Abstract
Objective
The development of acute kidney injury in patients with sepsis is associated with worse outcomes. Identifying those at risk for septic acute kidney injury could help to inform clinical decision making. We derived and tested a multibiomarker-based model to estimate the risk of septic acute kidney injury in children with septic shock.
Design
Candidate serum protein septic acute kidney injury biomarkers were identified from previous transcriptomic studies. Model derivation involved measuring these biomarkers in serum samples from 241 subjects with septic shock obtained during the first 24 hours of admission and then using a Classification and Regression Tree approach to estimate the probability of septic acute kidney injury 3 days after the onset of septic shock, defined as at least two-fold increase from baseline serum creatinine. The model was then tested in a separate cohort of 200 subjects.
Setting
Multiple PICUs in the United States.
Interventions
None other than standard care.
Measurements and Main Results
The decision tree included a first-level decision node based on day 1 septic acute kidney injury status and five subsequent biomarker-based decision nodes. The area under the curve for the tree was 0.95 (CI95, 0.91–0.99), with a sensitivity of 93% and a specificity of 88%. The tree was superior to day 1 septic acute kidney injury status alone for estimating day 3 septic acute kidney injury risk. In the test cohort, the tree had an area under the curve of 0.83 (0.72–0.95), with a sensitivity of 85% and a specificity of 77% and was also superior to day 1 septic acute kidney injury status alone for estimating day 3 septic acute kidney injury risk.
Conclusions
We have derived and tested a model to estimate the risk of septic acute kidney injury on day 3 of septic shock using a novel panel of biomarkers. The model had very good performance in a test cohort and has test characteristics supporting clinical utility and further prospective evaluation.
Keywords: biomarkers, decision tree, inflammation, kidney injury, modeling, sepsis
Acute kidney injury (AKI) is a common complication of sepsis, and the development of AKI increases sepsis-related mortality (1–3). Identifying patients at risk of developing septic AKI (SAKI) would identify patients in need of intervention or preventive measures, such as earlier initiation of renal replacement therapy, restrictive fluid management, and avoidance of nephrotoxic medications when clinically feasible. Biomarkers have the potential to identify such patients (3–8).
The pathogenesis of SAKI is complex and incompletely understood. Ischemia is a well-accepted mechanism for AKI, but it is not the unifying mechanism for sepsis since SAKI can occur in the absence of global kidney ischemia (9, 10). It has been proposed that SAKI results from a complex interplay between inflammation, oxidant stress, apoptosis, and microvascular alterations (3, 11). Consequently, biomarkers that reflect just one mechanistic pathway, such as primarily ischemic kidney insults, may be insufficient for predicting SAKI in all those at risk.
We have developed a multi-institutional clinical and biological repository of pediatric septic shock and have conducted extensive genome-wide expression studies (12). These transcriptomic data have been leveraged for the discovery of novel therapeutic targets (13–15), gene expression–based subclasses of pediatric septic shock (16), and sepsis-related biomarkers (17–19). The latter effort included the identification of a 21-gene expression signature (messenger RNA) having predictive capacity for SAKI (20). With the goal of developing a clinical tool that can meet the time-sensitive diagnostic demands of critically ill patients, we determined if the protein products of five of these 21 class predictor genes have utility as biomarkers for SAKI.
METHODS
Study Subjects and Data Collection
The study protocol was approved by the institutional review boards of each participating institution. Children 10 years old or younger admitted to the PICU and meeting pediatric-specific consensus criteria for septic shock were eligible for enrollment (15). After informed consent from parents or legal guardians, serum samples were obtained within 24 hours of initial presentation to the PICU with septic shock. Clinical and laboratory data were collected daily while in the PICU. Mortality was tracked for 28 days after enrollment. Illness severity was measured using Pediatric Risk of Mortality (PRISM) scores (21), and septic shock-related mortality probability at study entry was estimated using the updated Pediatric Sepsis Biomarker Risk Model (PERSEVERE) (18).
The derivation cohort subjects included 175 subjects previously reported (20), plus an additional 66 subjects prospectively enrolled since our previous report and randomly selected from our database. The test cohort consisted of 200 different prospectively enrolled subjects and randomly selected from our database. None of the study subjects had known preexisting renal disease.
SAKI Definition
SAKI was defined based on a modification of the Kidney Disease Improving Global Outcomes (KDIGO) stage 2 AKI classification, which is at least two-fold increase from baseline serum creatinine (22). We focused on KDIGO stage 2 AKI because this was the definition used in our original study that identified the candidate SAKI biomarkers (20), and we made an a priori plan to use the same definition in the current study. Because we did not have baseline serum creatinine data for the study subjects, our modification was to use age-based normal values as surrogates, as previously reported (20, 23).
Biomarker Selection and Measurement
We previously reported a messenger RNA expression signature of 21 genes with predictive capacity for SAKI (20). In the feasibility stage of the current study, we were only able to identify suitable immunoassay reagents to measure the protein products of five of the previously identified genes in serum samples. The resulting five candidate serum protein biomarkers were elastase 2 (ELA2), fibroblast growth factor 13 (FGF13), matrix metalloproteinase 8 (MMP8), olfactomedin 4 (OLFM4), and proteinase 3 (PRTN3). The serum concentrations of the candidate biomarkers were measured using a multiplex magnetic bead platform (MILLIPLEX MAP) designed for this project by the EMD Millipore (Billerica, MA). Biomarker concentrations were measured in a Luminex 100/200 System (Luminex, Austin, TX), according the manufacturers’ specifications. The median biomarker concentrations with interquartile ranges are provided in a Supplemental Table 1 (Supplemental Digital Content 1, http://links.lww.com/CCM/B324).
Statistical Analysis and Modeling Procedures
Initially, data are described using medians, interquartile ranges, frequencies, and percentages. Comparisons between groups used the Wilcoxon rank-sum test for age and PRISM score, the unpaired t test for PERSEVERE Mortality Probability, and the chi-square test, or Fisher exact test if needed, for gender and 28-day mortality. Descriptive statistics and comparisons used SigmaStat Software (Systat Software, San Jose, CA).
The primary outcome variable for the modeling was the presence of SAKI on day 3 after presentation with septic shock. Classification and Regression Tree (CART) analysis was used to estimate the probability of day 3 SAKI (Salford Predictive Modeler v7.0; Salford Systems, San Diego, CA) (24). The predictor variables included the five candidate biomarkers, the presence of SAKI on day 1 of septic shock, age, and gender. Weighting of cases and the addition of cost for misclassification were not used in the modeling procedures. The code used to generate the model is available from the authors. Performance of the derived model is reported using diagnostic test statistics with 95% CIs computed using the score method as implemented by the VassarStats Website for Statistical Computation (25).
Areas under the receiver operating characteristic curves were compared using the method of Hanley and McNeil (26) for nonindependent samples. The net reclassification improvement (NRI) was used to estimate the incremental predictive ability of the biomarker-based model compared to day 1 SAKI status alone (27). The NRI was computed using the R-package Hmisc (28).
RESULTS
Model Derivation
Table 1 shows the demographics and clinical characteristics of the derivation cohort (n = 241). Twenty-eight subjects (12%) had SAKI on day 3 of septic shock. Compared with the subjects without SAKI, the subjects with SAKI had a higher mortality rate, a higher median PRISM score, and a higher PERSEVERE-based mortality probability, and a greater proportion were male. No other differences were noted.
Table 1.
Clinical and Demographic Data for the Derivation Cohort
| Variable | All | No AKI | AKI |
|---|---|---|---|
| n | 241 | 213 | 28 |
| Median age, yr (IQR) | 2.5 (0.8–5.9) | 2.5 (0.8–5.9) | 2.8 (1.2–7.0) |
| Males, n (%) | 150 (62) | 127 (60) | 23 (82)a |
| 28-d mortality, n (%) | 17 (9) | 8 (4) | 11 (39)a |
| Median Pediatric Risk of Mortality score (IQR) | 12 (7–21) | 12 (6–18) | 24 (19–30)b |
| Pediatric Sepsis Biomarker Risk Model mortality probability (95% CI) | 11.0 (8.9–13.3) | 8.8 (6.7–10.9) | 27.3 (18.5–36.1)c |
| Gram-negative bacteria, n (%) | 51 (21) | 43 (20) | 8 (29) |
| Gram-positive bacteria, n (%) | 57 (24) | 51 (24) | 6 (21) |
| Other pathogen isolated, n (%) | 21 (9) | 18 (8) | 3 (11) |
| No pathogen identified, n (%) | 112 (46) | 101 (47) | 11 (39) |
| Comorbidity, n (%) | 82 (34) | 71 (33) | 11 (39) |
AKI = acute kidney injury, IQR = interquartile range.
p < 0.05 vs no AKI cohort, chi-square test.
p < 0.05 vs no AKI, rank-sum test.
p < 0.05 vs no AKI, t test.
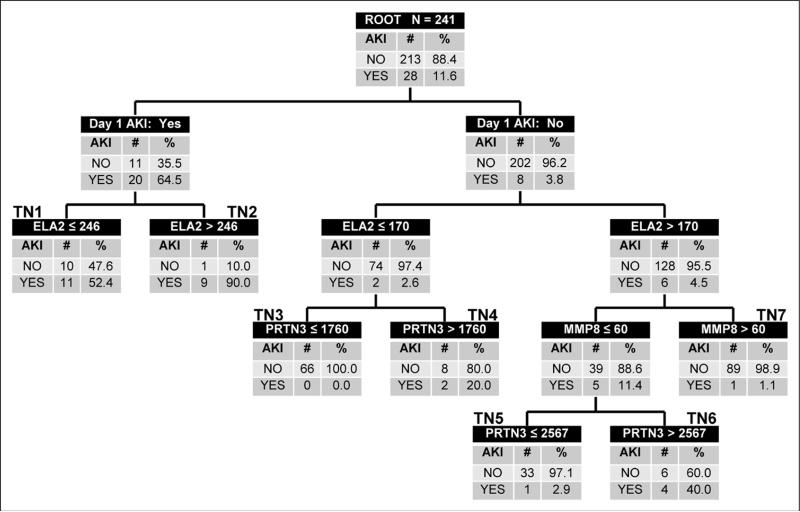
Figure 1 shows the derived decision tree. The top node of the decision tree, the root node, provides the total number of subjects and the number and proportion of subjects with and without SAKI on day 3 of septic shock. Subjects in the root node are subsequently allocated to daughter nodes based on the results of binary recursive partitioning, as determined by the CART methodology. Each daughter node provides the criterion for deciding subsequent partitions, along with the number and proportion of subjects with and without SAKI on day 3 of septic shock. Terminal nodes reflect the final assignment of risk to an individual case and are annotated with bold numbers above each terminal node in the tree.
Figure 1.
Classification tree from the derivation cohort (n = 241). The classification tree consists of six decision points and 12 daughter nodes. The classification tree includes three of the five candidate septic acute kidney injury (AKI) biomarkers: elastase 2 (ELA2), matrix metallopeptidase 8 (MMP8), and proteinase 3 (PRTN3). The serum concentrations of all candidate SAKI biomarkers are provided in ng/mL. The classification tree also includes the presence/absence of day 1 SAKI as defined in the Methods section. The root node provides the total number of patients in the derivation cohort and the number of subjects with and without SAKI on day 3 of septic shock, with the respective rates. Each daughter node provides the respective decision rule criterion and the number of subjects with and without SAKI on day 3 of septic shock. The numbers above daughter nodes designate terminal nodes (TN). TN3, TN5, and TN7 are considered low-risk terminal nodes; TN1, TN4, and TN6 are considered intermediate-risk nodes; and TN2 is considered a high-risk terminal node.
The first-level decision node was based on day 1 SAKI status. Further accuracy for estimating the risk of day 3 SAKI was achieved with three of the five candidate biomarkers (ELA2, MMP8, and PRTN3). FGF13, OLFM4, age, and gender did not improve predictive accuracy. There were three low probability terminal nodes (0.0–2.9% probability; terminal nodes 3, 5, and 7), three intermediate probability terminal nodes (20.0–52.4% probability; terminal nodes 1, 4, and 6), and one high probability terminal node (90.0% probability; terminal node 2) for SAKI on day 3 of septic shock. Among the 190 subjects classified as low probability, two subjects (1%) had SAKI on day 3 of septic shock. Among the 51 subjects classified as intermediate to high probability, 26 (51%) had SAKI on day 3 of septic shock.
For calculating the diagnostic test characteristics using a two-by-two contingency table, all subjects in the low probability terminal nodes are predicted to not have SAKI, whereas all subjects in the intermediate and high probability terminal nodes are predicted to have SAKI on day 3 of septic shock. Table 2 shows the diagnostic test characteristics of the derived decision tree.
Table 2.
Test Characteristics of the Decision Tree With 95% CIs
| Variable | Derivation Cohort | Test Cohort |
|---|---|---|
| No. of subjects | 241 | 200 |
| No. of true positives | 26 | 17 |
| No. of true negatives | 188 | 139 |
| No. of false positives | 25 | 41 |
| No. of false negatives | 2 | 3 |
| Sensitivity | 93 (75–99) | 85 (61–96) |
| Specificity | 88 (83–92) | 77 (70–83) |
| Positive predictive value | 51 (37–65) | 29 (18–43) |
| Negative predictive value | 99 (96–100) | 98 (93–99) |
| Positive likelihood ratio | 7.9 (5.4–11.6) | 3.7 (2.7–5.2) |
| Negative likelihood ratio | 0.08 (0.02–0.3) | 0.2 (0.07–0.6) |
| Area under the curve | 0.95 (0.91–0.99) | 0.83 (0.72–0.95) |
Since 20 of 28 subjects with SAKI (71%) on day 3 of septic shock also had SAKI on day 1 of septic shock, and 20 of 31 subjects with SAKI (65%) on day 1 of septic shock still had SAKI on day 3 of septic shock, we compared the predictive capacity of the decision tree to that of SAKI status on day 1 of septic shock alone. SAKI status on day 1 of septic shock by itself had an area under the receiver operating curve of 0.83 (CI95, 0.73–0.93), which was significantly less than that of the decision tree (0.95 [0.91–0.99]; p = 0.001). When adding the information from the decision tree to the information provided by SAKI status on day 1 of septic shock alone, the NRI was 0.93 (CI95, 0.56–1.31; p < 0.001).
Testing the Model
The test cohort consisted of 200 subjects, of whom 20 (10%) had SAKI on day 3 of septic shock. Table 3 provides the clinical and demographic data for the test cohort. Compared with the derivation cohort, the test cohort had a lower proportion of males. No other differences were noted. Within the test cohort, the subjects with SAKI had a higher mortality rate, a higher median PRISM score, and a higher PERSEVERE-based mortality probability, compared with the subjects without SAKI. No other differences were noted.
Table 3.
Clinical and Demographic Data for the Test Cohort
| Variable | All | No AKI | AKI |
|---|---|---|---|
| n | 200 | 180 | 20 |
| Median age, yr (IQR) | 2.6 (0.8–5.7) | 2.7 (0.8–5.7) | 2.2 (1.2–4.8) |
| Males, n (%) | 105 (53)a | 96 (53) | 9 (45) |
| 28-d mortality, n (%) | 28 (14) | 19 (11) | 9 (45)b |
| Median Pediatric Risk of Mortality score (IQR) | 13 (9–19) | 12 (9–18) | 19 (11–34)c |
| Pediatric Sepsis Biomarker Risk Model mortality probability (95% CI) | 11.6 (9.5–13.7) | 10.8 (8.7–12.9) | 18.7 (10.6–26.8)d |
| Gram-negative bacteria, n (%) | 45 (23) | 40 (22) | 5 (25) |
| Gram-positive bacteria, n (%) | 44 (22) | 38 (21) | 6 (30) |
| Other pathogen isolated, n (%) | 17 (9) | 16 (9) | 1 (5) |
| No pathogen identified, n (%) | 94 (47) | 86 (48) | 8 (40) |
| Comorbidity, n (%) | 73 (37) | 68 (38) | 5 (25) |
AKI = acute kidney injury, IQR = interquartile range.
p < 0.05 vs derivation cohort.
p < 0.05 vs no AKI cohort, chi-square test.
p < 0.05 vs no AKI, rank-sum test.
p < 0.05 vs no AKI, t test.
The test cohort subjects were classified according to the derived model without any modifications (Supplemental Fig. 1, Supplemental Digital Content 2, http://links.lww.com/CCM/B325). Among the 142 subjects classified as low probability, three subjects (2%) had SAKI on day 3 of septic shock. Among the 58 subjects classified as intermediate to high probability, 17 (29%) had SAKI on day 3 of septic shock. Table 2 shows the diagnostic test characteristics of the decision tree in the test cohort.
We next compared the predictive capacity of the decision tree to that of SAKI status on day 1 of septic shock alone in the test cohort. SAKI status on day 1 of septic shock by itself had an area under the receiver operating curve of 0.73 (CI95, 0.59–0.87) in the test cohort, which was significantly less than that of the decision tree (0.82 [0.72–0.95]; p = 0.044). When adding the information from the decision tree to the information provided by SAKI status on day 1 of septic shock alone in the test cohort, the NRI was 0.69 (CI95, 0.24–1.14; p = 0.003).
Comparison to Mortality Prediction Models
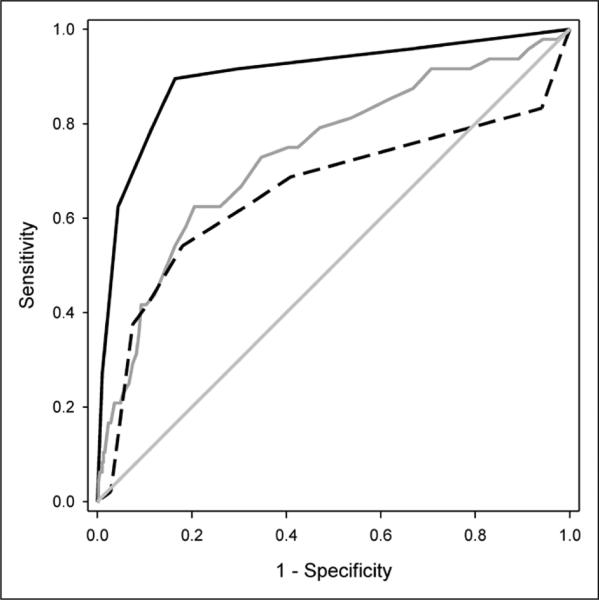
Since AKI is associated with increased mortality in patients with sepsis, it is possible that the candidate SAKI biomarkers are generic biomarkers of illness severity, rather than being biomarkers of SAKI risk per se. To test this possibility, we compared the performance of the SAKI risk model in the combined derivation and test cohorts to that of two mortality risk models, PRISM (21) and PERSEVERE (18), for estimating the risk of SAKI on day 3 of septic shock. As shown in Figure 2, the area under the receiver operating curve for the SAKI model was superior to that of PRISM and PERSEVERE.
Figure 2.
Comparison of the receiver operating characteristic (ROC) curves for the septic acute kidney injury (SAKI) risk model, Pediatric Risk of Mortality (PRISM), and Pediatric Sepsis Biomarker Risk Model (PERSEVERE), for estimating the risk of SAKI on day 3 of septic shock (combined derivation and test cohorts, n = 441). The ROC curve for the SAKI risk model (solid black line) yielded an area under the curve of 0.90 (CI95, 0.85–0.96), which was superior to that of PRISM (solid gray line; 0.73; CI95, 0.66–0.82; p < 0.001) and PERSEVERE (dashed black line; 0.66; CI95, 0.55–0.76; p < 0.0001) for estimating the risk of SAKI on day 3 of septic shock.
DISCUSSION
We have derived and tested a multibiomarker-based model to estimate the probability of SAKI 3 days after the onset of septic shock. In the derivation cohort, the model had excellent performance with an area under the curve (AUC) of 0.95 and a sensitivity of 93%. We found the model was additive to the information provided by day 1 SAKI status alone. The model had very good performance in the test cohort, with an AUC of 0.83 and a sensitivity of 85%. Similar to the derivation cohort, the model added to the information provided by day 1 SAKI status alone in the test cohort. A recently reported study of cell-cycle arrest biomarkers for AKI in critically ill adults included a sample among which 24% had sepsis as a primary diagnosis (8). The current model demonstrated higher specificity while retaining similar sensitivity and positive predictive values.
We have previously used a CART approach to develop multibiomarker risk models related to sepsis (17, 18). We favor this approach because it has the potential to reveal interactions between predictor variables that may not be evident using other modeling approaches (24). In the test cohort, the intermediate- and high-risk terminal nodes identified a cohort of patients with a rate of SAKI of 29%, whereas the low-risk terminal nodes identified a cohort with a rate of SAKI of 2%. This dichotomous interpretation demonstrates that the model can partition a heterogeneous cohort of patients into two broad groups having an approximately 15-fold difference in SAKI risk. A more comprehensive interpretation allows for assigning risk based on the respective terminal nodes, and this partitions patients across a clinically relevant range of SAKI probabilities.
The candidate biomarkers that went into the model were selected objectively based on extensive transcriptomic studies focused on discovery of SAKI biomarkers (20). This is an important strength of our study. We noted that the model performed significantly better than a physiology-based mortality risk scoring system (PRISM) and a multibiomarker-based mortality risk model for sepsis (PERSEVERE) for estimating the risk of SAKI 3 days after the onset of septic shock. This indirectly suggests that the candidate SAKI biomarkers are intrinsically associated with AKI risk in patients with septic shock, rather than being generic biomarkers of illness severity.
The three biomarkers in the model are proteases, primarily derived from neutrophils. These general characteristics are consistent with the concept that inflammatory mechanisms play an important role in the development of SAKI. MMP8 was previously reported as a candidate biomarker for detecting renal allograft rejection (29). Genetic ablation or pharmacologic inhibition of MMP8 delays renal recovery in mice subjected to renal ischemia (30), which is consistent with the MMP8-based decision node in the model. PRTN3 serves as the autoantigen for Wegener's granulomatosis, an autoimmune systemic vasculitide that commonly leads to kidney injury (31), and this is also consistent with the two PRTN3-based decision nodes in the model. Neutrophil extracellular traps containing PRTN3 and ELA2 are detectable in kidney biopsies from patients with autoimmune glomerulonephritis (32). Incorporation of MMP8 and ELA2 with clinical data used to calculate a “renal angina index” has been shown to predict AKI in critically ill children (33). We note that an ELA2-based decision node in the model may allow for distinguishing subjects who initially present with SAKI, but are more or less likely to have improved kidney function by day 3 of septic shock. Collectively, these data support mechanistic links between the candidate biomarkers and the pathophysiology of SAKI, which warrant further exploration.
We note the limitations of our study. We could not directly determine baseline creatinine values for the study subjects and consequently used median, age-dependent normal values as a surrogate. We also did not have the option of estimating baseline creatinine using a body surface area-based formula because we did not have height data for the subjects. This could have led to misclassified patients with or without AKI because some proportion may have had a preexisting kidney injury, as suggested by the presence of a comorbid condition in about one third of the study subjects and a relatively low prevalence of SAKI. An extensive adjudication process for determining AKI status in critically ill patients, as recently used (8, 34), may be superior as a criterion standard, but we contend that our definition is pragmatic in that it is reflective of patients admitted to an ICU with septic shock. We made an a priori decision to focus on day 3 of septic shock and KDIGO AKI stage 2 as the primary outcome variable. It might be argued that different time points and outcome definitions have differing clinical relevance, and it is possible that the results of our modeling would change if we had focused on a different time point or a different stage of AKI. Differences in model structure would be informative in understanding the mechanism of injury, but a single model with a different time point or endpoint would be no less limited in clinical applicability. It is reasonable to conclude that there is clinical significance in a patient with sepsis who has a doubling of their baseline creatinine 3 days after initial presentation.
Clinical care was not standardized in this observational study. Therefore, it is possible that the development of SAKI was confounded by variability of clinical processes, independent of biomarker data. The five SAKI biomarkers were selected from 21 candidate genes, based on the availability of suitable immune-assay reagents to measure their protein products in the serum compartment. These technical limitations eliminated other previously reported candidate genes, which could possibly have better predictive performance (20). Finally, our a priori analysis plan was confined to testing the validity of a multibiomarker panel in predicting day 3 SAKI. Whether adding other potential biomarkers or clinical information improve this model requires further investigation.
In summary, we have derived and tested a model to estimate the risk of SAKI on day 3 of septic shock using a novel panel of biomarkers. The model had very good performance in a test cohort and has test characteristics supporting clinical utility. Prospective evaluation of the updated model is warranted to further assess clinical utility, and the model may provide insight into the pathobiology of SAKI.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following clinical research coordinators for enrolling patients at the various study sites: Debra Spears, Jenny Bush, Mary Ann De Liberto, Adelle Barreiro, Trisha Williams, Amber Hughes, Michelle Goldsworthy, Christi Rider, Mary Ellen Riodan, Veronica DeLeon, Hyacinth Bryant, Tiffany Patterson, Ofelia Vargas, Monica Weber, Lauren Hoadley, Heather Anthony, Lisa Steele, Angela Doucette, and Katherine Woods. We thank Robert A. Oster, PhD, University of Alabama at Birmingham, for providing an independent statistical review of the article.
Dr. Wong conceived and developed the study, obtained funding for the study, conducted the analysis, and edited the article. Dr. Cvijanovich, Dr. Anas, Dr. Allen, Dr. Thomas, Dr. Bigham, Dr. Weiss, Ms. Fitzgerald, Dr. Checchia, Dr. Meyer, Dr. Shanley, Dr. Quasney, Dr. Hall, Dr. Gedeit, Dr. Freishtat, Dr. Nowak, Dr. Shekhar, Dr. Gertz, and Dr. Dawson enrolled subjects at the participating institutions, provided clinical data and biological samples, and edited the article. Ms. Howard and Ms. Frank maintained the clinical database and coordinated all interinstitutional research activity. Ms. Harmon maintained the biological repository and processed all biological samples. Mr. Lahni conducted all biomarker assays. Ms. Hart assisted with statistical analysis. Dr. Lindsell developed the study, assisted with analysis, and edited the article.
Supported, in part, by an Institutional Clinical and Translational Science Award, National Institutes of Health (NIH) and National Center for Research Resources (8UL1 TR000077).
Dr. Wong received support for article research from the NIH. His institution received grant support from the NIH and has biomarker patents (not related to the current work). Dr. Cvijanovich is employed by the Children's Critical Care Medical Group; lectured for the American Society for Parenteral and Enteral Research (speaker at annual conference 2014); and received support for article research from the NIH. Her institution received grant support from the Cincinnati Children's Hospital Medical Center (per patient fee via subcontract) and the U.S. Army Medical Research and Material Command (award no. W81XWH-BAA-11-1). Dr. Anas served as a board member for Children's Hospital of Orange County (CHOC) Health Alliance, consulted for CHOC Children's, is employed by CHOC Children's, and has retirement stock. Dr. Allen received support for article research from the NIH. His institution received grant support (subcontract for NIH grant). Dr. Thomas served as an Advisory Board Member for and Ikaria and received support for article research from the NIH. His institution received grant support from the Children's Hospital of Cincinnati (subcontract grant). Dr. Weiss is employed by The Children's Hospital of Philadelphia and received royalties from Up-To-Date. He and his institution received grant support from the NICHD (K12HD047349). Ms. Fitzgerald C/F and her institution received grant support from the NIH (grant nos. RO1GM064619, RO1GM099773, and R01GM108025). Dr. Meyer received support for article research from the NIH. His institution received grant support from the NIH. Dr. Shanley served as a board member for the International Pediatric Research Foundation (Trustee of Executive Committee), consulted for External Advisory Boards (U-Cinn, U-Minn, Case West), and received support for article research from the NIH. His institution received grant support from the NIH (Clinical and Translational Science Award principal investigator). Dr. Hall lectured for the Society of Critical Care Medicine and received support for article research from the NIH. His institution received grant support from the NIH. Dr. Freishtat received support for article research from the NIH. Dr. Gertz is employed by and received support for travel from the Hackensack University Medical Center. She received support for article research from the NIH. Her institution received grant support from the Hackensack University Medical Center. Dr. Dawson received support for article research from the NIH. Ms. Howard received support for article research from the NIH. Her institution received grant support from the NIH. Mr. Lahni received support for article research from the NIH. Ms. Frank received support for article research from the NIH. Her institution received grant support from the NIH, and she has disclosed employment, has a patent, and received support for travel. Dr. Lindsell received support for article research from the NIH. His institution received grant support from the NIH (He is a coinvestigator on NIH-funded grants supporting the work developing biomarkers and risk models in sepsis and coinvestigator on grants submitted to NIH exploring biomarkers and risk stratification in sepsis. The grants are not directly related to acute kidney injury.) and has patents with the U.S. Patent Office (Dr. Lindsell is a coinventor on patents for Pediatric Sepsis Biomarker Risk Model, the mortality risk model in pediatric sepsis, and its temporal version). The remaining authors have disclosed that they do not have any potential conflicts of interest.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (http://journals.lww.com/ccmjournal).
REFERENCES
- 1.Bagshaw SM, George C. Bellomo R; ANZICS Database Management Committee: Early acute kidney injury and sepsis: A multicentre evaluation. Crit Care. 2008;12:R47. doi: 10.1186/cc6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta RL, Bouchard J, Soroko SB, et al. Program to Improve Care in Acute Renal Disease (PICARD) Study Group: Sepsis as a cause and consequence of acute kidney injury: Program to improve care in acute renal disease. Intensive Care Med. 2011;37:241–248. doi: 10.1007/s00134-010-2089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrell ED, Kellum JA, Pastor-Soler NM, et al. Septic acute kidney injury: Molecular mechanisms and the importance of stratification and targeting therapy. Crit Care. 2014;18:501. doi: 10.1186/s13054-014-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCullough PA, Shaw AD, Haase M, et al. Diagnosis of acute kidney injury using functional and injury biomarkers: Workgroup statements from the tenth Acute Dialysis Quality Initiative Consensus Conference. Contrib Nephrol. 2013;182:13–29. doi: 10.1159/000349963. [DOI] [PubMed] [Google Scholar]
- 5.Marino R, Struck J, Hartmann O, et al. Diagnostic and short-term prognostic utility of plasma pro-enkephalin (pro-ENK) for acute kidney injury in patients admitted with sepsis in the emergency department. J Nephrol. 2014 Dec 9; doi: 10.1007/s40620-014-0163-z. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Martensson J, Glassford NJ, Jones S, et al. Urinary neutrophil gelatinase-associated lipocalin to hepcidin ratio as a biomarker of acute kidney injury in intensive care unit patients. Minerva Anestesiol. 2014 Dec 5; Epub ahead of print. [PubMed] [Google Scholar]
- 7.Fjell CD, Thair S, Hsu JL, et al. Cytokines and signaling molecules predict clinical outcomes in sepsis. PLoS One. 2013;8:e79207. doi: 10.1371/journal.pone.0079207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bihorac A, Chawla LS, Shaw AD, et al. Validation of cell-cycle arrest biomarkers for acute kidney injury using clinical adjudication. Am J Respir Crit Care Med. 2014;189:932–939. doi: 10.1164/rccm.201401-0077OC. [DOI] [PubMed] [Google Scholar]
- 9.Prowle J, Bagshaw SM, Bellomo R. Renal blood flow, fractional excretion of sodium and acute kidney injury: Time for a new paradigm? Curr Opin Crit Care. 2012;18:585–592. doi: 10.1097/MCC.0b013e328358d480. [DOI] [PubMed] [Google Scholar]
- 10.Romanovsky A, Morgan C, Bagshaw SM. Pathophysiology and management of septic acute kidney injury. Pediatr Nephrol. 2014;29:1–12. doi: 10.1007/s00467-013-2427-6. [DOI] [PubMed] [Google Scholar]
- 11.Gomez H, Ince C, De Backer D, et al. A unified theory of sepsis-induced acute kidney injury: Inflammation, microcirculatory dysfunction, bioenergetics, and the tubular cell adaptation to injury. Shock. 2014;41:3–11. doi: 10.1097/SHK.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013;73:564–569. doi: 10.1038/pr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong HR, Cvijanovich NZ, Allen GL, et al. Corticosteroids are associated with repression of adaptive immunity gene programs in pediatric septic shock. Am J Respir Crit Care Med. 2014;189:940–946. doi: 10.1164/rccm.201401-0171OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solan PD, Dunsmore KE, Denenberg AG, et al. A novel role for matrix metalloproteinase-8 in sepsis. Crit Care Med. 2012;40:379–387. doi: 10.1097/CCM.0b013e318232e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong HR, Shanley TP, Sakthivel B, et al. Genomics of Pediatric SIRS/Septic Shock Investigators: Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong HR, Cvijanovich NZ, Anas N, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. 2014;191:309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong HR, Lindsell CJ, Pettilä V, et al. A multibiomarker-based outcome risk stratification model for adult septic shock. Crit Care Med. 2014;42:781–789. doi: 10.1097/CCM.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong HR, Salisbury S, Xiao Q, et al. The Pediatric Sepsis Biomarker Risk Model. Crit Care. 2012;16:R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HR, Cvijanovich N, Wheeler DS, et al. Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med. 2008;178:276–282. doi: 10.1164/rccm.200801-131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu RK, Standage SW, Cvijanovich NZ, et al. Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit Care. 2011;15:R273. doi: 10.1186/cc10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III–Acute Physiology Score (PRISM III-APS): A method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr. 1997;131:575–581. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- 22.Kidney Disease: Improving Global Outcomes (KDIGO) Acute Kidney Injury Work Group KDIGO clinical practice guideline for acute kidney injury. Kidney Inter Suppl. 2012;2:1–138. [Google Scholar]
- 23.Finney H, Newman DJ, Thakkar H, et al. Reference ranges for plasma cystatin C and creatinine measurements in premature infants, neonates, and older children. Arch Dis Child. 2000;82:71–75. doi: 10.1136/adc.82.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller R, Möckel M. Logistic regression and CART in the analysis of multimarker studies. Clin Chim Acta. 2008;394:1–6. doi: 10.1016/j.cca.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 25. [November 19, 2014];VassarStats: Website for Statistical Computation. Available at: http://faculty.vassar.edu/lowry/VassarStats.html.
- 26.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148:839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 27.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Available at: http://biostat.mc.vanderbilt.edu/wiki/Main/Hmisc.
- 29.Metzger J, Chatzikyrkou C, Broecker V, et al. Diagnosis of subclinical and clinical acute T-cell-mediated rejection in renal transplant patients by urinary proteome analysis. Proteomics Clin Appl. 2011;5:322–333. doi: 10.1002/prca.201000153. [DOI] [PubMed] [Google Scholar]
- 30.Basu RK, Donaworth E, Siroky B, et al. Loss of matrix metalloproteinase-8 is associated with worsened recovery after ischemic kidney injury. Ren Fail. 2015;37:469–475. doi: 10.3109/0886022X.2014.996842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csernok E, Gross WL. Current understanding of the pathogenesis of granulomatosis with polyangiitis (Wegener's). Expert Rev Clin Immunol. 2013;9:641–648. doi: 10.1586/1744666X.2013.811052. [DOI] [PubMed] [Google Scholar]
- 32.Hakkim A, Fürnrohr BG, Amann K, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu RK, Wang Y, Wong HR, et al. Incorporation of biomarkers with the renal angina index for prediction of severe AKI in critically ill children. Clin J Am Soc Nephrol. 2014;9:654–662. doi: 10.2215/CJN.09720913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zappitelli M, Parikh CR, Akcan-Arikan A, et al. Ascertainment and epidemiology of acute kidney injury varies with definition interpretation. Clin J Am Soc Nephrol. 2008;3:948–954. doi: 10.2215/CJN.05431207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.