Abstract
Background
Genetic diversity among rice cultivars from Bangladesh and North East India was assessed using a custom 384-SNP microarray assay. A total of 511 cultivars were obtained from several sources, choosing landraces likely to be from the aus subpopulation and modern improved cultivars from Bangladesh. Cultivars from the OryzaSNP set and Rice Diversity Panel 1 (RDP1) were also included for reference.
Results
The population analysis program STRUCTURE was used to infer putative population groups in the panel, revealing four groups: indica (76 cultivars), japonica (55) and two distinct groups within the aus subpopulation (aus-1 = 99, aus-2 = 151). Principal Component Analysis was used to confirm the four population groups identified by STRUCTURE. The analysis revealed cultivars that belonged to neither aus-1 nor aus-2 but which are clearly aus based on the combined probabilities of their membership of the two aus groups which have been termed aus-admix (96). Information obtained from the panel of 511 cultivars was used to assign rice groups to 74 additional landraces obtained from Assam and West Bengal. While both the aus-1 and aus-2 groups were represented approximately equally in India, aus-2 (which includes cultivar N 22) was more common in Bangladesh, but was not found at all in West Bengal.
Conclusions
Examining the distribution of landrace names within theaus-1 and aus-2 groups suggests that aus-1 is associated with the term “boro”, a word used to describe a winter growing season in Bangladesh and Assam. The information described here has been used to select a population of 300 cultivars for Genome Wide Association studies of the aus rice subpopulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-015-0068-z) contains supplementary material, which is available to authorized users.
Keywords: Rice, SNP, Aus, Boro, Genotype, Ecotype
Background
Rice (Oryza sativa L.) has been cultivated as a crop for at least 8000 years (Maclean et al.,2002) and currently 50 % of the world’s population is dependent on rice as their staple diet. Within rice germplasm there is considerable genetic diversity that reflects its domestication and long breeding history and, most notably, two subspecies of rice are recognised: indica and japonica. There are a number of different theories about the domestication of rice. Molina et al. (2011) present evidence that rice was domesticated from Oryza rufipogon at a single point in time while in the review by Khush (1997) independent origins of domestications for japonica (in China) and indica (in North East India) are suggested. Recently, Huang et al. (2012) argue that japonica rice was domesticated from a population of O. rufipogon in southern China, but that the indica sub-species originated from a crossing of japonica and wild rice as rice cultivation spread into South and South East Asia.
The first widely adopted molecular classification of rice groups was based on isozyme analysis (Glaszmann, 1987), in which six varietal groups were identified. These are usually referred to as: I = indica, II = aus, III = ashina, IV = rayada, V = aromatic, VI = japonica (from Glaszmann, 1987 as modified by, for example, Wang et al.,2013). Subsequent DNA analysis has identified five subpopulation of rice within the two Oryza sub-species. The aus subpopulation was identified within indica, and the temperate, tropical and aromatic subpopulations were identified within japonica (Garris et al.,2005). These five subpopulations are now commonly used to describe rice cultivars.
Glaszmann’s group II (aus), which was originally considered to be exclusive to South and West Asia, was described as having a short life cycle and grown under a range of conditions from fully irrigated to upland (Glaszmann, 1987). This work suggested that group II rice encompasses the aus ecotype but also includes some boro ecotype rice. Normally, the term ‘boro’ refers to a growing season in Bangladesh and Assam during December-May (GRiSP, 2013) and also to genetically diverse cultivars grown during this season (Parsons et al.,1999).”aus/ahu” refers to a growing season in Bangladesh and Assam during April-August (GRiSP, 2013) and also to the rice cultivars grown during this season, which are broadcast sown. These aus/ahu rice cultivars are insensitive to photoperiod and are drought tolerant (Khush, 1997). The geographical term aus and the genetic term aus can cause confusion because there is considerable overlap between boro and aus cultivars at the genetic level, which means the geographical distinction between them is blurred. Most importantly, cultivars grown during the boro or aus seasons may not all be genetically ‘aus’ cultivars. The recently announced 3,000 rice genomes project (Li et al. 2014) refers to a group of aus/boro genotypes, which is the genetic group normally referred to as aus. A total of 208 accessions out of the 3,000 rice genomes are classified as aus/boro, based on 200,000 SNP markers. A subsequent phylogenetic analysis of all 3,000 genome sequences using 376,000 SNP markers also revealed a genetically related group called aus (Alexandrov et al.,2015). In this paper, we refer to genotypes from this group as ‘aus’ using italics to refer to genetic terms and ‘aus’ in normal font for similar geographic or ecotype terms.
A recent analysis of 409 rice cultivars indicated that lines from the aus group originate predominantly from areas in Bangladesh and India (Ali et al.,2011). The genetic diversity of the aus and boro ecotypes is large and includes a number of cultivars known for their adaptation to different environments. Cultivar FR 13A, for example, is the flood tolerant donor of the submergence tolerance gene Sub1 (Xu et al.,2006); Kasalath is the efficient phosphorus uptake donor of the phosphorus starvation tolerance gene Pstol1 (Gamuyao et al.,2012); Dular is a rice cultivar that has increased drought resistance, associated with greater root length and root density (Henry et al.,2011); Rayada also has a large root length and high root density (Henry et al.,2011); Black Gora is a rice cultivar with high seedling vigour (Redoña and Mackill, 1996) and deep roots (Shrestha et al.,2014; Al-Shugeairy et al.,2014) and; N 22 is a heat tolerant rice cultivar (Jagadish et al.,2008).
A number of previous studies have examined the genetic diversity of rice cultivars from Bangladesh (Parsons et al.,1999; Sajib et al.,2012; Hassan et al.,2012). However these studies involved relatively small numbers of genetic markers. Studies of global populations of rice (Ali et al.,2011) using a larger number of markers to capture global rice genetic diversity are limited by the relatively small number of cultivars obtained from specific geographic regions. The aim of this study is to examine the genetic variation of about 500 landraces from the aus and boro ecotypes of rice that fall within the aus genetic group of rice based on 384 SNP markers and also use the information obtained to identify the genetic background of field-grown cultivars collected from farmers in West Bengal and Assam in India. The cultivars investigated here will be used to establish a panel of aus cultivars for a subsequent genome wide association (GWA) mapping study within a genetically and geographically distinct group of rice genotypes.
Results
The design of the SNP chip was intended to achieve an approximately even spread of markers across the entire rice genome. Before the analysis, data for 58 of the 384 SNPs were removed because they displayed a high degree of heterozygosity on some microarray plates but not on others and were therefore considered to be unreliable. This resulted in a total of 326 markers included in the analysis which remain approximately evenly placed across the rice genome although some gaps are apparent, for example at the middle of chromosomes 4 and 7, and also at the top of chromosome 10 (Fig. 1).
Fig. 1.
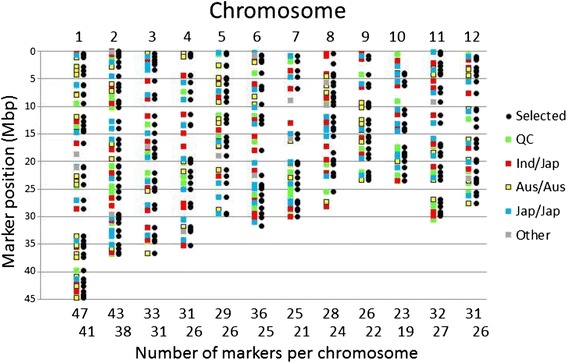
Physical position of the 384 SNPs according to the category of their predicted polymorphic discrimination. Based on data from the Rice Diversity website, where QC is the “Quality Control” set which can “Assign accessions into one of five O. sativa subgroups” and at the bottom of each chromosome is the number of markers on that chromosome (Selected SNPs, to the right, are the 326 SNP remaining for genetic analysis after filtering of poor quality SNP-calls on any one plate used in this study)
The STRUCTURE population analysis of Panel A (511 cultivars) indicated that there were 4 distinct population groups present using the Evano Delta-K method as shown in Additional file 1: Figure S1 (Online Resource 4). The group memberships for each cultivar in Panel A are summarised in Additional file 2: Figure S2 (Online Resource 5). Based on a priori knowledge of the rice subpopulations that the exemplar cultivars belonged to shown in Additional file 3: Table S1 (Online Resource 1) one of the STRUCTURE groups was determined to be japonica (jap) and another to be indica (ind). However, the two remaining groups both included cultivars that, in previous studies, have been shown to belong to the aus subpopulation and were therefore designated aus-1 and aus-2. Cultivars were allocated to four groups: ind (indica), jap (japonica), aus-1 and aus-2. Some of the remaining cultivars were clearly from the aus subpopulation based on the sum of their probabilities of membership to the two aus groups and these have been classified as aus-admix. All other cultivars are considered as admix. The largest number of cultivars in Panel A (OryzaSNP cultivars excluded) belong to aus-2 (150 cultivars) with the japonica group having only 50 cultivars (Fig. 2a).
Fig. 2.
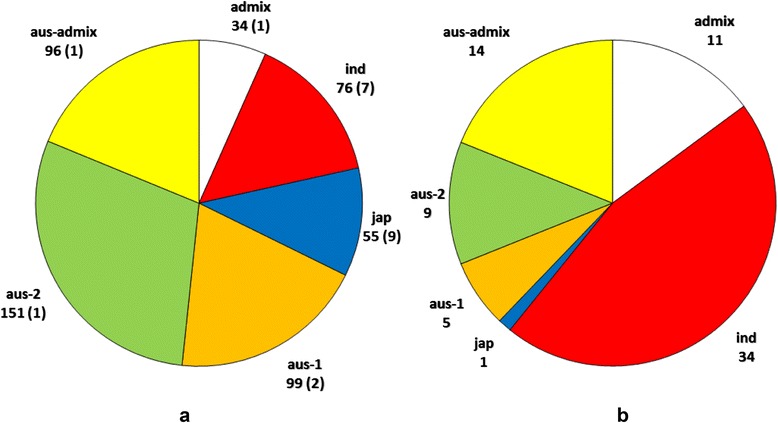
Distribution of cultivars in (a) Panel A (511 cultivars) and (b) Panel B (74 cultivars). The cultivars in Panel b were obtained from Assam Agricultural University and Calcutta University. Cultivars were assigned to rice groups on the basis of exemplars with > = 80 % probability using STRUCTURE (white = admix, red = indica, blue = japonica, orange = aus-1, green = aus-2, yellow = aus-admix; Numbers in brackets = OryzaSNP cultivars)
Using PCA it was possible to separate out the 511 cultivars in Panel A into groups that support the STRUCTURE analysis (Fig. 3). Both PCA axis 1 and PCA axis 2 separate the japonica, indica and aus subpopulations (Fig. 3a). It was also possible to separate out the aus cultivars into the two groups when PCA 1 was plotted against PCA 3 (Fig. 3b). The upper group in this PCA plot was arbitrarily named “aus-1” (orange in Fig. 3) and the lower one “aus-2” (green in Fig. 2). There are 96 cultivars that lie directly between the aus-1 and aus-2 groups in Fig. 3b which have been classified as aus-admix. These do not belong exclusively to either the aus-1 or aus-2 group based on a threshold of 80 % probability of their group membership in the STRUCTURE analysis, but the combined probabilities for their aus-1 and aus-2 group memberships exceeds the 80 % threshold, indicating that these are aus cultivars. The observation that four distinct groups of cultivars were inferred by STRUCTURE is supported by the NJ tree produced from a multi-FASTA alignment of the Panel A SNP data (Fig. 4). The indica and japonica groups are distinct while the aus-1 and aus-2 groups broadly separate from each other. A “nexml” format file of the NJ tree shown in Fig. 4 that can be visualised using Dendroscope is provided as Online Resource 6.
Fig. 3.
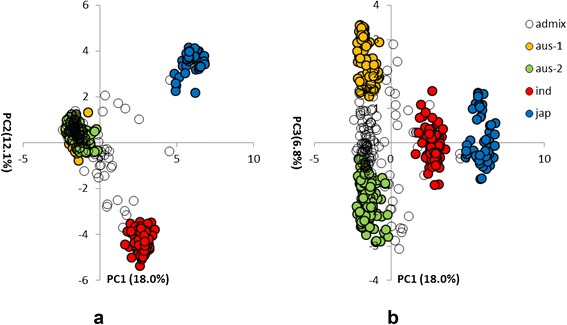
Principal Component Analysis of 511 cultivars in Panel A. (a ) PC1/PC2 showing three clusters corresponding to the main rice groups indica, japonica, aus; (b) PC1/PC3 showing sub-groups within the aus cluster. Cultivars were assigned to rice groups on the basis of exemplars with > = 80 % probability using STRUCTURE (red = indica, blue = japonica, orange = aus-1, green = aus-2, white = admix (either aus-admix or admix)
Fig. 4.
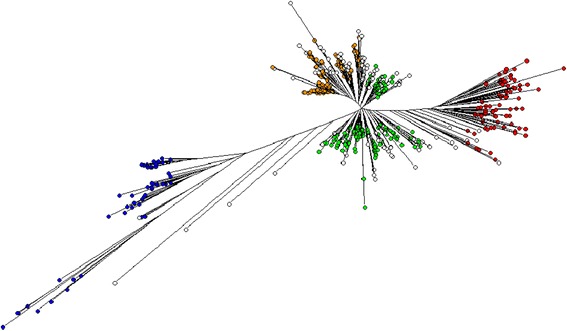
Neighbour-Joining tree (50 % consensus of 100 bootstraps) of 511 cultivars in Panel A. Cultivars were classified into rice groups based on > = 80 % probability of STRUCTURE group membership: Japonica (blue), indica (red), aus group1 (orange), aus group2 (green), aus-admix and admix (white) with < 80 % probability of belonging to a single designated STRUCTURE group. A nexml file of this NJ tree that can be visualised in Dendroscope is provided as Additional file 6
Cultivars definitively belonging to one of the four identified rice groups (indica, japonica, aus-2 or aus-1) at 80 % probability were selected to create a STRUCTURE ‘training’ set of cultivars with pre-defined populations that allowed an assessment to be made about the genetic composition of landraces and improved cultivars obtained from Assam and West Bengal (Panel B). The results are illustrated by the NJ tree shown in Fig. 5. The majority of the West Bengal landraces were identified as indica (23 out of 35) as are the two landraces from both Orissa and Bihar, while four are aus-2, six are aus-admix and three are admix (one a japonica-like and two indica-like admix based on where they cluster in the NJ tree). No West Bengal landraces were identified as either japonica or aus-1. For the Assam landraces: 12 out of 39 were indica, one was japonica, nine were aus-1, one was aus-2, eight were aus-admix and eight were admix (three japonica-like, three aus-like and two indica-like based on their position in the NJ tree).
Fig. 5.
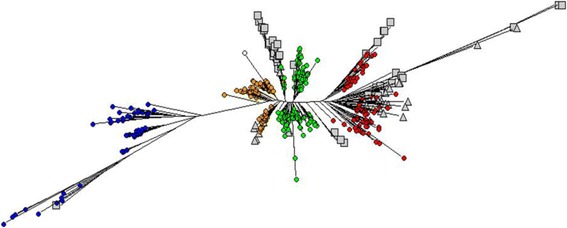
Neighbour-Joining tree (50 % consensus of 100 bootstraps) of 74 cultivars in Panel B + training set. The Assam and West Bengal cultivars in Panel B were classified into rice groups using a training set with ≥ 80 % probability of STRUCTURE group membership selected from all 511 AWD cultivars. Training set: Japonica (blue), indica (red), aus group1 (orange), aus group2 (green), admix (white), Assam (grey circles), West Bengal (grey squares). A nexml file of this NJ tree that can be visualised in Dendroscope is provided as Additional file 7
Discussion
Many molecular methods have been developed to assess genetic diversity and the technology improves at a rapid rate. In the study of rice diversity, the first systematic molecular approach was that of Glaszmann (1987). During the 1990s RFLP (e.g. Wang et al. 1995) and PCR-based methods such as RAPD (e.g. Virk et al.,1996), AFLPs (Virk et al.,2000) and microsatellite markers (Panaud et al.1996) were developed. Since the availability of large amounts of sequencing information, such as that provided by the OryzaSNP project (McNally et al.,2009), highly-multiplexed SNP assay methods have been developed as techniques based on resequencing are beginning to emerge (Huang et al. 2010, Ebana et al. 2010). This study exploits the work led by the activities of the Rice Diversity Project (www.ricediversity.org; Thomson et al. 2012), to develop a targeted 384-SNP BeadXpress assay which is able to differentiate rice cultivars on a genetic basis into the known rice diversity subpopulations. Here, we carefully selected a combination of known SNP-markers to achieve two objectives: First to obtain good classification of rice cultivars into the recognised rice subpopulation, but also to have high resolution within the aus subpopulation of the indica subspecies. This was achieved by having evenly distributed SNPs throughout the genome (Fig. 1), especially SNPs that were predicted to differentiate aus cultivars as revealed in the 44 k SNP data (Zhao et al.,2011). While several of the SNPs could not be used because they were non-polymorphic or provided low quality data, 326 SNPs were available for diversity analysis. The number of SNPs which were polymorphic within indica, japonica and aus accessions (as classified here) was 191, 229 and 142 respectively.
The results clearly indicate that our custom designed SNP assay achieved its primary objective of being able to correctly allocate known rice cultivars to the indica, aus, and japonica subpopulation (note that too few temperate japonicas were included in Panel A to allow subdivision of the japonica subspecies into two groups and, likewise, too few aromatic cultivars were included to allow that group to be recognised). Of all the exemplar cultivars used here with rice subpopulation known a-priori only one was not correctly allocated to its recognised rice group: the OryzaSNP cultivar Minghui 63, which was recognised as an indica, but at less than the 80 % threshold we used and was therefore classified as an admix including 20 % aus-2 group membership. Regarding the secondary objective of the SNP assay design, it is also clear that high resolution has been achieved within the aus subpopulation of Panel A because two distinct groups of aus cultivars were observed. This is the first time, to our knowledge, that a genetic study of rice has revealed such population structure within the aus subpopulation of rice. We speculate that this is due to a good choice of SNP-markers in our array design and because of the large number of accessions selected that were revealed to be aus (345 of the 511 cultivars used here). It is interesting to note that this subdivision of the aus subpopulation was not detected in the 3 K rice genomes project using 20 million SNPs, but only 208 aus cultivars (Alexandrov et al.,2015).
Examining the classification of cultivars into the rice groups presented in Additional file 3: Table S1 (Online Resource 1) indicates that some cultivars that are named “aus” are not genetically aus. Thus 10 cultivars with the term “aus” either at the start or end of their name are indica and two are japonica. This presumably reflects the distinction between the genetic use of the term and the geographic use discussed earlier. Of the 38 cultivars selected from the Assam Rice Collection (these are 6,630 accessions collected between 1965 and 1972 and sent to IRRI (Singh and Singh, 2000)), one was a japonica, two were indica, seven were admix, 14 were aus-2, six were aus-1 while seven were aus-admix. Thus the great majority of cultivars we selected from this collection appear to be aus in nature.
Examining the allocation of aus cultivars into the two aus sub-groups reveals that some well-characterised rice cultivars appear in separate aus sub-groups: The drought and heat tolerant cultivar N 22 (Lenka et al.,2011; Jagadish et al.,2010) appears in aus-2, while FR 13 (the donor of the Sub1 submergence tolerance gene (Xu et al.,2006)), Rayada (a Bangladesh deep water rice used in Garris et al. (2005)) and Kasalath (the donor of Pstol, the phosphorus starvation tolerance gene (Gamuyao et al.,2012)) all appear in aus-1. Other well-characterised aus cultivars are classified as aus-admix in this study, including Black Gora (a deep rooted check cultivar used by Shrestha et al. (2014)), Dular (a donor of drought tolerance) and the recently sequenced aus reference cultivar DJ 123 (Schatz et al.,2014).
The analysis of the 511 cultivars in Panel A provided a training set that revealed an insight into the genetic composition of the 74 improved varieties and landraces from Bangladesh. Most noticeable is the fact that the improved variety BR 16 is an aus-2 cultivar whereas the others (BR3, BR6, BRRI Dhan 28, 29, 45 and 47, BINA Dhan 5, 6, 8 and 50 and Iratom 24) are all indica as might be expected.
It is interesting to note that the landraces which can be allocated to specific regions (276 from Bangladesh, 77 from Assam (including 39 from Panel B and 38 from the Assam Rice Collection) and 31 from West Bengal), are not equally allocated into the rice subpopulations of indica, japonica and aus. The japonicas appear to be absent in cultivars selected from West Bengal (none) and are rare in cultivars from Assam (one), but japonicas are not uncommon in Bangladesh (32/276). Also potentially interesting is the relative rarity of aus-1 in Bangladesh (61 aus-1, 109 aus-2 and 49 aus-admix) and Assam (1 aus-1, 9 aus-2 and 8 aus-admix), and the absence of aus-2 in cultivars from West Bengal (4 aus-1 and 6 aus-admix). Of the 168 landraces from India that are not part of the Assam Rice Collection, aus-1 and aus-2 are approximately equally represented (32 aus-1, 28 aus-2 and 38 aus-admix). This suggests that the aus-1 and aus-2 sub-groups are geographically distinct, and might indicate that aus-1 originates from West Bengal.
A final observation is that the 60 cultivars used here whose name contains the term “boro” (all but 9 from Bangladesh) are predominantly aus-1. Of these “boro”-named landraces, 34 are aus-1, 13 are japonica, six are indica, four are admix, two are aus-admix but only one is aus-2. In fact, more than half of the aus-1 cultivars from Bangladesh have the term “boro” in their names, but they account for less than 16 % of all the aus cultivars from Bangladesh studied here. This strongly suggests that selection of cultivars suitable for the boro growing season has caused a differentiation between the aus-1 and aus-2 sub-groups identified in this study and that aus-1 is the genetic type of aus suited to production during the boro season.
Conclusions
Rice cultivars named by ecotype or growing season are not always of similar genotype and at least two geographically distinct groups of the aus genotype exist within the aus subpopulation of rice.
Methods
Plant material
Two different panels of rice cultivars were used in this study: Panel A was selected to allow the development of a GWA mapping population focused on aus cultivars from Bangladesh and North East India and consisted of 511 cultivars from a number of sources: A total 411 accessions were selected from the IRRI gene-bank. These were selected because they originate (or were collected) from either Bangladesh or India and are considered to be aus cultivars. In addition to these cultivars, 31 aus accessions from Bangladesh or India were selected from the Rice Diversity Panel 1 (Zhao et al.,2011). A total of 27 known aus accessions originating from Bangladesh or India were also obtained from the USDA core rice collection (Yan et al.,2010). A further 41 cultivars were collected by Bangladesh Agricultural University from sources in Bangladesh, including both landraces and improved cultivars. In addition to these cultivars, all of which originate from the North East of India or Bangladesh, Panel A also included 19 accessions from the OryzaSNP panel (McNally et al.,2009), which were selected to represent the wider genetic diversity of rice. Panel B was selected to provide information about the genetic diversity of a further 74 landraces from the same region: 35 cultivars were collected by Calcutta University from West Bengal (31), Bihar (2) and Orissa (2) in India; 39 cultivars were collected by Assam Agricultural University from Assam in India. A full list of the rice cultivars selected for Panel A, including the germplasm source and country of origin or collection, is presented in Additional file 3: Table S1 (Online Resource 1); A full list of the cultivars selected for Panel B is presented in Additional file 4: Table S2 (Online Resource 2).
DNA extraction
The 511 cultivars selected for Panel A were grown from seed under controlled conditions and DNA was then extracted from fresh leaf tissue using the DNeasy Plant kit (Qiagen). DNA from the 74 cultivars from Assam and West Bengal selected for Panel B was extracted from dried leaf tissue using a modification of the method described by Deshmukh et al. (2007) to include a genomic DNA precipitation step.
Genotyping
A 384 SNP Illumina GoldenGate array was designed by combining SNP probes selected from data available at the Rice Diversity website (http://ricediversity.org/data/): 83 probes were selected from RiceOPA1.0 (Quality Control), 100 from RiceOPA2.1 (indica/indica), 9 from RiceOPA3.1 (indica/japonica), 74 from RiceOPA4.0 (japonica/japonica), 9 from RiceOPA5.0 (indica/O. rufipogon), 4 from RiceOPA6.0 (japonica/O. rufipogon) and 105 from RiceOPA7.0 (indica/japonica). The aim of combining these probes was to produce a probe-set with a mixture of abilities to detect polymorphism between widely different accessions such as the indica and japonica sub-species, and to differentiate within more closely related groups such as the aus or japonica accessions. Each SNP probe was classified into three main groups; those that should discriminate between indicas and japonicas, within japonicas, or within aus. The SNP array was also designed so that there was an approximately even spread of markers of different categories across each genome (Fig. 1). Additional file 5: Table S3 (Online Resource 3) describes the classification of all probes used with their flanking sequences, SNP and physical position. Genomic DNA from each of the cultivars was extracted and 500 ng of each sample placed in 96 well plates used for the 384 SNP Illumina GoldenGate oligo pool assay (Illumina Inc.) using the BeadXpress platform, according to the manufacturer's protocol. Allele calls were performed using the “GenTrain” clustering algorithm available in Genome Studio v2011.1 (Illumina Inc.). Each SNP-call was checked manually in Genome Studio for quality and accuracy because rice is predominantly inbred, but the SNP detection algorithm used by Genome Studio was originally developed for out-breeders and by default searches for clusters of three SNP markers (two homozygotes and one heterozygote). Any allele calls below a threshold of 0.02 NormR were discarded.
Data analysis
Poor quality SNP markers were excluded from the analysis by removing any markers where there was statistical evidence that plates varied in the amount of heterozygosity detected. This removed a total of 58 SNP markers as shown in Additional file 5: Table S3 (Online Resource 3). The MCMC (Markov Chain Monte Carlo) population analysis program “STRUCTURE” (Pritchard et al.,2000; Falush et al.,2003) was used to infer underlying population structure in the SNP data using an initial burn-in of 5,000 iterations, followed by a run length of 50,000 iterations. The population structure in the data was investigated using putative population ‘K’ values ranging from 2-10 (10 replicates per K value) with a STRUCTURE model including admixture and correlated allele frequencies. STRUCTURE Harvester (Earl and VonHoldt 2012) was used to establish an optimum K value from the results of the exploratory STRUCTURE analysis using the Evanno “Delta-K” method (Evanno et al.,2005). An optimum K value of 4 groups was established for Panel A (511 cultivars) as shown in Additional file 1: Figure S1 (Online Resource 4). The SNP data was then re-analysed using STRUCTURE with a putative K value of 4 groups, burn-in of 10,000 iterations and run length of 100,000 iterations to generate a STRUCTURE ‘Q’ matrix. Major modes in the STRUCTURE output were identified using CLUMPAK (Kopelman et al.,2015) and are presented in Additional file 2: Figure S2a (Online Resource 5).
Rice subpopulations (indica, japonica or aus) were assigned to each of the four groups identified by STRUCTURE, based on a number of ‘exemplar’ cultivars from the OryzaSNP set and Rice Diversity Panel 1 for which the rice group was known a-priori as presented in Additional file 3: Table S1 (Online Resource 1). Two of the groups identified by STRUCTURE contained exemplars from the aus rice group and were arbitrarily named aus-1 and aus-2. Cultivars were only assigned to a group if the probability of their group membership determined by STRUCTURE was ≥ 80 %. Cultivars with < 80 % probability of a single group membership were classified as ‘admix’ unless their combined aus group memberships (aus-1 + aus-2) was ≥ 80 %. These cultivars were classified as aus-admix. The quantitative genetics program TASSEL (Bradbury et al., 2007) was used to perform Principal Component Analysis (PCA) of the SNP data, after filtering and imputation of missing SNPs using default parameters. The TreeBest program from “TreeFam” (Ruan et al.,2008) was used to obtain a 50 % consensus tree bootstrapped from 100 NJ (Neighbour-Joining) trees based on multi-FASTA alignments of SNPs. The NJ Trees were plotted using Dendroscope (Huson et al.,2007). The groups identified by STRUCTURE were used to assign coloured labels to groups on the PCA plots (Fig. 2) and NJ trees (Figs. 3 and 4). The Dendroscope nexml format files used to plot the NJ trees presented in Figs. 4 and 5 are provided as Additional file 6 and 7 in the supplementary material.
Prior population information about the exemplar cultivars was not used in the STRUCTURE model used to analyse Panel A (511 cultivars). However, cultivars with ≥ 80 % probability of single group membership identified in the analysis were used to create a training-set for analysis of Panel B (74 cultivars) with greater genetic diversity than the relatively small number of exemplar cultivars included in analysis of Panel A. Prior population information about the training set was included in the STRUCTURE model used to analyse Panel B.
Acknowledgements
This work was funded by BBSRC research project BB/J00336/1. FS and a part of the proportion of the cost of the Illumina genotyping was funded by a Beachell-Borlag International Fellowship. The authors would like to acknowledge the help of Dr MK Sarmah in collecting seed samples of the landraces and improved cultivars from Assam used in this study and Dr. Ma. Elizabeth B. Naredo and Ms. Sheila Mae Q. Mercado for handling of IRGC accessions and preparation of DNAs for genotyping. All rice seeds used here were obtained with MTA agreements and seed and dry leaves imported into the UK under import licence IMP⁄SOIL⁄18⁄2009 issued by Science and Advice for Scottish Agriculture.
Additional files
Optimum K value of 4 groups established for Panel A (511 cultivars). Results obtained from STRUCTURE were analysed by the Evanno ‘Delta-K’ method using STRUCTURE Harvester. (DOCX 59 kb)
Major modes detected by STRUCTURE. Modes for cultivars in (a) Panel A (511 AWD cultivars) and (b) Panel B (74 Indian cultivars) for K = 4 from the Evano method were obtained using CLUMPAK with search method LargeKGreedy, MCL cluster size threshold = 0.1 and cut-off = 0.50. (DOCX 62 kb)
Accessions of panel A with STRUCTURE group membership and PCA results. (XLSX 97 kb)
Accessions of Panel B with STRUCTURE group membership. (XLSX 18 kb)
SNPs in the 384 Illumina GoldenGate array (from Rice Diversity website http://ricediversity.org/). (XLSX 54 kb)
Dendroscope nexml format file used to plot the Neighbour-Joining tree shown in Fig 4. (NEXML 453 kb)
Dendroscope nexml format file used to plot the Neighbour-Joining tree shown in Fig 5. (NEXML 400 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Project conception (AHP, GJN, MRI). Microarray design (AHP, GJN, KLM, PH, FS). Seed collection (RS, TD, KLM). DNA extraction (GJN, SD). Rice cultivation from seed and sample collection (MH, MRI, RS). Microarray SNP assay (MM, PEH). Bioinformatics (AJT, RS). Data interpretation (AJT, GJN, AHP). Manuscript preparation (AJT, GJN, AHP, MRI). All authors read and approved the final manuscript.
References
- Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–7. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali ML, McClung AM, Jia MH, Kimball JA, McCouch SR, Eizenga GC. A rice diversity panel evaluated for genetic and agro-morphological diversity between subpopulations and its geographic distribution. Crop Sci. 2011;51:2021–2035. doi: 10.2135/cropsci2010.06.0359. [DOI] [Google Scholar]
- Al-Shugeairy Z, Islam MS, Shrestha R, Al-Ogaidi F, Norton GJ, Price AH. High throughput screening of rooting depth in rice using buried herbicide. Ann Appl Biol. 2014;165:96–107. doi: 10.1111/aab.12118. [DOI] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Deshmukh VP, Thakare PV, Chaudhari US, Gawande PA. A simple method for isolation of genomic DNA from fresh and dry leaves of Terminalia arjuna (Roxb.) Wight and Argot. Electron J Biotechnol. 2007;10:468–472. doi: 10.2225/vol10-issue3-fulltext-5. [DOI] [Google Scholar]
- Earl DA, VonHoldt BM. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Ebana K, Yonemaru J, Fukuoka S, Iwata H, Kanamori H, Namiki N, Nagasaki H, Yano M. Genetic structure revealed by a whole-genome single-nucleotide polymorphism survey of diverse accessions of cultivated Asian rice (Oryza sativa L.) Breed Sci. 2010;60:390–397. doi: 10.1270/jsbbs.60.390. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature. 2012;488:535–539. doi: 10.1038/nature11346. [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- GRiSP (Global Rice Science Partnership) Rice almanac. 4. Los Baños: International Rice Research Institute; 2013. p. 121. [Google Scholar]
- Hassan MM, Shamsuddin AKM, Islam MM, Khatun K, Halder J. Analysis of Genetic Diversity and Population Structure of Some Bangladeshi Rice Landraces and HYV. J Sci Res. 2012;4:757–767. doi: 10.3329/jsr.v4i3.10416. [DOI] [Google Scholar]
- Henry A, Gowda VRP, Torres RO, McNally KL, Serraj R. Variation in root system architecture and drought response in rice (Oryza sativa): Phenotyping of the OryzaSNP panel in rainfed lowland fields. F Crop Res. 2011;120:205–214. doi: 10.1016/j.fcr.2010.10.003. [DOI] [Google Scholar]
- Huang X, Xinghua W, Tao S, Qiang Z, Qi F, Yan Z, Canyang L, et al. Genome-Wide Association Studies of 14 Agronomic Traits in Rice Landraces. Nature Genetics. 2010;42(11):961–67. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish SVK, Craufurd PQ, Wheeler TR. Phenotyping parents of mapping populations of rice for heat tolerance during anthesis. Crop Sci. 2008;48:1140–1146. doi: 10.2135/cropsci2007.10.0559. [DOI] [Google Scholar]
- Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.) J Exp Bot. 2010;61:143–156. doi: 10.1093/jxb/erp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 1997;35:25–34. doi: 10.1023/A:1005810616885. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA and Mayrose I (2015) Clumpak : a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour n/a–n/a. doi: 10.1111/1755-0998.12387 [DOI] [PMC free article] [PubMed]
- Lenka SK, Katiyar A, Chinnusamy V, Bansal KC. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J. 2011;9:315–327. doi: 10.1111/j.1467-7652.2010.00560.x. [DOI] [PubMed] [Google Scholar]
- Li J-Y, Wang J, Zeigler RS. The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience. 2014;3:8. doi: 10.1186/2047-217X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean J, Dawe D, Hardy B, Hettel G. Rice Almanac. Third. Wallingford: CABI Publishing; 2002. [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci U S A. 2011;108:8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons BJ, Newbury HJ, Jackson MT, Ford-Lloyd BV. The genetic structure and conservation of aus, aman and boro rices from Bangladesh. Genet Resour Crop Evol. 1999;46:587–598. doi: 10.1023/A:1008749532171. [DOI] [Google Scholar]
- Panaud O, Chen X, McCouch SR. Development of Microsatellite Markers and Characterization of Simple Sequence Length Polymorphism (SSLP) in Rice (Oryza Sativa L.) MGG Molecular & General Genetics. 1996;252(5):597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redoña ED, Mackill DJ. Genetic variation for seedling vigor traits in rice. Crop Sci. 1996;36:285–290. doi: 10.2135/cropsci1996.0011183X003600020012x. [DOI] [Google Scholar]
- Ruan J, Li H, Chen Z, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–40. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajib AM, Hossain M, Ali S. SSR marker-based molecular characterization and genetic diversity analysis of aromatic landreces of rice (Oryza sativa L) J BioSci Biotech. 2012;1:107–116. [Google Scholar]
- Schatz MC, Maron LG, Stein JC, Wences AH, Gurtowski J, Biggers E, Lee H, Kramer M, Antoniou E, Ghiban E, et al. Whole genome de novo assemblies of three divergent strains of rice (Oryza sativa) document novel gene space of aus and indica. Genome Biol. 2014;15:506. doi: 10.1186/s13059-014-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R, Al-Shugeairy Z, Al-Ogaidi F, Munasinghe M, Radermacher M, Vandenhirtz J, Price AH. Comparing simple root phenotyping methods on a core set of rice genotypes. Plant Biol. 2014;16:632–642. doi: 10.1111/plb.12096. [DOI] [PubMed] [Google Scholar]
- Singh VP and Singh RK (2000) Rainfed rice: a sourcebook of best practices and strategies in eastern India. International Rice Research Institute. ISBN: 81-86789-02-2
- The 3 000 rice genomes project The 3,000 rice genomes project. Gigascience. 2014;3:7. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, Zhao K, Wright M, McNally KL, Rey J, Tung CW, Reynolds A, Scheffler B, Eizenga G, McClung A, et al. High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol Breed. 2012;29:875–886. doi: 10.1007/s11032-011-9663-x. [DOI] [Google Scholar]
- Virk PS, Ford-Lloyd BV, Jackson MT, Pooni HS, Clemeno TP, Newbury HJ. Predicting quantitative variation within rice germplasm using molecular markers. Heredity (Edinb) 1996;76:296–304. doi: 10.1038/hdy.1996.43. [DOI] [Google Scholar]
- Virk PS, Zhu J, Newbury HJ, Bryan GJ, Jackson MT, Ford-Lloyd BV. Effectiveness of different classes of molecular marker for classifying and revealing variation in rice (Oryza sativa) germplasm. Euphytica. 2000;112:275–284. doi: 10.1023/A:1003952720758. [DOI] [Google Scholar]
- Wang ZY, Second G, Tanksley SD. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet. 1995;83:565–581. doi: 10.1007/BF00226900. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhu ZF, Tan L, Liu FX, Fu YC, Sun CQ, Cai HW. Complexity of indica-japonica varietal differentiation in Bangladesh rice landraces revealed by microsatellite markers. Breeding Science. 2013;63:227–232. doi: 10.1270/jsbbs.63.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Yan W, Agrama H, Jia M, Fjellstrom R, McClung A. Geographic description of genetic diversity and relationships in the USDA rice world collection. Crop Sci. 2010;50:2406–2417. doi: 10.2135/cropsci2010.02.0096. [DOI] [Google Scholar]
- Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, Tyagi W, Ali ML, Tung CW, Reynolds A, et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One. 2010;5:e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]


