existence of defense priming. Furthermore, presence of immune priming should also be associated with molecular changes underpinning the phenomenology, which is another fundamental investigation we conducted in this study.
Results
Host response to bacterial pathogen improves upon repeated pathogen encounter
To test the hypothesis that a sub-lethal exposure of cnidarians to a pathogen induces a defense response that is memorized and expressed in an accelerated manner upon subsequent exposure, the host sea anemone Exaiptasia pallida (formerly Aiptasia pallida) was used as a cnidarian model (Fig. 1). E. pallida is easily reared and grown in the laboratory and these anemones closely resemble coral species in their associations with the same symbiotic dinoflagellate genus, Symbiodinium, and many of the same bacterial species24,25. These characteristics make the sea anemone an adequate system for asking biological questions of relevance for coral reef systems. The clonal line of sea anemones (CC7, originally obtained from Dr. John Pringle’s lab at Stanford University) was used for these experiments and has been reared in the laboratory for more than six years. Using the clonal line of anemones was advantageous as it removed any potential physiological and disease resistance variability that could be associated with unknown genetic differences among individual anemones found in a natural population. Moreover, the clonal anemones harbor the same symbiotic dinoflagellate type of Symbiodinium A4 (sensu: cp23S rRNA genotyping26), indicating that the photo-physiology of these clonal anemones is likely the same. Since experimental anemones were reared under the same environmental conditions for the last six years, the nutritional and physiological status of all anemones were presumed to be the same. The known coral bacterial pathogen Vibrio coralliilyticus was used as the infectious agent to elicit a defense response in the sea anemone. It is a major coral pathogen known to cause coral bleaching27 and white syndrome in Acropora corals28. It has also been shown to cause disease and mortality in Exaiptasia pallida29. The anemone responds to V. coralliilyticus with darkening of the tissues and retraction of tentacles, followed by complete disintegration of polyp tissues29,30. The disease progression pattern is consistent with the behavior of necrotizing pathogens29.
Figure 1. The sea anemone, Exaiptasia pallida, utilized in the immunological studies as a Cnidarian model system.

To assess the response of E. pallida anemones to repetitive encounters with the infectious agent, we first determined a sub-lethal exposure of the bacterial pathogen V. coralliilyticus that would allow priming of the host without causing mortality. It was determined that a concentration of 1 × 108 CFU ml−1 of this bacterial agent causes stress and mortality in E. pallidaanemones after four days of exposure (Supplementary Fig. S1A online). Within a ten-day bacterial exposure, mortality ranged from 60 to 90% in the anemones (Supplementary Fig. S1A online). However, if anemones were removed from the bacterial challenge, washed, and placed in pathogen-free seawater after the third day of pathogen exposure, anemones would recover and show 100% survivorship comparable to unexposed (control) anemones (Supplementary Fig. S2 online). Based on these results, a sub-lethal challenge of a three-day pathogen exposure at 1 × 108 CFU ml−1 was used for the immune priming experiments. The bacterial challenges were conducted at 30 °C as it has been shown the virulence in this pathogen increases at temperatures above 28 °C27,31. We demonstrated that this experimental temperature was not a factor of mortality during the bacterial exposure trials (Supplementary Fig. S1B online).
Following these trials, three experiments were performed to evaluate the existence of a priming response. Anemones were first subjected to a sub-lethal exposure of V. coralliilyticus followed by resting periods (pathogen-free recovering time from the sub-lethal challenge) of either two, four, or six weeks before exposing the sea anemones again to a lethal exposure (ten day pathogen challenge). It is important to note that none of the anemones died during the resting period or prior to the lethal challenge. The response and survivorship of these anemones (primed group) were compared to anemones that were exposed to a lethal challenge but without prior sub-lethal exposure (non-primed group), and also to a control group in which anemones were never exposed to sub-lethal or lethal bacterial challenges. The results showed that anemones that had previously encountered the pathogen (primed) had a higher survivorship than those anemones that encountered the pathogen for the first time (Fig. 2; Kaplan Meier; Mantel – Cox Post hoc test, p = 0.0001). The survivorship rate appeared to vary as a function of the lapsed time between the two consecutive pathogen exposures. Anemones exposed to the lethal challenge two and four weeks after the sub-lethal exposure presented seven- and five fold increases in survival, respectively, compared to the non-primed anemones (Fig. 2A-B; Kaplan-Meier; Mantel - Cox Post hoc test; Two weeks, p = 0.031; Four weeks, p = 0.039). However, the experimental group of anemones challenged six weeks after the sub-lethal exposure showed a 1.4 fold increase in survivorship that was not statistically significant (Fig. 2C; Kaplan-Meier; Mantel - Cox Post hoc test, p > 0.05). The improved response of the anemones to repeated encounters of the pathogen suggests the existence of transient defense priming that lasts for up to four weeks.
Figure 2. Kaplan-Meier survival plots for Exaiptasia pallida during 10-day lethal challenge to the pathogen Vibrio coralliilyticus following: (A) two weeks; (B) four weeks; and (C) six weeks recovery period post priming with a sub-lethal exposure.
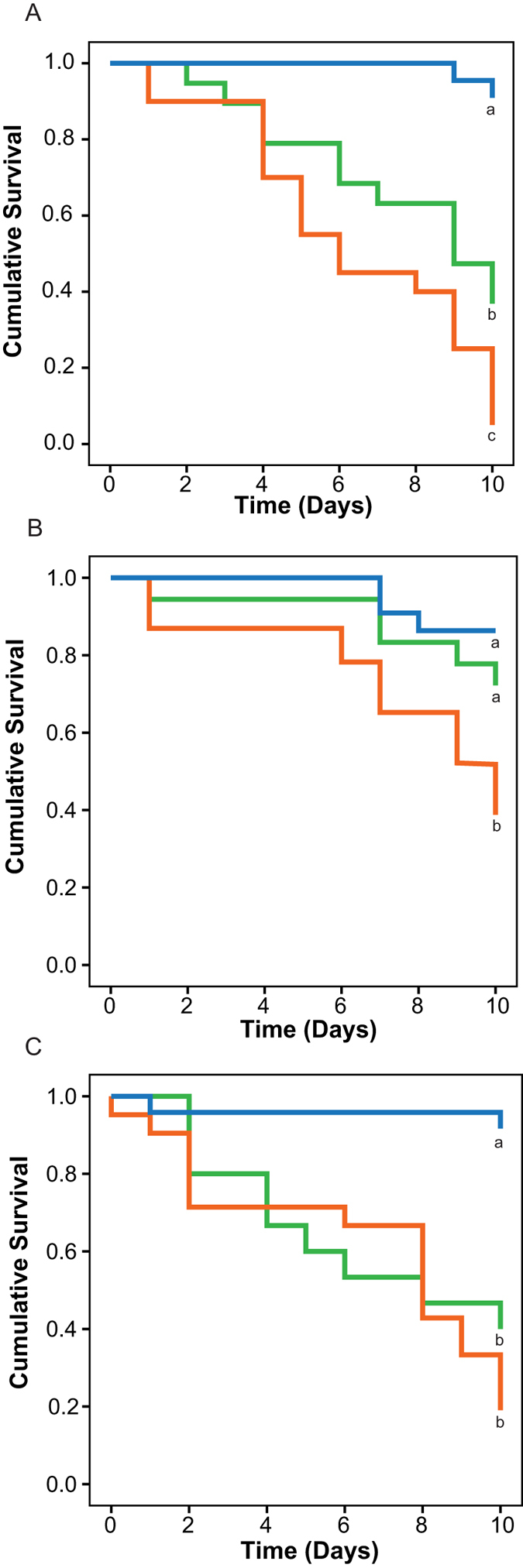
Green lines indicate those anemones that were primed with a sub-lethal exposure prior to the lethal challenge; orange lines represent anemones that were exposed only to the 10-day lethal challenge with no prior priming, and blue lines indicate control anemones that were not exposed at all to the pathogen. Different letters next to the graphed lines indicate statistically significant difference among the treatment at p < 0.05 (Kaplan-Meier; Mantel-Cox Post hoc test) N = 20 per treatment.
To determine whether the improved response in the primed anemones was not due to a chronic infection that continued over the experimental period, the pathogen load on the subjected anemones after the sub-lethal exposure was quantified using quantitative PCR (qPCR). Results from the assay indicated that by day four of the recovery period, V. coralliilyticus was no longer detectable in E. pallida (Supplementary Fig. S3A online; ANOVA, p < 0.05), suggesting that the improved response to pathogen upon repetitive encounters was due to a priming phenomenon and not to chronic infection. The same qPCR assay was used to confirm that V. coralliilyticus was present in E. pallida hosts throughout the lethal experiment, which showed considerable presence of pathogen load throughout the lethal exposure (Supplementary Fig. S3B online).
Proteomic analysis to dissect the molecular changes associated with the immune priming response
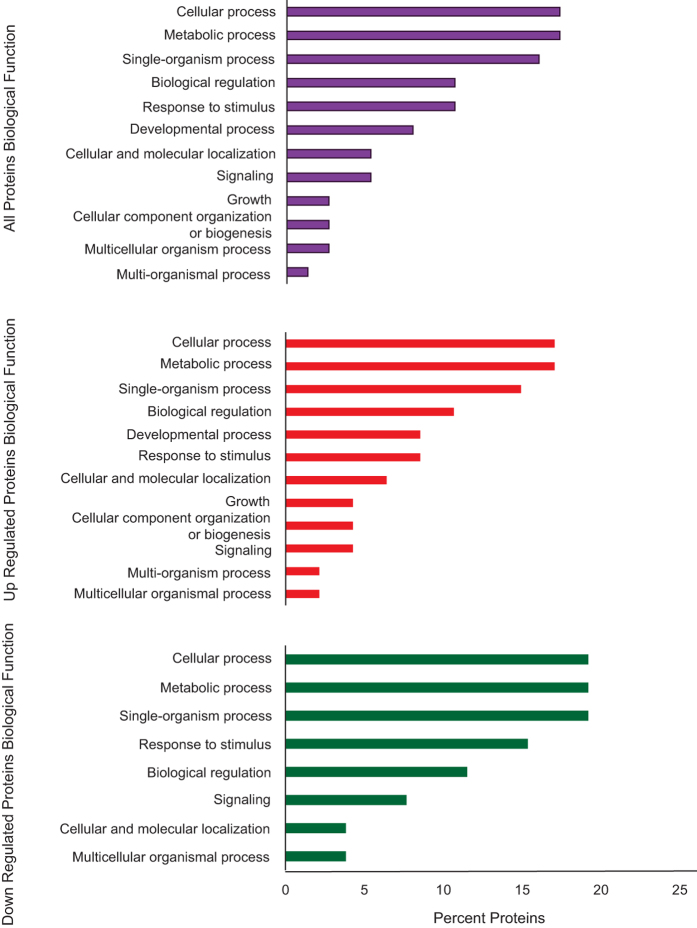
Further exploration of the immunological priming phenomenon took place at the protein level by analyzing samples collected four weeks following the post-priming phase and immediately before the lethal exposure. A two-dimensional fluorescence gel electrophoresis combined with mass spectrometry was used for the analysis. Based on the replicated 2D fluorescent in-gel analysis, a total of 1400 spots were detected. From this proteome, 39 spots (2.79%) with at least 1.3-fold change were identified as being differentially expressed between primed and non-primed anemones four weeks after the sub-lethal exposure to the primed anemones (Biological Variation Analysis, BVA, p < 0.05). Among these proteins, 16 were up-regulated, and 23 proteins were down-regulated in the primed anemones (Fig. 3). Of the 39 identified spots, the protein identities of 32 spots were determined using MALDI-TOF mass spectrometry and proteomic database comparisons with high confidence (Confidence Interval >95%; table 1). The protein profiles showed surprising complexity as some of these spots represented multiple isoforms of proteins varying in molecular mass and/or charge (Supplementary Fig. S4 online). The differentially expressed proteins identified through this method were involved in 27 different biological processes. The most represented biological processes were metabolic process (n = 14, GO: 0008152), cellular process (n = 13, GO: 0009987), response to stimulus (n = 8, GO: 0050896), and single organism process (n = 12, GO: 0044699) (Fig. 4, Supplementary Table S1 online). The representation of biological processes was similar between up-regulated and down-regulated proteins except the categories of developmental processes and cellular component organization and biogenesis, which were only represented by up-regulated proteins.
Figure 3. Histogram of the differentially expressed proteins as a function of fold change from Exaiptasia pallida anemones four weeks post priming in comparison to naïve anemones never exposed to the pathogen.
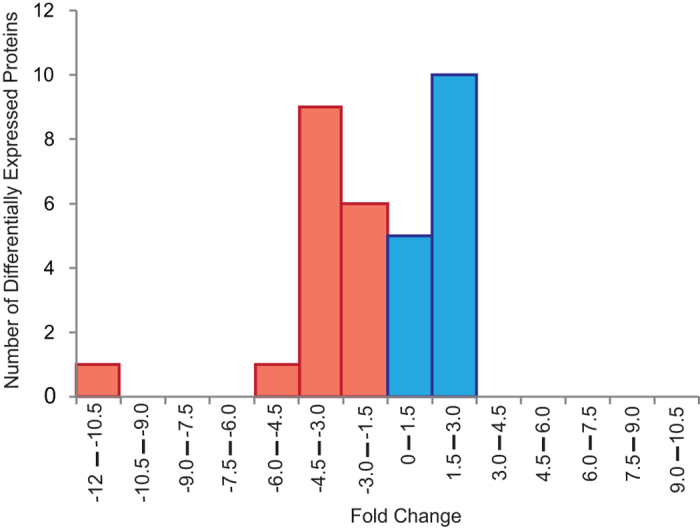
Blue bars indicate proteins that were differentially up-regulated in the anemones previously primed with a sub-lethal exposure to the pathogen; and red bars indicates down-regulated proteins in the same treatment.
Table 1.
| 2D Gel | Protein Identification | Exaiptasia | Protein | Protein | Fold | P |
|---|---|---|---|---|---|---|
| Spot | Genome | MW | value | |||
| Number | Gene ID | (Da) | PI | Change | T test | |
| 2 | Heat Shock Protein (70 KDa) | AIPGENE12496 | 74,685 | 5.6 | 2.02 | 0.091 |
| 6 | Signal Recognition Particle | AIPGENE24594 | 72,108 | 6.4 | 1.88 | 0.12 |
| 84 | MRP Protein | AIPGENE1468 | 95,674 | 9.3 | 1.72 | 0.029 |
| 32 | Calumenin-A | AIPGENE25880 | 35,247 | 4.5 | 1.69 | 0.015 |
| 34 | Glutamate Receptor | AIPGENE24462 | 101,286 | 9.2 | 1.63 | 0.0081 |
| 50 | Myosin Heavy Chain | AIPGENE8264 | 220,878 | 5.4 | 1.63 | 0.022 |
| 27 | Aminopeptidase | AIPGENE26826 | 53,551 | 6.4 | 1.59 | 0.0046 |
| 3 | Zona Pellucida | AIPGENE843 | 47,035 | 5.0 | 1.57 | 0.044 |
| 11 | Heat Shock Protein (60 KDa) | AIPGENE15267 | 62,708 | 5.3 | 1.57 | 0.051 |
| 13 | Moesin/ezrin/radixin | AIPGENE9804 | 66,884 | 5.8 | 1.56 | 0.064 |
| 43 | Fructose-Bisphosphate Aldolase | AIPGENE2871 | 38,732 | 7.6 | 1.50 | 0.055 |
| 61 | Heat Shock Protein (70 KDa) | AIPGENE12775 | 41,848 | 5.4 | 1.47 | 0.067 |
| 57 | Voltage Dependent Anion Selective Channel | AIPGENE3808 | 35,188 | 9.1 | 1.45 | 0.027 |
| 82 | Rho GDP-Dissociation Inhibitor | AIPGENE26434 | 22,251 | 4.8 | 1.32 | 0.12 |
| 65 | Cathepsin | AIPGENE26157 | 36,124 | 6.7 | 1.25 | 0.041 |
| 42 | Aspartate Aminotransferase | AIPGENE19338 | 45,969 | 6.8 | −1.47 | 0.0043 |
| 87 | Zinc Finger Protein | AIPGENE28354 | 68,856 | 8.8 | −1.48 | 0.015 |
| 35 | Cysteine Desulfurase | AIPGENE22361 | 54,104 | 6.2 | −1.48 | 0.091 |
| 39 | Fumarylacetoacetase | AIPGENE8362 | 46,370 | 6.2 | −1.49 | 0.013 |
| 15 | Selenium Binding Protein | AIPGENE13749 | 53,877 | 5.9 | −1.50 | 0.0025 |
| 28 | Cysteine Desulfurase | AIPGENE22361 | 54,104 | 6.2 | −1.50 | 0.0046 |
| 14 | Bleomycin Hydrolase | AIPGENE9335 | 55,335 | 5.8 | −1.53 | 0.015 |
| 41 | Pancreatic Triacylglycerol Lipase | AIPGENE19570 | 38,517 | 8.7 | −1.55 | 0.025 |
| 71 | Heat Shock Protein (70 KDa) | AIPGENE8252 | 41,986 | 5.3 | −1.58 | 0.055 |
| 46 | Calumenin B | AIPGENE9938 | 38,837 | 5.5 | −1.59 | 0.0044 |
| 38 | Cysteine Desulfurase | AIPGENE22361 | 54,104 | 6.2 | −1.69 | 0.0021 |
| 56 | Hemicentin | AIPGENE28714 | 49,371 | 6.6 | −1.70 | 0.011 |
| 55 | Thyroglobulin | AIPGENE20635 | 314,272 | 8.6 | −1.85 | 0.0039 |
| 74 | Nuclear Receptor | AIPGENE629 | 49,435 | 5.4 | −1.91 | 0.046 |
| 70 | Heat Shock Protein (70 KDa) | AIPGENE8252 | 41,986 | 5.3 | −2.67 | 0.0018 |
| 68 | Cyclic AMP and cGMP Phosphodiesterase | AIPGENE4644 | 51,903 | 5.8 | −4.05 | 0.017 |
| 69 | Heat Shock Protein (70 KDa) | AIPGENE8252 | 41,986 | 5.3 | −9.73 | 0.0037 |
Identification of the 32 proteins subjected to Mass Spectrometry analysis. 2D Gel Spot Number: Number as indicated on the 2D-DIGE gel on supplemental Fig. 4; Protein Identification: Identity assigned by BLAST data base searches; Exaiptasia Genome Gene ID: Gene ID is based on the gene mapping identification for the Exaiptasia genome (aiptasia.reefgenomics.org73); Protein MW: Molecular weight of the protein; Protein PI: Isoelectric point of the protein; Fold Change: The fold change is based on protein expression of primed anemones versus non-primed (naïve) anemones; P Value T test: The significance probability value based on statistical T test.
Figure 4. Representation of biological processes Gene Ontology (GO) terms for the 32 Exaiptasia pallida characterized proteins.
The results are summarized in three subgroups: all GO terms, GO terms from up-regulated and down-regulated proteins.
Of the proteins identified in this study, heat shock protein 70 was the most highly up-regulated protein in primed E. pallida with a 2.02-fold-higher expression in primed as opposed to control anemones (Table 1). Interestingly, a second protein also identified as heat shock protein 70, based on amino acid sequence of the generated mass-spec polypeptides was the most highly down-regulated protein with a 9.73 fold decrease in expression in primed anemones when compared to controls. However the size of this protein on the gels does not correspond to a 70 kda protein (Supplementary Fig. S4 online) indicating that this protein might be a smaller Heat Shock Protein.
Discussion
The sea anemone, Exaiptasia pallida, showed susceptibility to bacterial challenge with Vibrio coralliilyticus similar to coral species affected by the same bacterial pathogen at seawater temperatures 3 to 5 °C warmer than ambient temperatures27,31. However, the survivorship of the challenged anemones depended on previous pathogen exposure. Anemones that encountered the pathogen in sub-lethal conditions prior to lethal exposures showed higher survivorship than naïve anemones encountering the pathogen for the first time. Such findings suggest the potential presence of a protective priming defense mechanism in some members of the phylum Cnidaria. The priming response was also shown to be short-lived, lasting up to one month. The improved response of primed compared to non-primed anemones was not the result of a sustained immune response due to a chronic infection from the sub-lethal exposure, as the pathogen V. corallilyticus was cleared by the sea anemones four days after the termination of the sub-lethal exposure and several weeks before the second pathogen exposure. This is critical as a chronic infection in which a low level response of the immune system continues actively combating an infection32 and cause a more rapid secondary response in a subsequent pathogen challenge due to an already engaged immune system of the attacked host9. It is important to note that it is possible that even though V. corallilyticus presence decreases beyond detection, the presence of large concentrations of Vibrio at the onset of the experiment may have impacted the other microbial species associated with the anemone that may function as beneficial symbionts. Further studies are required to explore this possibility. Our findings demonstrate that the cnidarian defense system is functionally capable of unexpectedly durable induced protection. This suggests that selective pressures that triggered the evolution of immunological priming have a signature from early diverging animals.
Immunological priming has been documented in other invertebrates including crustaceans and insects5,6,7,8,9,10. In these cases, the improved response to the pathogens upon multiple encounters was also shown to be short-lived. For instance, the social bumble bee, Bombus terrestris, gain increased protection against pathogens upon a secondary exposure that lasted up to 27 days9. In this case, the priming of this duration would compare with an average life span of around 4 weeks for adult B. terrestris workers in the field. It highlights the clear ecological, and thus evolutionary, benefits of immunological priming in these organisms. In the case of cnidarians, such as anthozoans that can live for hundreds of years, immunological priming that confers a lasting protection of a month might appear to have a low ecological and evolutionary value. Interestingly, a similar timeframe of priming has also been described within the context of allorecognition in cnidarians. For example, specific memory of tissue transplantation immunity has been demonstrated to last four weeks in the coral Montipora verrucosa13, and eight weeks in the gorgonian, Swiftia exerta15. Yet some corals, such as the hydrocoral Millepora dichotoma appear not to possess a memory component to allorecognition33. The three above examples show a varying memory response to previously encountered allografts. In the cases that do show allomemory, it appears also to be relatively short-lived. While the mechanisms used in alloimmunity by cnidarians are unknown, it is possible that common ground might exist between the mechanisms modulating the processes of allografting and pathogen recognition. Regardless of the similarity between the mechanisms, the question remains as to what would be the advantage for a long-lived organism to have such a short-lived priming of their defense system. We speculate that short-lasting priming of the defense response could be ecologically relevant if pathogen encounters are concentrated and restricted to particular seasons (short period of time) characterized by high stress. In such seasons when pathogens are more active and virulent, cnidarians could be ecologically and evolutionary benefited if they have the capability to remember pathogenic encounters during the duration of the high stress season. This would allow maximizing the allocation of energy towards immunological priming when it is most needed since it has been shown to be energetically taxing on other organismal processes34. In recent years we have learned that sustained high temperatures above the average seasonal maximum are often related to an increase of disease outbreaks21,23,27,35,36,37. For example, Bruno and collaborators (2007) showed a highly significant relationship between the frequencies of warm temperature anomalies and the occurrence of white syndrome in Pacific reef-building corals. High temperatures during the summer months have also been associated with the activation of virulence among bacterial pathogens that are common residents in the coral reef environment27,38,39. Moreover, the high virulence and disease prevalence is seasonal in the majority of cases and ceases as the water temperature declines at the end of the summer17,21. Therefore, it is conceivable that the short duration of immunological priming described for Exaiptasia pallida mirrors the short time window when pathogens are seasonally active and during which they could be encountered repetitively by the sea anemone. This strategy would allow the sea anemone to reallocate energy use to other vital physiological needs during times where pathogens are less infectious. Additionally, the duration of priming might also be pathogen-specific. For example, in mice different pathogens elicit varying durations of priming and memory40. Pathogens that are encountered more often by a given organism could cause longer priming/memory. Further investigations are needed to understand whether different pathogens trigger longer immunological priming, and also to reveal how pathogen-specific the priming phenomenon is in the defense system of cnidarians, which could shed light about the existence of immunological memory.
Proteomic analysis of immune priming in E. pallida
The phenomenological data presented above indicate that there are responses in cnidarians that might be trained by past experience and increase upon a second exposure. This adds to the growing notion that invertebrates have extremely plastic immune effectors that can generate novel and functional immune response changes in relation to past experience. In the past the logical fallacy that because an organism lacks B and T cells, the organism will also lack an adaptive immune response has hindered our appreciation of the capability of basal metazoans to possess immunological priming and memory41. These organisms could generate a trained immune response in another way as has been documented for insects42,43. In our study we attempted to characterize molecular changes correlated with the priming phenomenon using a comparative proteomic approach to start dissecting the mechanism underlying an inducible enhanced immunity in cnidarians.
Statistically significant differences in proteomic profiles between naïve sea anemones and those primed after pathogen exposure suggests a clear molecular signature associated with immunological priming in cnidarians. The group of differentially expressed genes was diverse, suggesting that the molecular regulation of the priming defense is governed by changes in multiple cellular processes. None of these proteins were identified as antimicrobial peptides, which are the molecules normally produced during the actual fighting and clearing of pathogen infections in cnidarians44,45,46. In other invertebrates such as bumblebees, antimicrobial peptides are produced immediately after the bacterial challenge and then subside thereafter when the pathogen load decreases9. Therefore, the lack of differentially expressed antimicrobial peptides was expected as the proteomic analysis was conducted in primed anemones at least three weeks after the pathogen from the sub-lethal exposure was cleared. Consequently, the changes in protein expression detected in the primed anemones four weeks after the first pathogen exposure seem to be related the phenomenon of immunological priming rather than to pathogen clearance.
Gene ontology analysis indicated that many of the differentially produced proteins linked to immune defense priming were grouped into metabolic processes pathways, a pattern that has also been detected from transcriptomic analyses in corals affected with disease signs47,48. An example of a metabolic protein is the Fructose-Bisphosphate Aldolase protein that is an enzyme involved in glycolysis, a metabolic process that assures the production of energy required for a large number of other metabolic processes. We detected a higher amount of this protein in primed anemones, suggesting an enhanced metabolic function in primed animals. Previous studies have also shown a correlation between increased expression of metabolic enzymes in the response to secondary exposure of pathogens49. Recently, it was proposed that a shift of central glucose metabolism from oxidative phosphorylation to aerobic glycolysis (the “Warburg effect”) is the metabolic basis for trained immunity (i.e. the memory characteristics of the innate immune system recently described in vertebrates50), providing the energy and metabolic substrates for the increased activation of trained immune cells51. Further experimental and physiological studies are needed to investigate whether an increase of glycolysis is indeed a fundamental process in primed “trained” immunity in early-diverging metazoans, such as cnidarians. Findings from these future research avenues will provide an appreciation of the evolutionary origin for the key role of metabolism in innate host defense.
Many of the identified proteins in this study show homology to immune genes functionally characterized in other organisms. Although we need to be cautious when borrowing functionality of these proteins based on homology to other organisms52, their expression in this study bolsters the idea that they may be involved in the immune response of E. pallida. In the context of cnidarian immunology, we discuss key changes in protein production involved in following functional groups: stress response, ion transport and proteolysis.
Several proteins involved in stress response were detected in association with defense priming: two putative heat shock protein 70 (HSP70) orthologs and one heat shock protein 60 (HSP60) were up-regulated whereas three small heat shock proteins (~20 kDa) were down-regulated. It is well-known that the up-regulation of HSP synthesis provides resistance to toxic stresses such as heat shock53,54; however, their involvement has also been shown in response to many other environmental and biological insults, such as pathogenic infections55. Recently, we also documented transcriptional up-regulation of a HSP70 gene in the scleractinian coral, Acropora millepora, within hours of exposure to bacterial pathogens56. There is still no clear understanding of the molecular mechanisms involving HSP in response to pathogenic infection and whether its action in the immune defense is at the intracellular and/or at the extracellular level. However, it has been suggested that many HSPs have the property of damage associated molecular patterns (DAMPs) as they can bind to exposed hydrophobic residues of a wide spectrum of polypeptides57. HSPs could play a critical role in mediating innate immunity by activating Toll-like receptor (TLR) signaling due to their status as DAMPs and thus induce cytokine-mediated inflammatory responses. For instance, some evidence indicates that extracellular HSP70 can interact with TLR4 under a number of pathological situations58,59. Furthermore, HSP70 has been implicated in immunity stimulation either by antibody-independent activation of the complement immune system60 or by enhancing the expression of the prophenoloxidase system61. Under the hypothesis of DAMP-acting HSPs, it is possible that a higher synthesis of HSPs in primed organism, such as in the case of the sea anemone from this study, could allow for a faster response at the detection of infection-associated danger through interaction of TLR-HSP-DAMP, and thus induce a quicker inflammatory response upon a new exposure to pathogens (see model at Fig. 5). This model is supported by studies conducted on the brine shrimp Artemia showing that heat-induced accumulation of HSP70 appears to protect crustacean from pathogenic infection by Vibrio campbellii62. Current findings have shown that direct delivery of HSP vaccines to crustaceans improve their defense response and success to fight infections of pathogens61,62,63. For example, feeding with E. coli YS2 over-producing DnaK, the prokaryotic equivalent of Hsp70, enhances gnotobiotic Artemia larvae survival approximately two- to three-fold upon challenge with pathogenic V. campbellii63.
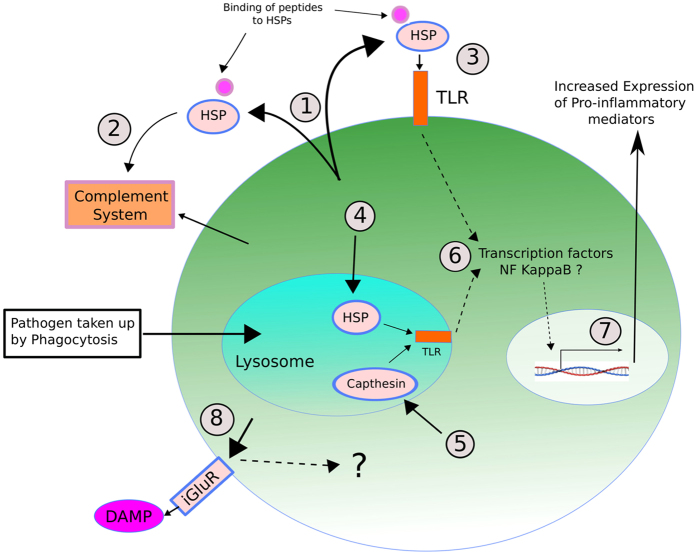
Figure 5.
Proposed model of Heat Shock Proteins (HSP),Capthesin and Glutamate Receptor (iGluR) roles in cnidarian molecular defense priming: (1) HSP are up regulated and some are extracellularly secreted where bind to peptides and act as DAMPs; (2) as DAMPs, HSP help with a faster activation of the innate complement system, and/or (3) interact and cause a quicker activation of outer host cell membrane TLRs; (4) intracellularly, up-regulated HSP proteins can be delivered into lysosomes in which they can also interact and activate endosomal cell membrane TLRs; (5) higher production of Capthesin are delivered into lysosomes in which they can also interact and activate endosomal cell membrane TLRs; (6) activated TLRs either from the outer membrane or endosomal membranes will trigger cell signaling pathways that will converge in the activation of transcription factors (likely NF-kappa β) that will ultimately induce the expression of immune-related genes (7) resulting in the production of potential pro-inflammatory molecules; (8) Higher expression of iGluR expressed on the outer membrane will also facilitate a faster sensing of potential DAMPs upon secondary exposure of pathogens.
Other proteins detected in this study were putatively characterized as being involved in ion transport. One of particular attention was the inotropic glutamate receptor (iGluR)-like protein found to be up-regulated in the primed anemones. iGluRs are ligand-gated ion channels best known for their role in fast excitatory neurotransmission in vertebrate and invertebrate nervous systems. However, new findings have shown that many homologs of these receptors are implicated in other biological process. A novel family of iGluR-related genes from insects (referred to as Ionotropic Receptors, IR) have been characterized as chemosensory receptors and are involved in olfaction and gustation processes64,65. The findings that these receptors are present across diverse groups of organisms from bacteria, plants and animals also suggests that this receptor family represents an evolutionarily ancient mechanism for sensing both internal and external chemical cues64. Of great interest are the findings showing that plant iGluRs are implicated in sensing a broad range of amino acids as part of the defense mechanism against infectious agents66. Recent studies examining the wound response and disease susceptibility in Arabidopsis thaliana Glutamate-Like Receptors (GLR) knockout mutants have provided evidence that some members of the GLR gene family encode important components of the plant’s defense response67. These discoveries are in line with our finding of higher production of iGluR-like proteins in primed Exaiptasia anemones. If involved in recognizing/sensing danger associated molecular components, a primed anemone with higher levels of expression of iGluRs would be better prepared to respond faster to infectious agents upon secondary exposures (Fig. 5).
Finally, we detected a higher production of immune-related proteolytic proteins including aminopeptidases and cathepsin, suggesting a potential enhancing key role of proteolysis in immune priming. For instance, capthepsins are key lytic enzymes and members of the proteases machinery packed in host lysosomes. These enzymes are not only involved in lysosome-contained pathogen degradation but also have been implicated in activating endosomal Toll-Like Receptors (TLR), which induce downstream cytokine-mediated pro-inflammatory responses68,69. Cathepsins generate a proteolytic cleavage, a prerequisite for TLR7 and TLR9 signaling70,71. Greater amounts of this protein in primed anemones implies that up-take of pathogens via phagocytosis will be digested and cleared faster through the phagosome-lysosome pathway. Additionally, their recognition by TLRs could be enhanced by a higher rate of proteolytic cleavage.
In summary, while immune priming has been found in several invertebrates6,9, this study discovered for the first time a similar phenomenon in an early diverging animal. Our findings support the notion that immunological priming may have evolved much earlier in the tree of life than previously thought. Additionally, the considerable amount of proteins that appear to be involved in the immune response of primed E. pallida suggests immunological priming in cnidarians is a more complex phenomenon than so far has been recognized. Furthermore, finding immunological priming in a sea anemone implicates the potential presence of the same mechanisms in other cnidarians such as corals. Future research addressing these mechanisms might be of crucial influence on developing restoration strategies for threatened and endangered coral reef species. As a potential outcome, “immunization” could become a tool to improve tolerance and survivorship of long-lived wild and re-introduced corals and thus, mitigate the deterioration of coral reef ecosystems.
Materials and Methods
Exaiptasia pallida Anemone Husbandry
Anemones used in these experiments were from the clonal CC7 population (John Pringle Lab, Stanford University) and were maintained in filtered artificial seawater at approximately 27 °C. Populations were kept on a day:night cycle of 12 h light:12 h dark with 30 to 60 μmole photons m−2 s−1 of light and fed freshly hatched brine shrimp nauplii twice a week. E. pallida used in this study were all approximately 3 mm in diameter and 10 mm high.
Pathogenic Bacterial Species and Culture Preparation
Vibrio coralliilyticus strain BAA 450 (ATCC) was used for the infection experiment. To recover the bacteria from the glycerol stocks, the frozen culture was streaked out and grown overnight on Marine Agar (Difco, USA) at 30 °C. The following day a single colony was picked with an inoculating loop and grown to logarithmic phase at 30 °C in Marine Broth-2216 (Difco, USA) while shaking at 100 rpm. Cultures were centrifuged at 10,000 g for five minutes, washed and re-suspended in sterile seawater. Bacterial cells were prepared to the designated final concentration based on a growth curve of V. coralliilyticus generated with optical density readings at 600 nm plotted against known bacterial culture concentration (CFU).
Determination of Vibrio coralliilyticus Concentration for Infection Trials
In order to determine an infective dose of the coral pathogen Vibrio coralliilyticus, ten-day infection trials were conducted on E. pallida anemones. Single anemones, acclimated to the experimental temperature (30 °C), were placed into single wells of a twelve-well culture dish containing filtered artificial seawater. During these trials, anemones (six per concentration) were challenged with three concentrations of the bacterium: 106, 107, or 108 CFU ml−1 at 30 °C. V. coralliilyticus becomes virulent at temperatures greater than 28 °C27,31. The inoculation of bacteria was conducted using via balneation56. Anemones were monitored daily over ten days to assess behavioral changes and mortality events.
Temperature Dependent Pathogen Virulence
To test if V. coralliilyticus showed similar temperature-dependent infectivity in E. pallida as it does in corals, infection trials were conducted at 25 °C and 30 °C for ten days. This experiment also allowed for the determination of the effect of experimental temperature on anemone survivorship. The concentration of V. coralliilyticus used for this experiment was 108 CFU ml−1 as this inoculum dose showed the most consistent infectivity pattern on anemones (Supplementary Fig. S1A online). Four treatments were conducted (+Bacteria at 25 °C, +Bacteria at 30 °C, −Bacteria at 25 °C, and −Bacteria at 30 °C). Each treatment contained a total of six anemones. The anemones were allocated in single wells of twelve-well tissue culture plates. The inoculation of bacteria on the +Bacteria treated anemones were conducted using the balneation technique56. Anemones were monitored daily over ten days to assess behavioral changes and mortality events.
Determination of Sub-lethal Pathogen Exposure
The sub-lethal exposure was not defined based on bacterial dose but based on the duration of exposure to the pathogen. From the infections experiments described above, the survivorship curves showed consistently that no anemone died during the first three days of pathogen exposure at a dose of 108 CFU ml−1 (Supplementary Fig. S1 online). Based on these results, an additional experiment was conducted aiming to confirm if a 3-day exposure at 108 CFU ml−1 to V. coralliilyticus could be considered a sub-lethal treatment. For this, anemones were exposed to V. coralliilyticus pathogen for a period of only three days, and placed in pathogen-free filtered seawater. The inoculation was carried out using via balneation56. The survivorship of these anemones was compared to a second group of anemones that remained under the bacterial exposure for a total of 10 days. Along with these two treatments, a control group of anemones were maintained in pathogen-free sea-water. The results from this experiment were fundamental to define the sub-lethal challenge used in subsequent priming experiments described below.
Priming Experiments on Exaiptasia pallida anemones
All experiments in this section were conducted at 30 °C and at a final concentration of 108 CFU ml−1 of V. coralliilyticus. Individual E. pallida, acclimated to the experimental temperature (30 °C) were placed into a single well of a twelve-well tissue culture plate containing filtered artificial seawater. Studies were conducted in three phases: sub-lethal exposure, recovery period, and lethal exposure. During the sub-lethal exposure, as indicated in the previous section, anemones (referred to as primed anemones) were challenged for three days with V. coralliilyticus (Supplementary Fig. S1B online). The sub-lethal exposures were followed by recovery periods, which varied as two, four, or six weeks. In this phase, anemones were removed from the sub-lethal exposure wells, transferred to pathogen-free seawater and left to recover during the designated recovery times. The final phase of the experiment consisted of the lethal exposure. From the previous experiment, it was determined that exposure to pathogen concentration of 108 CFU ml−1 between 4 and 10 days was considered a lethal challenge (Supplementary Fig. S1 online). During this phase the primed anemones were challenged again with V. coralliilyticus. Additionally, a second treatment (defined as non-primed E. pallida) was composed of naive anemones that were not subjected to sub-lethal exposure but challenged with V. coralliilyticus in the last phase of the experiment. The control anemones were kept at 30 °C for the entire experiment but were never challenged with the bacterium.
Quantitative PCR Assay for Determining Vibrio coralliilyticus Load on the Anemones
To determine the clearance dynamics of the V. coralliilyticus pathogen by the exposed anemones after the sub-lethal exposure time and during the lethal challenge, a quantitative real time PCR (qPCR) assay specific to test for pathogen presence was conducted. Anemone samples were collected immediately after finishing the sub-lethal exposure, and two and four days afterwards. During the lethal challenge (secondary exposure), anemones were collected at the following time points: one, three, seven and ten days after the start of the lethal exposure. For all the sampling times, three anemones were collected from each of the three treatments (primed, non-primed and controls). Total DNA was extracted from the collected anemones using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). DNA concentrations were estimated using the NanoDrop 2000c (Nano-Drop Technology, Wilmington, DE). V. coralliilyticus specific primers designed by Polson 2008 were used for the qPCR: 96F (5′-GTTRTCTGAACCTTCGGGGAACG-3′) and 1019R (5′- CTGTCTCCAGTCTCTTCTGAGG-3′)72. Reactions were conducted in 23 μl reactions with 12.5 μl of SYBR green master mix (BioRad, Hercules, CA) and 0.2 μM of each primer. The PCR conditions were as follows: 95 °C at 5 min followed by 40 cycles of 95 °C at 30 sec, 67 °C at 30 sec, and 72 °C at 60 sec. Dissociation curves were analyzed to confirm single product amplification at the end of qPCR runs. Samples were run in triplicate and mean values were used for calculations. Pathogen load on the infected anemones was expressed as relative proportion to amount of bacteria used at the initial inoculation (108 CFU ml−1). For the calculations, we used the following equation; 2(Cti–Cta); where Cti represented the mean amplification cycle value for the DNA extracted from the initial inoculum (108 CFU ml−1), and Cta represented the mean amplification cycle value for the DNA extracted from the infected anemone. A relative proportion of 1 means the pathogen load on an infected anemone corresponds to the same amount of pathogen found in concentration of 108 CFU m−1.
Statistical Analysis of Survivorship Data and Pathogen Load on Inoculated Anemones
For the priming experiments, Kaplan Meier estimators and survival plots were constructed for each of the three different recovery periods using the statistical software SPSS 21 (IBM). Post hoc comparisons using the Mantel–Cox test were further conducted to determine significant differences among the survival curves for each of the treatments. Analysis of variance (ANOVA) was performed in conjunction with a Tukey’s posthoc test to assess significance difference in the bacterial load on the anemones during different times after the sub-lethal challenge and during the lethal challenge using square root transformed data.
Proteomic Analysis
In order to determine the molecular changes underpinning immunological priming in E. pallida anemones, a proteomic analysis was conducted by Applied Biomics (Hayward, CA) according to the company’s standard protocol. In this analysis, the proteomic profiles of anemones primed with the pathogen during a sub-lethal exposure were compared to naïve anemones never exposed to the pathogen four weeks after the experimental anemones were subjected to the sub-lethal exposure. Fifteen anemones were collected from each of two treatments. These fifteen anemones were pooled in three groups of five anemones each and snap frozen in liquid nitrogen. The pooling of anemones was necessary since it allowed enough tissue material for extraction of proteins to run the proteomic analyses. Samples were sent to Applied to Biomics (Hayward, CA) for two-dimensional differential in-gel electrophoresis (2D DIGE) profiling and separation. The 2D DIGE gels were scanned using a Typhoon image scanner (GE Healthcare). The images were analyzed using Image Quant software (GE-Healthcare), and then subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.5 (GE-Healthcare). Protein differential expression ratio changes were obtained by in-gel DeCyder software analysis. Of the spots that were differentially expressed, 32 were subjected to isolation using an Ettan Spot Picker (GE Healthcare) followed by MALDI-TOF (MS) using a 5800 mass spectrometer (AB Sciex).
Proteins were identified by submitting the peptide mass and fragmentation spectra to GPS Explorer version 3.5 using the MASCOT search engine (Matrix Science) where the National Center for Biotechnology Information non-redundant (NCBInr) database and Exaiptasia pallida genome (aiptasia.reefgenomics.org73) were explored. Significant candidates had either protein score C.I.% or Ion C.I.% greater than 95. Once the proteins were identified, gene ontology analysis was conducted using Blast2GO. E-values less than 1 × 10−30 and a percent identity of 97% were used as a cut off for identifying the gene fragments.
Additional Information
How to cite this article: Brown, T. and Rodriguez-Lanetty, M. Defending against pathogens – immunological priming and its molecular basis in a sea anemone, cnidarian. Sci. Rep. 5, 17425; doi: 10.1038/srep17425 (2015).
Supplementary Material
Acknowledgments
We would like to thank Dr. John Pringle for providing the CC7 Exaiptasia pallida clonal line used in this study. We are grateful to Dr. John Pringle and Dr. Christian Voolstra for allowing us to use the E. pallida genome for the proteomic analysis. We are also grateful to Dr. Don Ennis and Dr. Anthony Bellantuono for helpful suggestions and discussions during the early anemone infection trials. We would like to thank IMaGeS lab members: Dan Merselis, Cindy Lewis, Katherine Dougan, Ellen Dow, Carolina Guillot, Christopher Otero, Melissa Morlote-Triana, and Leidy Gonzalez for comments on the manuscript. This research was funded by NSF-IOS (1453519) and NSF-OCE (0851123) grants awarded to MRL.
Footnotes
Author Contributions M.R.L. conceptualized the project. T.B. and M.R.L designed the experiments and wrote the paper. T.B. conducted the experiments and analyzed the data.
References
- Janeway C., Travers P., Walport M. & Capra J. Immunobiology: the immune system in health and disease. (ed. Elsevier Science Ltd./Garland Publishing) (1999). [Google Scholar]
- Beck G. & Habicht G. S. Immunity and the invertebrates - The fabulously complex immune systems of humans and other mammals evolved over hundreds of millions of years-in sometimes surprising ways. Sci. Am. 275, 60−+ (1996). [DOI] [PubMed] [Google Scholar]
- Loker E. S., Adema C. M., Zhang S. M. & Kepler T. B. Invertebrate immune systems-not homogeneous, not simple, not well understood. Immunol Rev 198, 10–24 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little T. J., Hultmark D. & Read A. F. Invertebrate immunity and the limits of mechanistic immunology. Nature Immunol 6, 651–654 (2005). [DOI] [PubMed] [Google Scholar]
- Kurtz J. Memory in the innate and adaptive immune systems. Microbes and Infect. 6, 1410–1417 (2004). [DOI] [PubMed] [Google Scholar]
- Kurtz J. & Franz K. Evidence for memory in invertebrate immunity. Nature 425, 37–38 (2003). [DOI] [PubMed] [Google Scholar]
- Little T. J., O’Connor B., Colegrave N., Watt K. & Read A. F. Maternal transfer of strain-specific immunity in an invertebrate. Curr. Biol. 13, 489–492 (2003). [DOI] [PubMed] [Google Scholar]
- Roth O., Sadd B. M., Schmid-Hempel P. & Kurtz J. Strain-specific priming of resistance in the red flour beetle, Tribolium castaneum. Proc. R. Soc. B 276, 145–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadd B. M. & Schmid-Hempel P. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr. Biol. 16, 1206–1210 (2006). [DOI] [PubMed] [Google Scholar]
- Johnson K. N., van Hulten M. C. W. & Barnes A. C. “Vaccination” of shrimp against viral pathogens: phenomenology and underlying mechanisms. Vaccine. 26, 4885–4892 (2008). [DOI] [PubMed] [Google Scholar]
- Hildemann W. H., Bigger C. H. & Jokiel P. L. Characteristics of immune memory in invertebrates. Am. Zool. 19, 911–911 (1979). [Google Scholar]
- Hildemann W. H., Jokiel P. L., Bigger C. H. & Johnston I. S. Allogeneic polymorphism and alloimmune memory in the coral, Montipora verrucosa. Transplantation 30, 297–301 (1980). [DOI] [PubMed] [Google Scholar]
- Hildemann W. H., Raison R. L., Cheung G., Hull C. J., Akaka L. & Okamoto J. Immunological specificity and memory in a scleractinian coral. Nature 270, 219–223 (1977). [DOI] [PubMed] [Google Scholar]
- Bigger C. H. Interspecific and intraspecific acrorhagial aggressive-behavior among sea anemones - a recognition of self and not-self. Biol. Bull. 159, 117–134 (1980). [Google Scholar]
- Saltercid L. & Bigger C. H. Alloimmunity in the gorgonian coral Swiftia exerta. Biol. Bull. 181, 127–134 (1991). [DOI] [PubMed] [Google Scholar]
- Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W. & Harvell C. D. Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562 (2009). [DOI] [PubMed] [Google Scholar]
- Richardson L. L. Coral diseases: what is really known? Trends Ecol. Evol. 13, 438–443 (1998). [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R. & Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- Sheridan C., Kramarsky-Winter E., Sweet M., Kushmaro A. & Leal M. C. Diseases in coral aquaculture: causes, implications and preventions. Aquaculture 396, 124–135 (2013). [Google Scholar]
- Harvell C. D. et al. Ecology - climate warming and disease risks for terrestrial and marine biota. Science. 296, 2158–2162 (2002). [DOI] [PubMed] [Google Scholar]
- Harvell D. et al. Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography. 20, 172–195 (2007). [Google Scholar]
- Hoegh-Guldberg O. et al. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007). [DOI] [PubMed] [Google Scholar]
- Burge C. A. et al. Climate change influences on marine infectious diseases: implications for management and society. Ann. Rev. Mar. Sci. 6, 249–277 (2014). [DOI] [PubMed] [Google Scholar]
- Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M. & Pringe J. R. Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376 (2008). [DOI] [PubMed] [Google Scholar]
- Otero C., Brown T. & Rodriguez-Lanetty M. Genomic analysis shows bacterial community shifts in Aiptasia pallida between environments. Proc. 43rd Benthic Ecology Meeting. (2014). Available at: http://imageslab.fiu.edu/sites/default/files/Otero_et_al_2014.pdf
- Santos S. R., Gutierrez-Rodriguez C. & Coffroth M. A. Phylogenetic identification of symbiotic dinoflagellates via length heteroplasmy in domain V of chloroplast large subunit (cp23S)-ribosomal DNA sequences. Mar. Biotechnol. 5, 130–140 (2003). [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y. et al. Vibrio coralliilyticus sp nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315 (2003). [DOI] [PubMed] [Google Scholar]
- Sussman M., Willis B. L., Victor S. & Bourne D. G. Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. Plos One. 3, e2393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza W. J., Krediet C. J., Meyer J. L., Canas G., Ritchie K. B. & Teplitski M. Outcomes of infections of sea anemone Aiptasia pallida with Vibrio spp. pathogenic to corals. Microb. Ecol. 68, 388–396 (2014). [DOI] [PubMed] [Google Scholar]
- Brown T. & Rodriguez-Lanetty M. Molecular mechanisms underpinning immunological memory in a basal metazoan (Cnidaria). Integrative and Comparative Biology. 55, E21–E21 (2015). [Google Scholar]
- Ben-Haim Y., Zicherman-Keren M. & Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D., Hussell T. & Dougan G. Chronic bacterial infections: living with unwanted guests. Nature Immunol. 3, 1026–1032 (2002). [DOI] [PubMed] [Google Scholar]
- Rinkevich B. Allorecognition and xenorecognition in reef corals: a decade of interactions. Hydrobiologia 530, 443–450 (2004). [Google Scholar]
- Contreras-Garduno J., Rodriguez M. C., Rodriguez M. H., Alvarado-Delgado A. & Lanz-Mendoza H. Cost of immune priming within generations: trade-off between infection and reproduction. Microbes Infect. 16, 261–267 (2014). [DOI] [PubMed] [Google Scholar]
- Bruno J. F. et al. Thermal stress and coral cover as drivers of coral disease outbreaks. Plos Biol. 5, 1220–1227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervino J. M. et al. Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Moreno D., Vargas I. S., Olson K. E. & Harrington L. C. Modeling dynamic introduction of Chikungunya Virus in the United States. PloS Negl. Trop. Dis. 6, e1918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. R., Kim K. & Harvell C. D. Temperature affects coral disease resistance and pathogen growth. Mar. Ecol. Prog. Ser. 329, 115–121 (2007). [Google Scholar]
- Miller A. W. & Richardson L. L. Fine structure analysis of black band disease (BBD) infected coral and coral exposed to the BBD toxins microcystin and sulfide. J Invertebr. Pathol. 109, 27–33 (2012). [DOI] [PubMed] [Google Scholar]
- Abdul-Careem M. F. et al. Genital HSV-2 infection induces short-term NK cell memory. Plos One. 7, e32821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziauddin J. & Schneider D. S. Where does innate immunity stop and adaptive immunity begin? Cell Host Microbe. 12, 394–395 (2012). [DOI] [PubMed] [Google Scholar]
- Dong Y. M., Taylor H. E. & Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. Plos Biol. 4, 1137–1146 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson F. L. et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309, 1874–1878 (2005). [DOI] [PubMed] [Google Scholar]
- Hemmrich G., Miller D. J. & Bosch T. C. G. The evolution of immunity: a low-life perspective. Trends Immunol. 28, 449–454 (2007). [DOI] [PubMed] [Google Scholar]
- Jesus Otero-Gonzalez A. et al. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. Faseb J. 24, 1320–1334 (2010). [DOI] [PubMed] [Google Scholar]
- Mydlarz L. D., Jones L. E. & Harvell C. D. Innate immunity environmental drivers and disease ecology of marine and freshwater invertebrates. Annu. Rev. Ecol. Evol. Syst. 37, 251–288 (2006). [Google Scholar]
- Libro S., Kaluziak S. T. & Vollmer S. V. RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis infected with white band disease. Plos One 8, e81821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon J. H. C., Beach-Letendre J., Weil E. & Mydlarz L. D. Relationship between phylogeny and immunity suggests older Caribbean coral lineages are more resistant to disease. Plos One. 9, e104787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Roth O., Philipp E. E. R., Saphoerster J., Rosenstiel P. & Reusch T. B. H. Specific immune priming in the invasive ctenophore Mnemiopsis leidyi. Biol. Lett 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M. G., Quintin J. & van der Meer J. W. M. Trained immunity: a memory for innate host defense. Cell Host Microbe. 9, 355–361 (2011). [DOI] [PubMed] [Google Scholar]
- Cheng S. C. et al. mTOR- and HIF-1 alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 345, 1579−+ (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich B. The ‘immunology trap’ of anthozoans. Invertebr. Surviv. J. 8, 153–161 (2011). [Google Scholar]
- Li G. C. & Werb Z. Correlation between synthesis of heat-shock proteins and development of thermotolerance in Chinese hamster fibroblasts. Proc. Natl. Acad. Sci. USA 79, 3218–3222 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. & Craig E. A. The heat-shock proteins. Annu. Rev. Genet. 22, 631–677 (1988). [DOI] [PubMed] [Google Scholar]
- Zugel U. & Kaufmann S. H. E.. Immune response against heat shock proteins in infectious diseases. Immunobiology. 201, 22–35 (1999). [DOI] [PubMed] [Google Scholar]
- Brown T., Bourne D. & Rodriguez-Lanetty M. Transcriptional activation of c3 and hsp70 as part of the immune response of Acropora millepora to bacterial challenges. Plos One 8, e67246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H. & Rock K. L. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8, 279–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez L. et al. Killing of normal melanocytes, combined with heat shock protein 70 and CD40L expression, cures large established melanomas. J. Immunol. 177, 4168–4177 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang K., Zhou B., Wang Y. Y., Rao L. & Zhang L. The TLR4 gene polymorphisms and susceptibility to cancer: a systematic review and meta-analysis. Eur. J. Cancer. 49, 946–954 (2013). [DOI] [PubMed] [Google Scholar]
- Prohaszka Z. et al. Heat shock protein 70 is a potent activator of the human complement system. Cell Stress Chaperones. 7, 17–22 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah K., Ranjan J., Sorgeloos P., MacRae T. H. & Bossier P. Priming the prophenoloxidase system of Artemia franciscana by heat shock proteins protects against Vibrio campbellii challenge. Fish Shellfish Immunol. 31, 134–141 (2011). [DOI] [PubMed] [Google Scholar]
- Sung Y. Y., Pineda C., MacRae T. H., Sorgeloos P. & Bossier P. Exposure of gnotobiotic Artemia franciscana larvae to abiotic stress promotes heat shock protein 70 synthesis and enhances resistance to pathogenic Vibrio campbellii. Cell Stress Chaperones. 13, 59–66 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. Y. et al. Ingestion of bacteria overproducing DnaK attenuates Vibrio infection of Artemia franciscana larvae. Cell Stress Chaperones. 14, 603–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C. & Vosshall L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V. et al. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. Plos Genet. 6, e1001064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B. G. & Roberts M. R. Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep. 6, 12703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S. A. R., Chauvin A., Pascaud F., Kellenberger S. & Farmer E. E. Glutamate receptor-like genes mediate leaf-to-leaf wound signalling. Nature. 500, 422−+ (2013). [DOI] [PubMed] [Google Scholar]
- Conus S. & Simon H.-U. Cathepsins and their involvement in immune responses. Swiss Med. Wkly. 140, 4–11 (2010). [DOI] [PubMed] [Google Scholar]
- Matsumoto F. et al. Cathepsins are required for toll-like receptor 9 responses. Biochem. Biophys. Res. Commun. 367, 693–699 (2008). [DOI] [PubMed] [Google Scholar]
- Ewald S. E. et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 456, 658–U688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y. M. & Ploegh H. L. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nature Immunology. 9, 1407–1414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson S. W., Higgins J. L. & Woodley C. M. PCR-based assay for detection of four coral pathogens. Proc. 11th Int. Coral Reef Symposium. 8, 247–251 (2008).
- Baumgarten et al. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. USA 112, 11893–11898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.