Abstract
High-grade serous carcinoma (HGSC) of the ovary is an aggressive and devastating neoplasm and the identification of novel therapeutic targets may result in a significant decrease in patient morbidity and mortality. Over the last few years, chromatin regulators have become attractive targets for cancer therapy. More specifically, bromodomain-containing protein 4 (BRD4), a protein that is associated with acetylated chromatin and transcriptional activation, has been shown to selectively regulate the transcription of key oncogenic drivers, such as CMYC, in several tumor types. The Cancer Genome Atlas (TCGA) Project has molecularly characterized the genome of ovarian serous carcinomas, which enabled us to study the association of genomic alterations of BRD4 with patient survival and clinicopathological characteristics. Our analysis using clinical and genomic data from the TCGA ovarian carcinoma samples revealed that somatic amplification of BRD4 (observed in 12% of the cases) was correlated with increased BRD4 mRNA levels and is significantly associated with worse overall and progression-free survival compared to wild-type cases. These findings support the hypothesis that future studies and trials investigating newly developed BRD4 inhibitors are required in a subset of patients with ovarian HGSC.
Keywords: bromodomain-containing protein 4, ovarian cancer, serous carcinoma, MYC, The Cancer Genome Atlas project
Introduction
Ovarian high-grade serous carcinoma (HGSC) is the most common type of epithelial ovarian carcinoma, with the highest mortality rate among gynecological malignancies. Following debulking surgery and adjuvant carboplatin and paclitaxel chemotherapy, a significantly high percentage of cases eventually recur, with resistance to systemic therapy. The availability of targeted therapies is currently limited and death typically occurs within 5 years after tumor recurrence and development of chemoresistance (1).
Recently, the Cancer Genome Atlas (TCGA) project has characterized the genomic landscape of HGSC and highlighted a prevalence of somatic tumor protein p53 gene mutations and numerous DNA amplifications and deletions (2). Similar to previous studies, the TCGA study additionally confirmed that a significant number of HGSCS exhibited genetic alterations in the breast cancer 1, early onset (BRCA1) and BRCA2, cyclin E and MYC pathways, with extensive gene copy number alterations. However, the absence of high-frequency oncogenic point mutations in drug target genes has precluded the development of targeted therapeutics for HGSC (2–5).
The goal of this study was to investigate whether novel therapeutic target pathways with available inhibitors may be associated with specific survival outcomes in patients with HGSC. In recent years, chromatin regulators, such as bromodomain-containing protein 4 (BRD4), a member of the bromodomain and extra terminal domain family, which is associated with acetylated chromatin and transcriptional activation, have become attractive targets for cancer therapy. BRD4 has been shown to selectively regulate the transcription of key oncogenic drivers, such as CMYC, in several tumor types (6–9), whereas more recent analyses using epigenetic datasets have demonstrated that BRD4 specifically targets cell-type specific enhancer sequences (i.e., super enhancers) (10). The aim of the present study, was to determine whether the BRD4 gene, which is a druggable gene product, is amplified in a subset of HGSCS, and whether its amplification is associated with worse prognosis and survival.
Patients and methods
Survival analysis
Clinical data from newly diagnosed patients with ovarian carcinoma from the TCGA study (2), specifically 489 cases with copy number alterations data, were integrated with BRD4 amplification data from the TCGA data portal. Kaplan-Meier overall and disease-free survival analysis was performed with the cBio Cancer Genomics Portal (http:cbioportal.org; Memorial Sloan Kettering Cancer Center, New York, NY, USA); significance was estimated using the log-rank test (11,12).
Review of clinical and pathological data
Pathology reports were downloaded from the Cancer Genome Atlas data portal control-access directory (https://tcga-data.nci.nih.gov/tcga) or via the cBio portal. Quality control hematoxylin and eosin images from frozen and permanent sections were analyzed via the TCGA BioSig website hosted at Lawrence Berkeley National Laboratory (http:tcga.lbl.gov:8080/biosig/tcgadownload.do) and via the Cancer Digital Slide Archive (http:cancer.digitalslidearchive.net; Emory University, Atlanta, GA, USA).
This study was conducted in accordance with the TCGA publication policy for ovarian carcinomas (http:cancergenome.nih.gov) and was approved by the Beth Israel Deaconess Medical Center HCC Institutional Review Board.
Results
Cases and survival
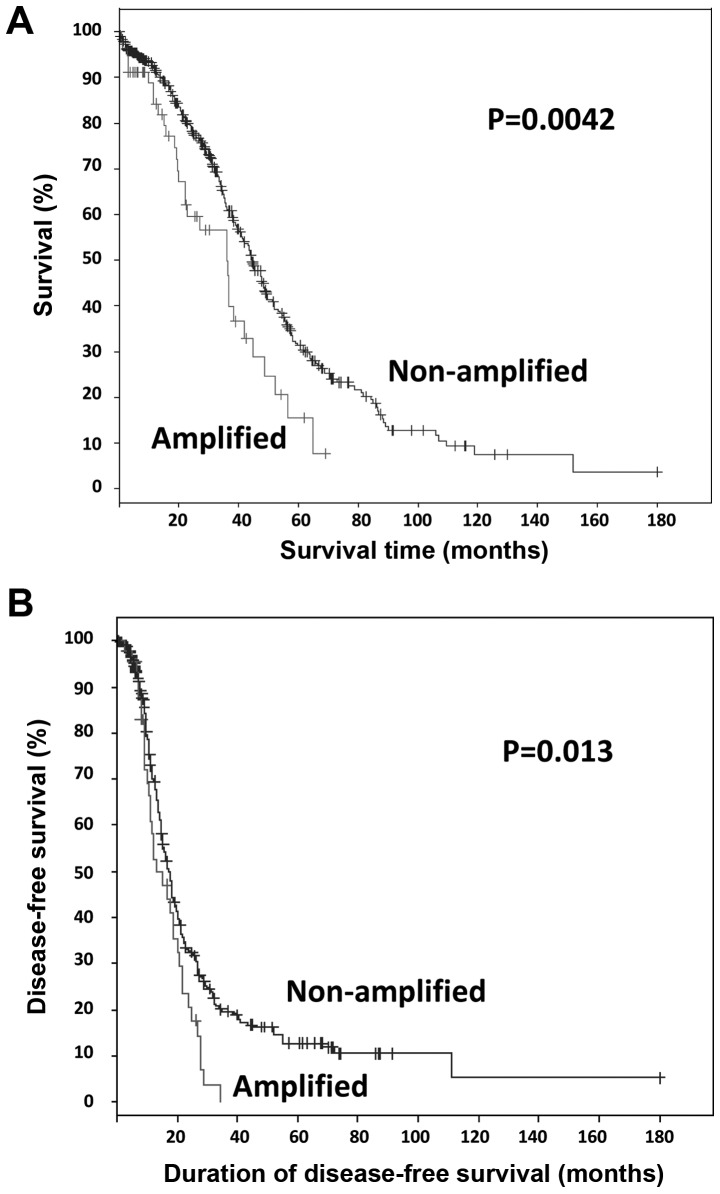
Clinical data from newly diagnosed patients with ovarian carcinoma from the TCGA study, specifically 489 cases with copy number alterations data, were integrated with BRD4 amplification data from the TCGA data portal via the cBio Cancer Genomics Portal (2,11,12). Somatic amplification of BRD4 was identified in 12% (57/489) of the cases. Survival analysis of all TCGA tumors with ovarian HGSC (n=489), revealed that patients with somatic BRD4 amplification exhibited significantly worse overall and progression-free survival compared with those with non-amplified BRD4 (log-rank test P=0.0042 and P=0.013, respectively; Fig. 1). The median number of months until relapse was 13 for cases with amplification, compared with 17 for cases without amplification. Patients with BRD4 amplification exhibited a median survival OF 36 months, compared with 44 months for patients without amplification.
Figure 1.
Survival Kaplan-Meier curves for bromodomain-containing protein 4 gene (BRD4)-amplified tumors. (A) Overall and (B) disease-free survival of the cancer genome atlas cases with copy number alteration data (n=489), with or without BRD4 amplification.
mRNA data
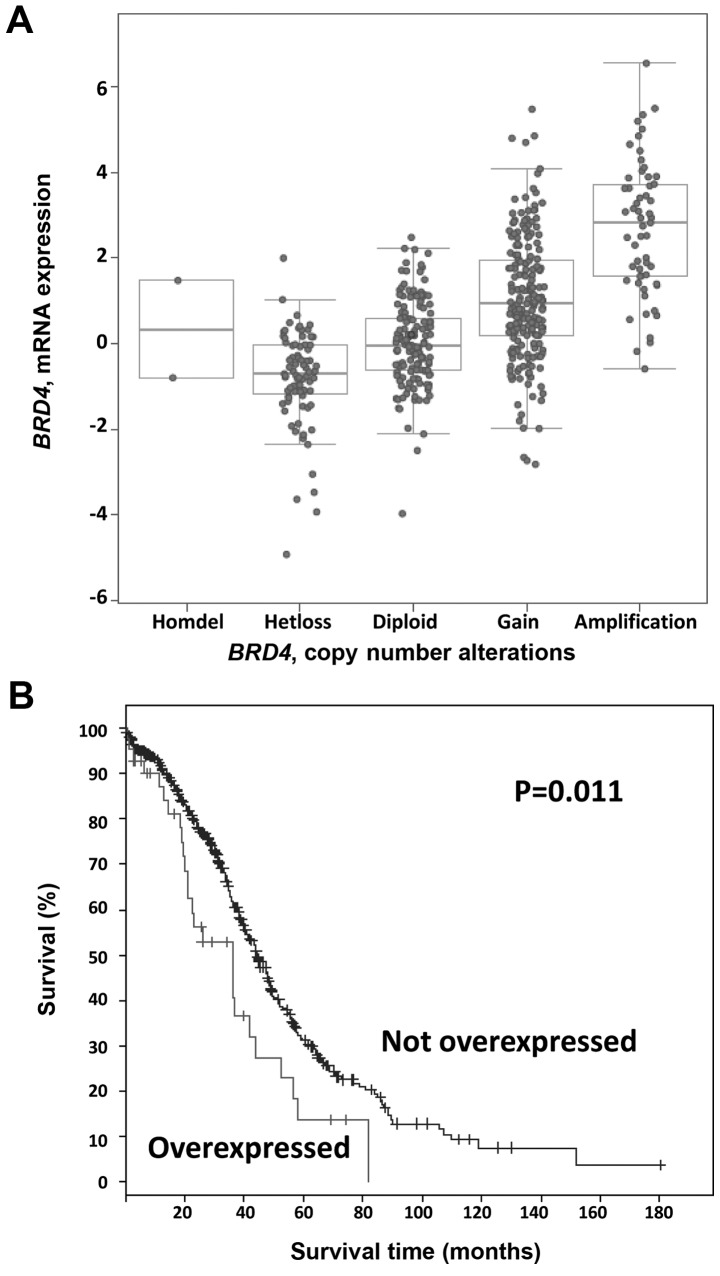
We next examined the mRNA data for these cases. As shown in Fig. 2A, BRD4 mRNA levels tended to increase with copy number. Using a stringent Z score threshold of +3.0 to define significant upregulation, BRD4 mRNA was increased in 9% (43/489) of the cases, and increased BRD4 mRNA levels were associated with worse overall survival (log-rank test P=0.01; Fig. 2B). By contrast, there were no significant differences in progression-free survival in cases with BRD4 mRNA upregulation (log-rank test P=0.59). Again using a Z score threshold of +3.0, in 45% of the amplified cases (26/57), BRD4 amplification resulted in mRNA upregulation. Conversely, 60% (26/43) of tumors with high mRNA levels harbored an underlying amplification.
Figure 2.
Bromodomain-containing protein 4 (BRD4) mRNA data. (A) BRD4 mRNA levels tended to increase with increased copy number. (B) Kaplan-Meier curve depicting overall survival with BRD4 mRNA upregulation, Z score threshold of +3.0 of BRD4 mRNA levels.
Genes differentially expressed in BRD4-amplified tumors
To elucidate the mechanism through which BRD4 amplification may affect patient survival, we assessed which genes may be differentially expressed in BRD4-amplified tumors, including CMYC or NMYC, which are known downstream targets of BRD4 (7,13,14). In the TCGA cohort, a subset of BRD4-amplified tumors exhibited high CMYC and NMYC mRNA levels; however, there was only a weak positive association between BRD4 copy number and CMYC mRNA (Pearson, 0.130; and Spearman, 0.072), or NMYC mRNA levels (Pearson, 0.119; and Spearman, 0.063) (data not shown). By contrast, there was a strong association between BRD4 amplification and increased mRNA levels of other genes located in close proximity to BRD4, such as WIZ, PRKACA, NOTCH3 and SMARCA4 (Table I), suggesting a potential collaborative regional amplification effect. Future studies with independent cohorts, cell culture and/or animal model systems are required to address the potential mechanisms through which BRD4 amplification contributes to the pathogenesis and prognosis of ovarian HGSCS.
Table I.
Differentially expressed genes (mRNA) in bromodomain-containing protein 4 gene-amplified tumors.
| Genes | Pearson | Spearman |
|---|---|---|
| WIZ | 0.79 | 0.80 |
| AKAP8 | 0.74 | 0.72 |
| CHERP | 0.69 | 0.70 |
| AKAP8L | 0.67 | 0.64 |
| SIN3B | 0.66 | 0.66 |
| DCAF15 | 0.66 | 0.58 |
| MYO9B | 0.65 | 0.65 |
| MED26 | 0.65 | 0.66 |
| PRKACA | 0.64 | 0.60 |
| CCDC130 | 0.62 | 0.58 |
| NACC1 | 0.61 | 0.60 |
| PKN1 | 0.60 | 0.55 |
| CC2D1A | 0.60 | 0.58 |
| AP1M1 | 0.59 | 0.58 |
| C19ORF44 | 0.59 | 0.57 |
| EPS15L1 | 0.59 | 0.59 |
| TECR | 0.58 | 0.56 |
| MAP1S | 0.57 | 0.56 |
| ZNF333 | 0.57 | 0.53 |
| NOTCH3 | 0.56 | 0.55 |
| SLC35E1 | 0.56 | 0.56 |
| TRMT1 | 0.55 | 0.50 |
| GTPBP3 | 0.55 | 0.53 |
| KRI1 | 0.55 | 0.53 |
| TNPO2 | 0.54 | 0.50 |
| JUND | 0.53 | 0.57 |
| RAD23A | 0.53 | 0.51 |
| SMARCA4 | 0.53 | 0.52 |
| TYK2 | 0.53 | 0.49 |
| ILF3 | 0.52 | 0.47 |
Discussion
BRD4 is a bromodomain protein and part of a chromatin remodeling complex, which regulates gene expression by binding acetylated chromatin at specific promoters and enhancers. Via its bromodomains, BRD4 recruits positive transcription elongation factor b and triggers the release of RNA polymerase II from promoters, thereby resulting in productive transcriptional elongation and active gene expression of genes such as CMYC (13,15).
We exploited the TCGA data portal on ovarian carcinomas to interrogate specific genetic events of druggable genes associated with survival and clinicopathological characteristics, as well as to generate hypotheses for future testing. We herein propose that BRD4 amplification in ovarian HGSCS may be associated with unfavorable prognosis and survival. However, due to the limitations of the TCGA study (i.e., relatively short follow-up time and lack of information on other comorbidities), these findings are exploratory and hypothesis-generating in nature rather than definitive. Future studies with independent patient cohorts are required to validate the TGCA findings.
Furthermore, the mechanism through which BRD4 amplification may affect patient prognosis and survival remains unclear. Amplification of BRD4 appears to segregate preferentially to the non-BRCAness molecular subgroup of ovarian HGSCS (16). A recently published study suggested that BRD4 inhibitors may be effective in a subset of ovarian HGSCS that exhibit high CMYC or MYCN levels (17). In the TCGA cohort, there was a weak positive association between BRD4 copy number alterations and CMYC or MYCN mRNA levels, suggesting that survival effects may be MYC-independent. By contrast, there was a strong association between BRD4 amplification and increased mRNA levels of other genes located in close proximity to BRD4, such as NOTCH3 and SMARCA4, suggesting a potential collaborative regional amplification effect. Supporting this hypothesis, SMARCA4 (a member of the SWI/SNF chromatin remodeling complex, also known as Brg1) has been found to occupy the same MYC enhancer sites as BRD4, thereby enhancing MYC expression (18). In addition, NOTCH1 has been shown to enhance MYC expression in T-cell leukemia independently of BRD4 (19). In ovarian HGSC, our finding that worse overall survival is also observed with increased BRD4 mRNA levels in addition to BRD4 amplification suggests that this survival effect may be specific to BRD4 function.
Finally, we hypothesized that BRD4-amplified tumors may exhibit increased sensitivity to newly developed BRD4 small-molecule inhibitors. Future studies, including animal xenografts or cell culture models, are required to further elucidate the mechanisms through which BRD4 amplification and overexpression may affect tumor progression and patient survival. In summary, we herein presented evidence that the BRD4 gene is amplified and overexpressed in a subset of HGSCS, and that its amplification and overexpression are associated with worse prognosis and survival. Our findings suggest that targeted inhibition of BRD4 with specific inhibitors should be tested in a subset of ovarian carcinoma patients.
References
- 1.Romero I, Bast RC., Jr Minireview: Human ovarian cancer: Biology, current management, and paths to personalizing therapy. Endocrinology. 2012;153:1593–1602. doi: 10.1210/en.2011-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network, corp-author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BastRC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: New opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal T, PermuthWey J, Betts JA, Krischer JP, Fiorica J, Arango H, LaPolla J, Hoffman M, Martino MA, Wakeley K, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 5.Etemadmoghadam D, AuYeung G, Wall M, Mitchell C, Kansara M, Loehrer E, Batzios C, George J, Ftouni S, Weir BA, et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin Cancer Res. 2013;19:5960–5971. doi: 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 6.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., III Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:p11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu LL, Tian M, Li X, Li JJ, Huang J, Ouyang L, Zhang Y, Liu B. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goundiam O, Gestraud P, Popova T, De la Motte Rouge T, Fourchotte V, Gentien D, Hupé P, Becette V, Houdayer C, RomanRoman S, et al. Histo-genomic stratification reveals the frequent amplification/overexpression of CCNE1 and BRD4 genes in non-BRCAness high grade ovarian carcinoma. Int J Cancer Apr. 2015;17 doi: 10.1002/ijc.29568. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 17.Baratta MG, Schinzel AC, Zwang Y, Bandopadhayay P, BowmanColin C, Kutt J, Curtis J, Piao H, Wong LC, Kung AL, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc Natl Acad Sci USA. 2015;112:232–237. doi: 10.1073/pnas.1422165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Whyte WA, ZepedaMendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, EckersleyMaslin MA, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.YashiroOhtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci USA. 2014;111:E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]