Abstract
Stem cell maintenance requires expression of genes essential for stemness and repression of differentiation genes. How this is achieved remains incompletely understood. Here we investigate the requirement for central components of heterochromatin, Heterochromatin Protein 1 (HP1) and the histone H3 lys9 methyltransferase Su(var)3-9, in the Drosophila male germline stem cell (GSC) self-renewal, a paradigm for studying adult stem cell behavior. We found that mutations or RNAi knock down of HP1 or Su(var)3-9 cause loss of GSCs, accompanied by defects in cell division or survival and premature expression of the differentiation gene bag of marbles (bam). Conversely, over-expressing HP1 increases GSC number in wildtype flies and, strikingly, restores fertility to the sterile hopscotch (hop) mutant flies that lack niche signals. These results suggest that the central components of heterochromatin play roles including repressing differentiation genes in Drosophila male GSC maintenance.
Stem cells possess unique epigenetic modifications and gene expression profiles that are conducive to their function: long-term maintenance of undifferentiated state yet poised for differentiation1,2,3. A salient feature of stem cells is that many developmentally important genes are epigenetically repressed. For instance, it has been shown that the Polycomb group proteins (PcGs) confer repressive chromatin modifications and are essential for maintenance of both embryonic and adult stem cells4,5,6. The molecular mechanisms controlling stem cells self-renewal are not completely understood, and different types of stem cells may use different strategies to repress differentiation genes. Heterochromatin formation, marked by histone H3 lys9 di- or tri-methylation (H3K9me2,3), is responsible for epigenetic gene repression in many developmental contexts7,8. Central components of heterochromatin comprise Heterochromatin Protein 1 (HP1) and histone H3 lys9 methyltransferases, including Su(var)3-9 and SETDB17,8. It has been reported that SETDB1 and its Drosophila homolog Eggless (Egg or dSETDB1) are essential for maintaining self-renewal of embryonic stem cells in mice and of adult germline stem cells in Drosophila, respectively9,10. In the Drosophila ovary, it has been shown that dSETDB1 and Su(var)3-9 sequentially function during GSC differentiation11. In planarians upon injury, HP1 is expressed and promotes self-renewal and triggers proliferation of adult stem cells for tissue regeneration12. However, it remains to be established that heterochromatin formation participates in stem cell self-renewal.
The male reproductive system in Drosophila provides an excellent model for understanding the fundamental mechanisms underlying stem cell regulation1,2,13 (Fig. S1A). At the apex of the testis, a group of post-mitotic somatic cells called hub cells comprise a key component of the male GSC niche, maintaining 8 to 12 germline stem cells (GSCs)14. GSCs and their primary derivatives, gonialblasts (GBs) and spermatogonia, express the germline specific protein Vasa; they are located adjacent to their niche – the hub cells, which express Fasciclin III (FasIII)1,2,13. GSCs are attached to the hub cells via adherens junctions. Hub cells express the cytokine-like ligand Unpaired (Upd), which activates the JAK/STAT (Hopscotch/STAT92E) pathway in the GSCs as well as the somatic cyst stem cells (CySCs), instructing their self-renewal. Upon dividing asymmetrically, a GSC produces two daughter cells. One of them retains contact with the hub and maintains stem cell identity, while the other is displaced from the hub to become a gonialblast, which begins transit-amplifying divisions as spermatogonia. Spermatogonia go through four synchronous mitotic divisions, resulting in 16 spermatocytes. Without JAK/STAT signaling, GSCs differentiate but do not self-renew, while ectopic JAK/STAT signaling greatly expands the stem cell population15,16. Further studies have indicated that JAK/STAT signaling primarily regulates self-renewal of somatic CySCs, which are essential for GSC self-renewal17,18. Despite these findings, the precise mechanisms controlling stem cell self-renewal remain incompletely understood.
Here, we investigated whether heterochromatin components are required for the maintenance of Drosophila male GSCs and examined the effects of overexpression or reduction of HP1 or Su(var)3-9, central heterochromatin components, on GSC numbers in Drosophila testes. Our results indicate that both HP1 or Su(var)3-9 are important for GSC self-renewal and for repressing differentiation genes, such as bam of marbles (bam), although they may also play general roles such as cell division and survival. Despite these general functions, our results suggest that proper heterochromatin formation might have a specific role in epigenetically repressing the expression of differentiation genes in GSCs, which is important for maintaining their self-renewal.
Results
Role of Heterochromatin components in GSC maintenance
To investigate the role of central heterochromatin components HP1 and Su(var)3-9 in GSC maintenance, we used genetic mosaics or RNAi-mediated knock down to determine the effects of loss of HP1 or Su(var)3-9 on GSCs. Heterochromatin Protein 1 (HP1) and the histone H3 lys9-specific methyltransferase Su(var)3-9 are central for heterochromatin formation, and altering HP1 or Su(var)3-9 levels directly impacts heterochromatin formation7. To identify GSCs, we carried out immunostaining with anti-alpha-spectrin antibodies to identify fusomes, with antibodies against the cell surface marker FasIII to mark hub cells (stromal cells), and with anti-Vasa antibodies to identify germ cells. GSCs can be identified as Vasa+ cells that are adjacent to the hub and that contain a dotted fusome.
We first generated clonal cells homozygous for a Su(var)3-9 null mutation, Su(var)3-92, using FLP/FRT-mediated mitotic recombination with a GFP marker, such that mutant cells can be identified as GFP– cells (see Methods). Su(var)3-92 is associated with a point mutation that abolishes the HMTase catalytic activity19. Su(var)3-92 homozygous flies were not viable, but became fully viable when a hsp70-Su(var)3-9+ transgene was expressed in the background (n > 100), ruling out the possibility that the Su(var)3-92 chromosome carried an unrelated lethal mutation. We thus used the Su(var)3-92 chromosome to carry out clonal analysis.
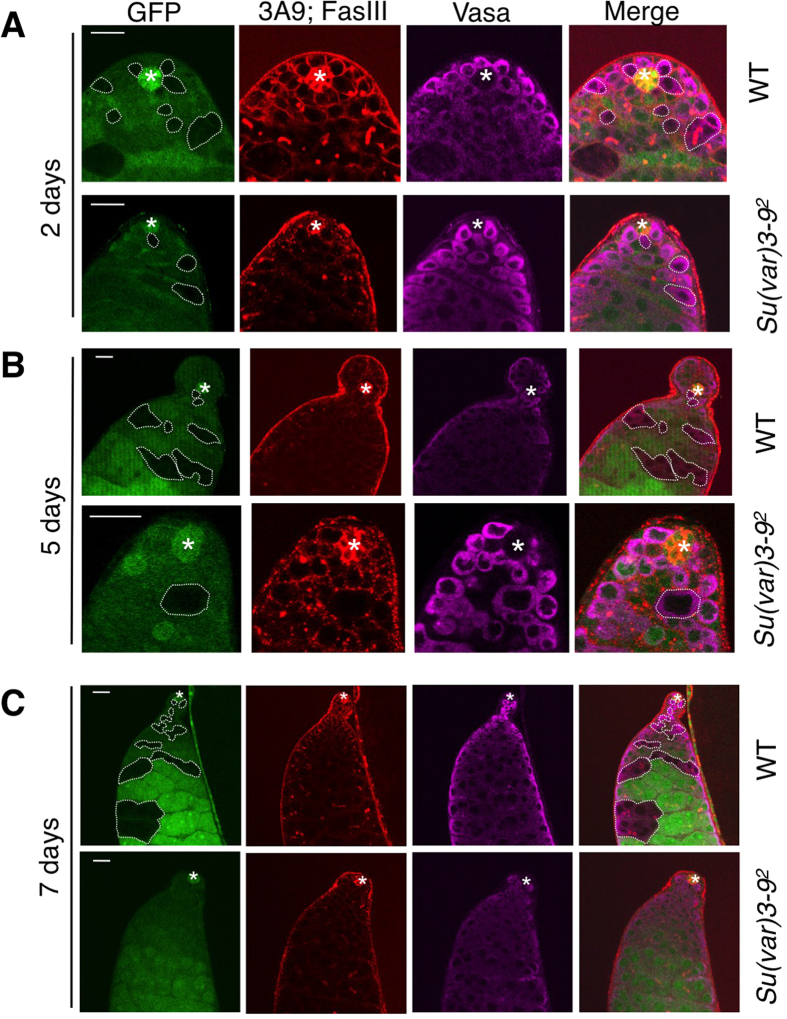
When examined 2 days after clone induction, wild-type control GSC, GB, spermatogonial clones were frequently found; clones of Su(var)3-92 homozygous GSCs, GBs, and spermatogonia were also found, albeit at a lower frequency (Fig. 1A; Table 1). However, when examined 5 and 7 days after clone induction, whereas wild-type control clones were frequently identified that included GSCs, GBs, spermatogonia, and spermatocytes, Su(var)3-92 clones were found at much lower frequency and usually not as GSCs but as spermatocytes only (Fig. 1B,C; Table 1), suggesting that Su(var)3-92 cells cannot remain as GSCs but can still undergo differentiation to give rise to spermatocytes. Consistent with the Su(var)3-9’s function as an H3K9 methyltransferase in heterochromatin formation and chromosomal compaction, Su(var)3-92 mutant cells exhibited increased nuclear size (Fig. 1B), suggesting a loss of chromosomal compaction, and a loss of the heterochromatin marker H3K9me3 (Fig. S1D). Interestingly, Su(var)3-92 mutant clones (Fig. 1B), as well as Su(var)3-9 RNAi-expressing clones (see below) contained fewer than 16 cells/cyst, suggesting that Su(var)3-9 may also play a role in spermatocyte division or differentiation.
Figure 1. Effects of Su(var)3-9 mutation on GSC maintenance.
Testes from male flies that had been subjected to induction of marked clones for loss-of-function of Su(var)3-9 were immunostained for Vasa (magenta cytoplasmic staining) and for fusomes and hub cells (both red). Scale bar = 20 μm. Testes were dissected after 2 (A), 5 (B), or 7 (C) days after clone induction. Wild-type control and Su(var)3-92 homozygous mutant clonal cells were marked by the absence of GFP (lack of green; circled by dotted lines). Note that wild-type control clones were always found at GSC positions (next to the hub) and as differentiated spermatogonia, and that Su(var)3-92 homozygous mutant clones were found at the GSC position only 2 day after clone induction, but not after 5 or 7 days, although GFP– spermatogonia were still found. Also note the increased nuclear size of the Su(var)3-92 homozygous cell (GFP–) in 5 days after clone induction.
Table 1. Su(var)3-9 is essential for GSC self-renewal.
| Su(var)3-92 mutant clones (GFP–) | |||
|---|---|---|---|
| Days after Induction | Clone Genotype | Testes with GSCclones/testes scored | testes scored |
| 2 | Wild type | 24/42 (57%) | 8/42 (19%) |
| Su(var)3-92 | 15/48 (31%) | 19/48 (40%) | |
| 5 | Wild type | 24/44 (55%) | 9/44 (20%) |
| Su(var)3-92 | 0/42 (0%) | 13/42 (31%) | |
| 7 | Wild type | 31/53 (58%) | 4/53 (7.5%) |
| Su(var)3-92 | 0/30 (0%) | 6/30 (20%) | |
| Su(var)3-9 RNAi clones (GFP+) | |||
| Days after Induction | Clone Genotype | Testes with GSCclones/testes scored | Testes with non-GSC clones/testes scored |
| 2 | Wild type | 62/67 (92%) | 5/67 (7.5%) |
| Su(var)3-9 RNAi | 4/74 (5.4%) | 44/74 (59%) | |
| 5 | Wild type | 44/51 (81%) | 6/51 (12%) |
| Su(var)3-9 RNAi | 3/65 (4.6%) | 18/65 (28%) | |
| 7 | Wild type | 35/46 (76%) | 10/46 (22%) |
| Su(var)3-9 RNAi | 1/27 (3.7%) | 5/27 (19%) | |
Mutant clones homozygous for Su(var)3-92 (loss-of-function allele) or wild-type (control) were identified as GFP– cells. Knock-down clones expressing Su(var)3-9 RNAi or wild-type control were identified as GFP+ cells. Testies with one or more mutant clones at the GSC position were counted as “testes with GCS clones”.
To confirm the role of Su(var)3-9 in GSC maintenance, we tested additional strong or null alleles, Su(var)3-917 and Su(var)3-9619. Testes from Su(var)3-917/6transheterozygous male survivors had no detectable heterochromatin marker H3K9me3 signal and were thinner (Fig. S1C). In addition, Su(var)3-917/6transheterozygous males lost their GSCs more precipitately than controls. By counting Vasa+ GSCs in 1- and 30-day-old males, we found that while 1- and 30-day-old control males had on average 7.9 ± 1.1 and 5.7 ± 1.2 GSCs/testis, respectively, Su(var)3-917/6 males had 7.4 ± 1.7 and 3.6 ± 1.3 GSCs/testis, respectively. 30-day-old Su(var)3-917/6 males had significantly fewer GSCs than their control siblings (p = 0.006; Student’s t-Test). These observations are consistent with the idea that Su(var)3-9 is important for male GSC maintenance.
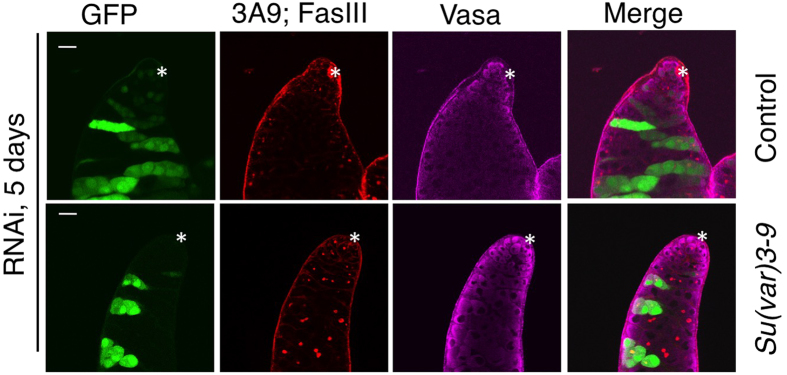
As an independent test for the requirement of Su(var)3-9 in GSC maintenance, we expressed Su(var)3-9 RNAi in random clonal cells that were marked by GFP (see Methods). We examined GFP+ cells 2, 5, and 7 days after clone induction. We found that while in the control experiment (no Su(var)3-9 RNAi expression), GFP+ clonal cells were abundantly found as both GSCs and differentiated cysts, GFP+ cells expressing Su(var)3-9 RNAi, however, were found as GSCs (at a lower frequency) only 2 and 5 days, but not 7 days after clone induction (Fig. 2; Table 1). Seven days after clone induction, GFP+ cells expressing Su(var)3-9 RNAi were found only as differentiated cysts (Fig. 2; Table 1). Taken together, the above two types of clonal studies suggest that loss of Su(var)3-9 caused loss of GSCs, and may also cause defects in GSC division or differentiation.
Figure 2. Effects of knocking down Su(var)3-9 by RNAi on GSC maintenance.
Testes from male flies expressing a Su(var)3-9 RNAi transgene with the germline specific nanos-Gal4 were immunostained for Vasa (magenta cytoplasmic staining) and for fusomes and hub cells (both red). Asterisk marks the position of the hub. Scale bar = 20 μm. In control testes (which did not express Su(var)3-9 RNAi) 5 days after clone induction, clonal GFP+ cells were found as both GSCs (next to the hub) and spermatogonia. Five days after Su(var)3-9 RNAi was expressed, no GFP+ GSCs were observed next to the hub, although GFP+ spermatogonia were still observed.
We also carried out similar experiments with a null mutation and RNAi transgenes targeting Su(var)205, which encodes HP1. We found that, similarly to Su(var)3-92 mutant clones, clones of Su(var)205 null GSCs (Su(var)2055) were found two days after clone induction, albeit at a lower frequency than wild-type control clones (Table S1). In contrast to Su(var)3-92clones, however, we never recovered any mature Su(var)2055 homozygous spermatocytes (Fig. S2A; Table S1), suggesting that complete loss of HP1 may cause lethality to GSCs. Similar results were obtained with expressing Su(var)205 RNAi in random clones (Fig. S2B). Since HP1 is not only essential for heterochromatin formation, but also has heterochromatin-independent functions, complete loss of HP1 or heterochromatin may cause cell death. Nevertheless, the above genetic mosaic studies using Su(var)3-9 and Su(var)205 mutations and their RNAi transgenes suggest that the major heterochromatin components, HP1 and Su(var)3-9, are essential for GSC maintenance, possibly playing differential roles in survival, division, and differentiation.
Role of HP1 and Su(var)3-9 in GSC-specific gene expression
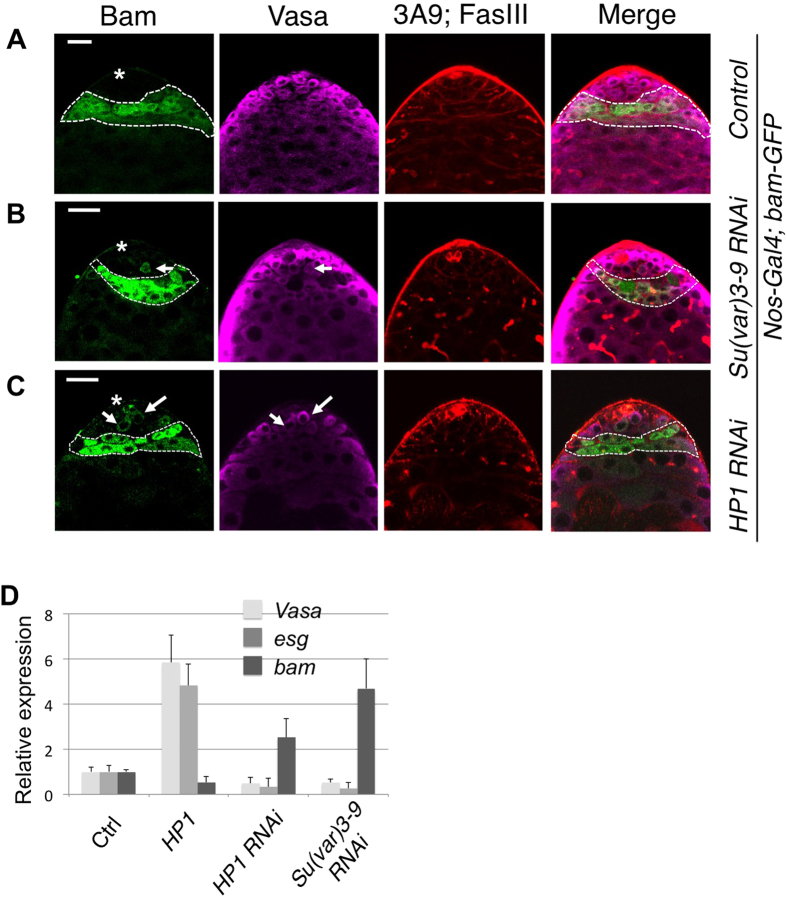
To understand whether HP1 and Su(var)3-9 play specific roles in GSC maintenance in addition to their general functions in cell division and survival, we examined changes in expression of a few known genes that are differentially expressed in GSCs. The differentiation marker gene bag of marbles (bam)20,21 is normally expressed in 4–16 cell differentiated spermatogonia cysts but not in GSCs and their immediate progeny, gonialblasts, resulting in a band of bam expression three cell diameters away from the hub20, as revealed by a bam-GFP reporter17 (Fig. 3A). We found that knocking down Su(var)3-9 by RNAi, driven by the germline specific nanos-Gal4, resulted in ectopic expression of bam-GFP in gonialblasts, as judged by the location of cell ectopically expressing bam-GFP (Fig. 3B), suggesting that these cells had undergone premature differentiation. HP1 RNAi knock-down also resulted in ectopic expression of bam-GFP in GSCs and gonialblasts, as cells immediately adjacent to the hub now expressed bam-GFP (Fig. 3C). In addition, knocking-down HP1 caused loss of the germline-specific marker Vasa in a fraction of germline cells, although some of these spermatogonia maintained bam-GFP expression (Fig. 3C, arrow). These results suggest that HP1 and Su(var)3-9 might be important for maintaining the unique expression profiles of GSCs and their progeny.
Figure 3. Loss of Su(var)3-9 and HP1 disrupts GSC specific gene expression.
(A-C) Testes from male flies of indicated genotypes were immunostained with mAb3A9 and anti-FasIII (both red) to reveal the fusome and hub cells, respectively, and with anti-Vasa (magenta). Green shows bam-GFP expression. Brackets indicate the band of bam-GFP cells normally detected in differentiated GSCs. The asterisk marks the hub. Scale bar = 20 μm. (A) A wild-type control testis, in which bam-GFP is expressed in spermatogonia three cell diameters away from the hub and is not expressed in GSCs and gonialblasts. (B) In a testis expressing Su(var)3-9 RNAi, bam-GFP is detected in cells next to the hub and also in those in gonialblast positions. The arrow points to one such cells. (C) In a testis expressing HP1 RNAi, bam-GFP is detected in cells in gonialblast positions and also weakly in cells next to the hub (arrows). The arrowhead points to a spermatogonia that expresses bam-GFP but has lost Vasa expression. (D) Individual testes of indicated genotypes were dissected and the tip region (see Methods) was used for qPCR experiments to quantify mRNA levels of rp49 (control), esg, Vasa, and bam. The results were normalized to rp49 and then to wild-type testis. Error bars represent standard deviations of three independent experiments.
To further investigate the role of HP1 and Su(var)3-9 in GSC gene expression, we performed qPCR experiments using testis tip regions (see Methods) to determine changes in expression of genes that are differentially expressed in GSCs. We found that HP1 over-expression caused an increase in the expression levels of germline or GSC-specific genes, esg and Vasa, and a decrease in those of the differentiation gene bam, while RNAi knockdown of HP1 and Su(var)3-9 had the opposite effects (Fig. 3D). Thus, heterochromatin components HP1 and Su(var)3-9 might be important for regulating differential expression of genes that are essential for GSC maintenance.
HP1 overexpression rescues GSC loss mutant phenotype
Next, we investigated whether the heterochromatin components HP1 and Su(var)3-9, when overexpressed, can promote GSC self-renewal. It has been shown that GSC self-renewal requires activation of JAK/STAT signaling by Upd secreted from the hub15,16. We investigated whether over-expressing HP1 could restore GSCs to the hop25 male survivors that do not have GSCs.
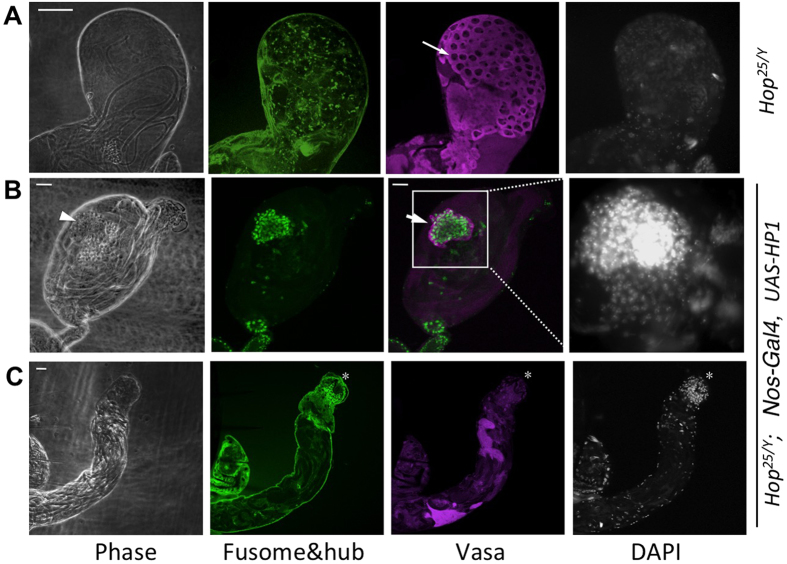
hop25 is a hypomorphic allele of hop; hop25 hemizygous males usually die, but occasionally escapers can be found that are completely sterile, with their testes lacking GSCs15,16. Indeed, testes of hop25 hemizygous male survivors lack esg-GFP+ cells or any small cells with condensed nuclei that are characteristic of GSCs and that can be distinguished by intense DAPI staining (Fig. 4A). Upon staining with antibodies that distinguish different cell types in the testis, we found that testes from hop25 hemizygous males contain exclusively large Vasa-positive germ cells (Fig. 4A, arrow). This is in contrast to a previous report that hop25 hemizygous testes had no germ cells16. In wild-type testes, large Vasa-positive germ cells contain branched fusomes. However, the large germ cells in hop25 hemizygous testes contain only dotted (not branched) fusomes (Fig. 4A) and were negative for the differentiation marker bam-GFP. We suggest that the large Vasa-positive (bam-negative) cells in hop25 hemizygous testes might represent primordial germ cells (PGCs) that were not induced to form GSCs due to lack of JAK/STAT signaling. Nonetheless, a few elongated spermatids were found in hop25 hemizygous testes, which could be derived from a few rare “escaper” GSCs in these testes (Fig. 4A, left), although hop25 hemizygous males are sterile.
Figure 4. HP1 overexpression rescues GSC loss mutants.
Testes from 3-day old hop25/Y male survivors of indicated genetic backgrounds were stained with DAPI, anti-Vasa, mAb3A9 and anti-FasIII, as indicated. (A) A hop25/Y; nos-Gal4 testis showing no sperm bundles, with all Vasa-positive germ cells exhibiting large nuclei (arrow) and dotted fusome but lacking intense DAPI signals. (B) A partially rescued hop25/Y; nos-Gal4/UAS-HP1 testis. Thick sperm bundles were seen (left). A fraction of Vasa-positive germ cells contain small DAPI-dense nuclei, dotted fusome, resembling GSCs (arrow), were observed. (C) A completely rescued testis from a hop25/Y; nos-Gal4/UAS-HP1 male survivor that had become fertile. The testis resembles that of wild-type flies, with mature long sperm bundles.
We expressed HP1 in hop25 hemizygous males using the nanos-Gal4 driver and examined their effects on the hop25 testes. We found that expressing HP1 rescued, to some extent, GSC formation in hop25 hemizygous testes, i.e., cells resembling GSCs were detected as small Vasa-positive cells that were brightly stained with DAPI in the testes (Fig. 4B). Consistent with a partial rescue of GSC formation, more thick bundles of differentiated spermatids were observed in these testes (Fig. 4B, left).
Strikingly, expressing HP1 was able to restore fertility to the otherwise sterile hop25 hemizygous males, such that a few hop25/Y; nosGal4/UAS-HP1 males sired progeny when crossed to wild-type virgin females (n = 4/13), while none of the hop25/Y; nosGal4/CyO control sibling males became fertile (n = 15). The presence of the hop25 and UAS-HP1 chromosomes in the rescued males was confirmed in the F1 generation. Without the HP1 transgene expression, no fertile hop25, nosGal4/Y male flies were ever found (n > 100; also see15,16). We dissected the fertile hop25/Y; nosGal4/UAS-HP1 males and found that, interestingly, some of them had asymmetric testes, with one testis having an elongated morphology containing a few sperm bundles (Fig. 4C), while the other retaining the “bulb” appearance of the typical hop25 hemizygous testis (see Fig. 4B). All the hop25/Y; nosGal4/CyO control sibling males examined had both testes exhibiting the “bulb” phenotype. Thus, over-expressing HP1 may promote GSC development or self-renewal even in the absence of adequate niche signals from the hub, enough for restoring fertility to the sterile hop25 hemizygous males.
Levels of heterochromatin components influence GSC number
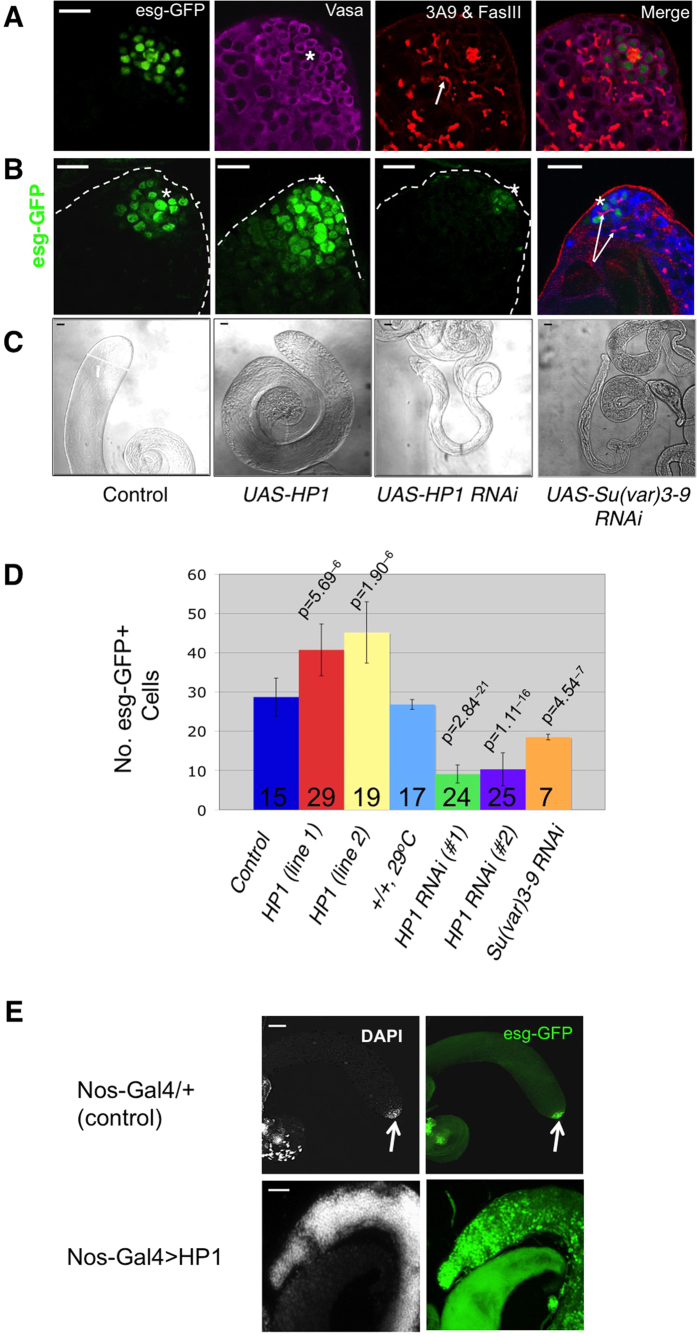
To further substantiate that HP1 and Su(var)3-9 are important for GSC maintenance, we tested the effects of altering their levels on Drosophila male GSC numbers. We used an esg-GFP enhancer trap line to estimate GSC number because it allows direct observation of GFP+ cells in dissected testes without immunostaining. esg-GFP is inserted in the escargot (esg) locus (esg-GFP; see Methods) and expresses high levels of GFP specifically in GSCs and gonialblasts (GBs), but not in CySCs (Fig. 5A, S3). esg encodes a transcription factor important for GSC function22,23, and is specifically expressed in many types of adult stem cells24,25,26. Although esg-GFP is also expressed in hub cells, these cells are clearly distinguishable from GSCs and GBs due to their morphology and unique localization (Fig. 5A). Although esg might also be expressed in CySCs, the esg-GFP used in this study was not detectable in CySCs (Fig. S3). An esg-lacZ enhancer trap line has previously been used to mark Drosophila male GSCs and GBs27. Therefore, GFP+ cells in the testis of esg-GFP males can be used to estimate the abundance of GSCs.
Figure 5. Levels of HP1 and Su(var)3-9 affect GSC number.
Scale bar = 20 μm. (A) The apex of a control testis expressing esg-GFP. Testes were immunostained with mAb3A9 and anti-FasIII (both red) to reveal the fusome and hub cells, respectively. GFP fluorescence (green) is detected in GSCs and GBs, as well as hub cells, but not in the somatic CySCs or CCs. The hub is marked with an asterisk. Germ cells (including GSCs) are positve for Vasa (magenta), which is a cytoplasmic protein specifically expressed in germ cells. GSCs usually contain spherical fusomes (red dots), and differentiated germline cysts are marked by the presence of branched fusomes (arrow). Scale bar = 20 μm. (B) Testis apex (outlined by dotted line) from 3-day old male flies expressing esg-GFP (green) and indicated transgenes (bottom) driven by nos-Gal4. The hub is marked with an asterisk. Branched fusomes are indicated (mAb3A9, red, arrows). (C) Effects of expressing indicated transgenes using nanos-Gal4 on the morphology of the testis. Representative images from 3-day old male flies are shown at the same scale. Note the HP1 or Su(var)3-9 knockdown testes have a smaller diameter than the wild-type control (left). (D) Quantification of esg-GFP+ cells of testes expressing indicated transgenes driven by nanos-Gal4. Numbers of testes analyzed are indicated; p: significance compared to control by Student’s T-test. (E) Testes were dissected from 40-day old adult nos-Gal4/+; esg-GFP/UAS-HP1 or control (nos-Gal4/+; esg-GFP/+) males and were scanned for GFP and DAPI. Note that the wild-type control testis has DAPI-dense and GFP+ cells present only at the tip region, while the nos-Gal4 > HP1 testis exhibits great expansion of esg-GFP+ and DAPI-dense cells.
To confirm that esg-GFP can be used for estimating GSCs, we carried out immunostaining with anti-spectrin antibodies to identify fusomes, with antibodies against the cell surface marker FasIII to mark hub cells, and with anti-Vasa antibodies to identify germ cells (Fig. 5A). We found that, except for hub cells, all GFP-positive cells also co-expressed germline marker Vasa (Fig. 5A, S3), consistent with the idea that esg-GFP marks GSCs and GBs. By analyzing serial optical sections using confocal microscopy, we found that there are on average 26.8 ± 2.6 GFP+ cells (excluding hub cells; n = 27 testes) in the testis of wild-type males carrying a copy of esg-GFP, corresponding to the number of GSCs and their immediate progeny (gonialblasts) present in the testis.
To assess the effects of altering HP1 and Su(var)3-9 on GSCs, we used the nanos-Gal4 driver to express transgenes in the germline, and examined 3-day old adult testes. We found that over-expressing HP1 caused an increase in the number of GFP+ cells, while expressing a HP1-RNAi or a Su(var)3-9 RNAi transgene dramatically reduced the GFP+ cell population and resulted in appearances of branched fusomes near the hub (Fig. 5B, arrows), and in shrinkage of the testis (Fig. 5C). In control and UAS-HP1 testes, GFP+ cells had dotted fusomes (not shown). These results support the idea that levels of HP1 and Su(var)3-9 influence GSC numbers.
To further determine to what extent HP1 overexpression can increase GSC number, we prolonged HP1 over-expression and examined the testes from adults 40 days after eclosion. We found that over-expressing HP1 in these conditions caused a great expansion of GSCs, such that the testis was, strikingly, filled with DAPI-dense, Vasa-positive GSCs and lacked any differentiated, elongated spermatid bundles (Fig. 5E). It has been previously reported that GSCs and their immediate daughter cells, gonialblasts (GBs), as well as Hub cells and CySCs contain highly condensed DNA, which is brightly stained with the DNA dye DAPI16,27. Wild-type control flies raised in parallel did not show over-population of GSCs, but rather exhibited reduced GSC number (Fig. 5E), consistent with previously reported GSC loss with aging28. These results suggest that HP1 over-expression promote GSC formation or maintenance, consistent with the above observation that over-expressing HP1 restored GSCs as well as fertility to hop25sterile flies that normally lack GSCs.
Discussion
Global epigenetic transcriptional repression, especially of differentiation genes, is a common feature of all stem cells. Heterochromatin formation has long been known as an important epigenetic gene repression mechanism7. However, the role of heterochromatin formation in stem cell self-renewal has not been systematically studied. To understand the molecular mechanisms that control stem cell self-renewal, we have genetically investigated the roles of two major heterochromatin components, HP1 and Su(var)3-9, in Drosophila male GSC self-renewal. We have shown that HP1 and Su(var)3-9 are each necessary for GSC maintenance, and that HP1 is sufficient for GSC self-renewal in certain contexts in the Drosophila testis.
We have examined the effects of mutations or RNAi knock down of HP1 or Su(var)3-9 on Drosophila male GSCs, and have found in each case there was loss of GSCs, suggesting that HP1 and Su(var)3-9 are each essential for GSC maintenance. Loss of GSC could result from defects in cell division, self-renewal, or survival. The phenotypes of mutant GSCs indicate that HP1 might be required for all the processes, whereas Su(var)3-9 seems to be required for GSC cell division and self-renewal, but not survival. This is not surprising since HP1 and Su(var)3-9 are required for maintenance of constitutive heterochromatin, which is essential for chromosomal compaction of genome stability7,8. A role of HP1 and Su(var)3-9 in GSC self-renewal is supported by the finding that knocking down HP1 or Su(var)3-9 in GSC causes premature expression of the differentiation marker bam (Fig. 3). More importantly, we found that over-expression of HP1 promotes GSC proliferation and is sufficient for restoring GSC to mutant flies that lack GCSs. This property of HP1 is reminiscent of planarian HP1, which is induced upon injury to promote regenerative proliferation of adult stem cells12.
Previous work has established that JAK/STAT signaling from hub cells is essential for maintenance of germline and somatic stem cells15,16,17,18. In somatic tissues, we have previously shown that JAK overactivation counteracts the functions of HP1 and Su(var)3-9 and reduces heterochromatin to promote tumorigenesis29. These findings appear paradoxical, as in GSCs JAK activation and HP1 both positively regulate proliferation. We suggest that GSCs might respond to JAK/STAT activation differently from somatic cells due to the presence of different levels of regulatory components. For instance, it has been shown that the JAK/STAT signaling inhibitor, Suppressor of Cytokine Signaling 36E (Socs36E) is required in somatic but not germline stem cells for self-renewal30. A possibility is that GSCs lack inhibitory molecules such as Socs36E and thus allows overproduction of STAT92E due to autoregulation31, leading to high levels of unphosphorylated STAT92E, which we have previously shown to promote heterochromatin formation31,32. In this scenario, HP1 and heterochromatin are downstream targets of JAK/STAT signaling. Consistent with this idea, we have found that HP1 overexpression can substitute for JAK signaling in GSC generation or maintenance and restore fertility to sterile hop mutant males. The precise mechanisms by which JAK signaling regulates heterochromatin for GSC maintenance await further investigation.
Since HP1 and Su(var)3-9 are central components of heterochromatin, which plays global roles in cellular functions including gene repression and chromosomal compaction, it is possible that the observed effects of HP1 or Su(var)3-9 on GSCs are indirect. Thus mutations in HP1 or Su(var)3-9 may affect expression of a large number of genes, indirectly causing derepression of differentiation genes, such as bam. While how bam is repressed in GSCs requires further investigation, we think it is plausible that heterochromatin formation contributes to repression of differentiation genes in GSCs. This idea is consistent with the finding that overexpression of HP1, which promotes heterochromatin formation, increases GSC number (this study and12) and restores GSC to mutant flies that lack niche signals.
Genes important for heterochromatin formation or epigenetic repression have been implicated in stem cell maintenance. It has previously been shown that another H3K9 specific methylatransferase SETDB1, and its Drosophila homolog Eggless (Egg or dSETDB1), is essential for maintaining self-renewal of embryonic stem cells in mice and of adult germline stem cells in Drosophila, respectively9,10. In line with our findings, it has been shown that planarian HP1 promotes self-renewal and triggers regenerative proliferation of adult stem cells upon injury12. In addition, it has been shown that during Drosophila oogenesis, the DNA-associated protein Stonewall (Stwl) is required for GSC maintenance possibly by heterochromatin-mediated epigenetic repression of differentiation genes33, and that constitutive DNA methylation, another epigenetic gene repression mechanism often associated with heterochromatin formation, is essential for mouse hematopoietic stem cell self-renewal34. Taken together, these reports suggest that global epigenetic gene repression such as heterochromatin formation might be a conserved mechanism for stem cell self-renewal.
On the other hand, the Polycomb Group (PcG) complexes are also important for epigenetic gene repression and chromosomal compaction, and components of PcG complexes have been in stem cell biology as well35,36. Studies have suggested that different PcG proteins may have specific functions in different types of stem cells. One of the major repressive marks in mammalian embryonic stem cells, H3K27 methylation, is catalyzed by a methyl transferase, EZH2 of Polycomb Repressive Complex 2 (PRC2), with the facility of other PRC2 components including EED, SUZ12 and RbAp46/4837,38. Recent studies have identified crosstalk between H3K9 and K27 methylation: the central components of PRC2, EZH2 and SUZ12, are required for HP1α stability; and binding of HP1α/β/γ to H3K9me3 is greatly enhanced in presence of H3K27me3, indicating a highly interactive relationship between heterochromatin components and polycomb complex39. Further exploration of this interaction in pluripotent stem cells, known to depend on chromatin based silencing of developmental gene expression, will be informative for understanding epigenetic mechanisms of stem cell maintenance.
Methods
Fly stocks and Genetics
All crosses were carried out at 25 °C on standard cornmeal/agar medium unless otherwise specified. Fly stocks of hop25, Su(var)20505, Su(var)20504, Su(var)3-92, nanos-Gal4, and hsp70-flp; Act5C > y+ > Gal4 UAS-GFP/CyO, and FRT82B, ubq-GFP were from the Bloomington Drosophila Stock Center (Bloomington, IN). RNAi lines of UAS-HP1 RNAi, and UAS-Su(var)3-9 RNAi were from Vienna Drosophila RNAi Center (VDRC; Vienna,Austria). Fly stocks of esg-GFP (YB0232; L. Cooley), Su(var)3-96 and Su(var)3-917 (Gary Karpen), and bam-GFP (S. DiNardo) were generous gifts. UAS-HP1 was constructed by inserting a Drosophila HP1 cDNA into Drosophila transformation vector pUAST. Standard techniques were used to obtain transgenic flies.
To express UAS-Gene in random clones using the “Flp-out” method40, hsp70-flp; Actin > y+ > Gal4 UAS-GFP flies were crossed to UAS-transgene flies, and the progeny were heat-shocked for 1 h at 37 °C. To generate GFP-marked Su(var)3-9 loss-of-function clones by the FLP/FRT-mediated methods, we crossed hsp70-flp; FRT82B Su(var)3-92/TM3 females to males of hsp70-flp; FRT82B ubiq-GFP to produce “twin-spot” clones41. The progeny were heat-shocked at 37 °C for 2 hrs at indicated developmental stages and examined at indicated times after clone induction.
Immunofluorescence and Western blotting
Mouse monoclonal anti-HP1 (C1A9; 1:50), anti-FasIII (7G10; 1:200; for hub cells), anti-α-spectrin (3A9; 1:10; for fusome) were from Developmental Studies Hybridoma Bank (University of Iowa). Rabbit antibodies against histone H3 (1:1000), H3K4me3 (1:500), and H3K9me3 (1:250) were from Upstate Biotechnology. These antibodies and rabbit anti-Vasa (1:1000; generous gifts from Ruth Lehmann) were used as primary antibodies and fluorescent (Molecular Probes) or HRP conjugated secondary antibodies were used in whole-mount immunostaining or Western blotting, respectively. Tissues were fixed in 4% paraformaldehyde and 0.3% Triton-X. Stained tissues were photographed with a Leica confocal microscope or a Zeiss epifluorescence microscope. Images were cropped and minimally processed with Adobe Photoshop.
Quantitative Real-Time PCR
Testes were dissected from individual 2-day old males of appropriate genotypes and were severed into tip and body regions in ice-cold Schneider medium. Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. The first strand complementary DNA (cDNA) was generated from 1 pair of testis-equivalent of purified total RNA using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)12-18. The cDNA was used as template for qPCR analysis using SYBR green based detection on a BioRad iCycler. Reactions were carried out in triplicate, and melting curves were examined to ensure single products. Results were quantified using the “delta-delta Ct” method to normalize to rp49 transcript levels and to control genotypes. Data shown are averages and standard deviations from at least three independent experiments. The following primer pairs were used.
rp49: TCCTACCAGCTTCAAGATGAC, CACGTTGTGCACCAGGAACT
bam: CCAATCGCGCAGACCAATTAGCAA, CGAGTGTGACAAGTTGCTTAAGGG
Vasa: CCCAAATGAACATAGGAGCGATCC, TTTCATCCGCATCAGCTGGTACCA
esg: TACCCATCATCACCATGCGCCTAT, TCCCGGCTGGCTAGTGTTTAGATT
Ubiq: CGTTCTCAATGGTATCGGATGGCT, CACTCTGTCCGACTACAACATCCA
Additional Information
How to cite this article: Xing, Y. and Li, W. X. Heterochromatin components in germline stem cell maintenance. Sci. Rep. 5, 17463; doi: 10.1038/srep17463 (2015).
Supplementary Material
Acknowledgments
We thank Kimberly Larson for excellent technical support. We thank Drs. S. DiNardo, Y. Yamashita, L. Cooley, R. Lehmann, and D. Bohmann for various Drosophila strains and reagents; the Developmental Hybridoma Bank (Iowa), the Vienna Drosophila RNAi Center, and the Bloomington Drosophila Stock Center for antibodies and fly stocks. This study was supported, in part, by grants from the National Institutes of Health and a Leukemia & Lymphoma Society Research Scholar Grant to W.X.L.
Footnotes
References
- Fuller M.T. & Spradling A.C. Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402–4 (2007). [DOI] [PubMed] [Google Scholar]
- Morrison S.J. & Spradling A.C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M. & Fisher A.G. Epigenetic signatures of stem-cell identity. Nat Rev Genet 8, 263–71 (2007). [DOI] [PubMed] [Google Scholar]
- Pietersen A.M. & van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 20, 201–7 (2008). [DOI] [PubMed] [Google Scholar]
- Lee T.I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A. et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–53 (2006). [DOI] [PubMed] [Google Scholar]
- Grewal S.I. & Elgin S.C. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12, 178–87 (2002). [DOI] [PubMed] [Google Scholar]
- Grewal S.I. & Jia S. Heterochromatin revisited. Nat Rev Genet 8, 35–46 (2007). [DOI] [PubMed] [Google Scholar]
- Bilodeau S., Kagey M.H., Frampton G.M., Rahl P.B. & Young R.A. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23, 2484–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Histone H3K9 trimethylase Eggless controls germline stem cell maintenance and differentiation. PLoS Genet 7, e1002426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. et al. dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS ONE 3, e2234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A. et al. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J Cell Biol 201, 409–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. The stem-cell niche theory: lessons from flies. Nat Rev Genet 3, 931–40 (2002). [DOI] [PubMed] [Google Scholar]
- Wallenfang M.R., Nayak R. & DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell 5, 297–304 (2006). [DOI] [PubMed] [Google Scholar]
- Kiger A.A., Jones D.L., Schulz C., Rogers M.B. & Fuller M.T. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–5. (2001). [DOI] [PubMed] [Google Scholar]
- Tulina N. & Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK- STAT signaling. Science 294, 2546–9. (2001). [DOI] [PubMed] [Google Scholar]
- Leatherman J.L. & Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44–54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman J.L. & Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol 12, 806–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A. et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18, 2973–83 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P., Matunis E. & DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124, 4361–71 (1997). [DOI] [PubMed] [Google Scholar]
- Schulz C. et al. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 167, 707–23 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A., Bernasconi L., Sergeev P., Cruz A. & Steinmann-Zwicky M. mgm 1, the earliest sex-specific germline marker in Drosophila, reflects expression of the gene esg in male stem cells. Int J Dev Biol 46, 159–66 (2002). [PubMed] [Google Scholar]
- Voog J. et al. Escargot restricts niche cell to stem cell conversion in the Drosophila testis. Cell Rep 7, 722–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C.A. & Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–9 (2006). [DOI] [PubMed] [Google Scholar]
- Ohlstein B. & Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–4 (2006). [DOI] [PubMed] [Google Scholar]
- Voog J., D’Alterio C. & Jones D.L. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 454, 1132–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran J., Brenner T.J. & DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature 407, 754–7 (2000). [DOI] [PubMed] [Google Scholar]
- Boyle M., Wong C., Rocha M. & Jones D.L. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1, 470–8 (2007). [DOI] [PubMed] [Google Scholar]
- Shi S. et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet 38, 1071–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis M. et al. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 326, 153–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol 18, 545–51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10, 489–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J.Z., Park J.K., Williams M. & McKearin D.M. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development 134, 1471–9 (2007). [DOI] [PubMed] [Google Scholar]
- Broske A.M. et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41, 1207–15 (2009). [DOI] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B. & Di Croce L. Polycomb complexes in stem cells and embryonic development. Development 140, 2525–34 (2013). [DOI] [PubMed] [Google Scholar]
- Richly H., Aloia L. & Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis 2, e204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu G.H. & Izpisua Belmonte J.C. Navigating the epigenetic landscape of pluripotent stem cells. Nat Rev Mol Cell Biol 13, 524–35 (2012). [DOI] [PubMed] [Google Scholar]
- Margueron R. & Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros J., Arnoult N., Stroobant V., Collet J.F. & Decottignies A. Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1alpha at chromatin. Mol Cell Biol 34, 3662–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y. & Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–71 (1997). [DOI] [PubMed] [Google Scholar]
- Xu T. & Rubin G.M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–37 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.