Abstract
Cytochrome P450-dependent ω-hydroxylation is a prototypic metabolic reaction of CYP4 family members that is important for the elimination and bioactivation of not only therapeutic drugs, but also endogenous compounds, principally fatty acids. Eicosanoids, derived from arachidonic acid, are key substrates in the latter category. Human CYP4 enzymes, mainly CYP4A11, CYP4F2, and CYP4F3B, hydroxylate arachidonic acid at the omega position to form 20-HETE, which has important effects in tumor progression and on angiogenesis and blood pressure regulation in the vasculature and kidney. CYP4F3A in myeloid tissue catalyzes the ω-hydroxylation of leukotriene B4 to 20-hydroxy leukotriene B4, an inactivation process that is critical for the regulation of the inflammatory response. Here, we review the enzymology, tissue distribution, and substrate selectivity of human CYP4 ω-hydroxylases and their roles as catalysts for the formation and termination of the biological effects of key eicosanoid metabolites in inflammation and cancer progression.
Keywords: Cytochrome P450, CYP4, Inflammation, Cancer, Omega-hydroxylases, Fatty acids, Eicosanoids, Leukotrienes, 20-HETE, Arachidonic acid
1 Introduction
Omega (ω)-hydroxylation is an oxidation reaction catalyzed by cytochrome P450 (CYP) monooxygenases that transforms the terminal methyl group of a hydrophobic aliphatic chain into a more polar alcohol metabolite. Fatty acid ω-hydroxylation is the pivotal catalytic step that initiates formation of mono- and dicarboxylic acids that are then catabolized through the β-oxidation pathway. The biological ω-hydroxylation pathway was first described more than 80 years ago for medium-chain fatty acids that were metabolized to urinary dicarboxylic acids of the same chain-length (Verkade et al., 1933). These linked enzymatic processes may prevent toxic buildup of some fatty acids in the body (Hardwick, 2008).
In the early 1960s, ω-hydroxylation was localized to the microsomal fraction and shown to be dependent on NADPH and molecular oxygen (Preiss & Bloch, 1964; Wakabayashi & Shimazono, 1963). Confirmation that fatty acid ω-hydroxylation was catalyzed by CYP followed from the successful separation and reconstitution of the P450, reductase and lipid components of the enzyme system that supported lauric acid ω-hydroxylation (Lu & Coon, 1968).
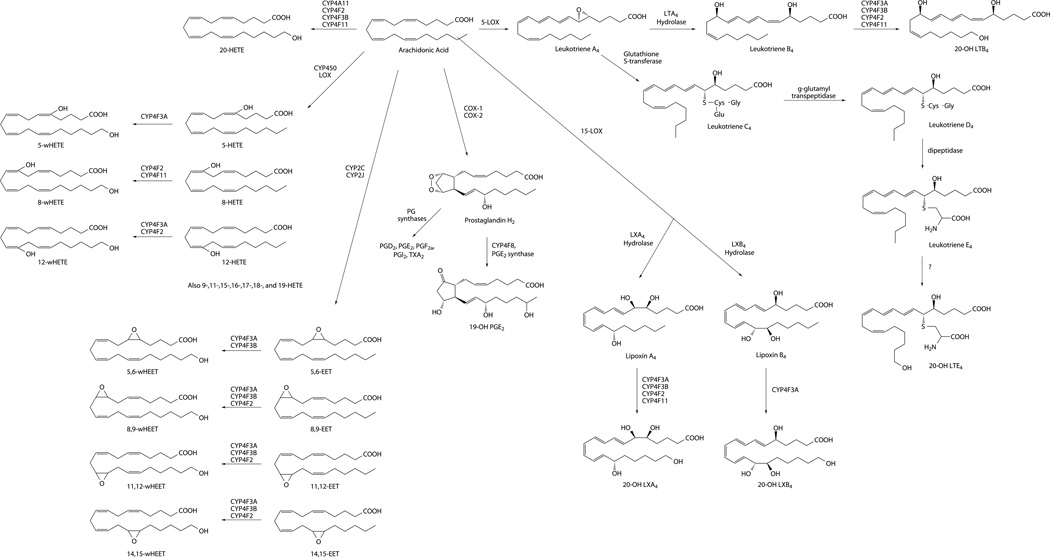
Although ω-hydroxylation is relatively a minor route in the overall catabolism of fatty acids (Draye & Vamecq, 1989), the pathway is essential for both the anabolism and catabolism of critical lipid mediators such as 20-hydroxyeicosatetraenoic acid (20-HETE) and leukotriene B4 (LTB4), respectively (Fig. 1). P450-dependent metabolism of arachidonic acid and LTB4 was established in the early 1980s (Bednar, Schwartzman, Ibraham, McGiff, & Mullane, 1984; Capdevila, Chacos, Werringloer, Prough, & Estabrook, 1981). Over the next 30 years, these ω-hydroxylation pathways have emerged as critical determinants of numerous disease processes, including inflammation and cancer progression, which are the focus of this review.
Figure 1.
Eicosanoid pathways for bioactive lipid anabolism and catabolism.
2 Physiological Roles, Multiplicity, Tissue Distribution, and Substrate Specificities of the CYP4 ω-Hydroxylases
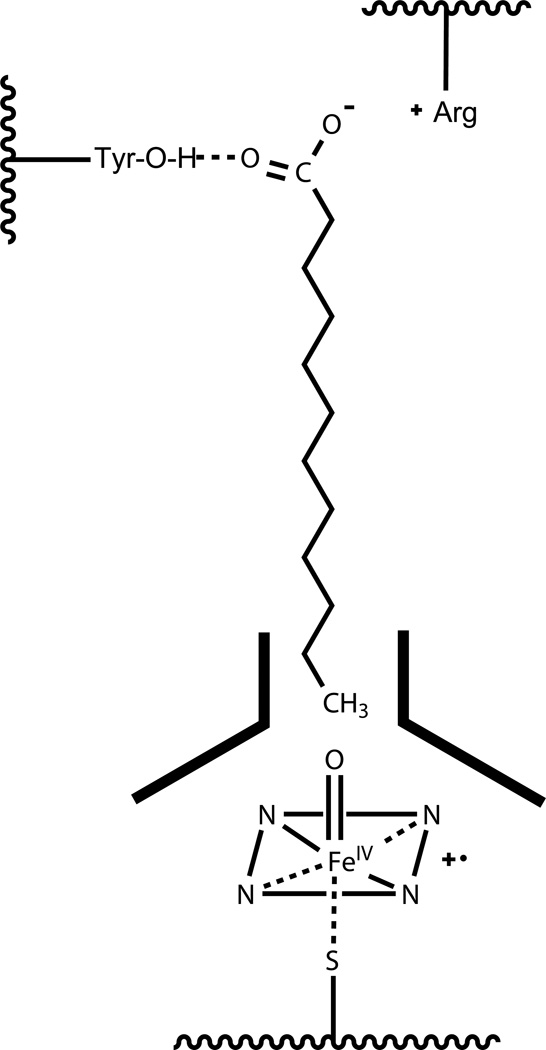
The CYP4 enzyme family play a primary physiological role in the ω-hydroxylation of endogenous fatty acids, exemplified by the eicosanoids. Several CYP4A and CYP4F enzymes generate the active signaling compound, 20-HETE, by ω-hydroxylation of arachidonic acid (Kroetz & Xu, 2005; Lasker et al., 2000; Powell, Wolf, Jin, & Lasker, 1998). On the other hand, inactivation of LTB4 via ω-hydroxylation is the defining function of neutrophil CYP4F3A (Kikuta et al., 1998). CYP4 enzymes also play important endogenous roles as ω-hydroxylases in vitamin E and vitamin K catabolism (Edson et al., 2013; McDonald, Rieder, Nakano, Hsia, & Rettie, 2009; Parker, Sontag, Swanson, & McCormick, 2004), and the metabolism of xenobiotics such as ebastine, terfenadine, pafuramidine, and fingolimod (Hashizume et al., 2002; Kovarik et al., 2009; Wang et al., 2006). The structural basis for selective ω-hydroxylation by CYP4 enzymes is thought to be a narrow channel near the heme (~ 4 Å for CYP4A1) that constrains access of only the terminal carbon to the active site iron-oxo species (He, Cryle, De Voss, & de Montellano, 2005; Lewis & Lake, 1999). A schematic of the proposed active site for CYP4A1 binding of lauric acid is shown in Fig. 2.
Figure 2.
Proposed structural requirements for selective ω-hydroxylation of fatty acids by CYP4A1.
In humans, the cytochrome P450 4 (CYP4) family consists of 12 genes and 13 enzymes divided into 6 subfamilies: CYP4A, CYP4B, CYP4F, CYP4V, CYP4X, and CYP4Z (Table 1). At the gene level, CYP4A, B, X, and Z are clustered on chromosome 1, while the CYP4F and CYP4V genes reside on chromosome 19 and 4, respectively (Nelson et al., 2004). While the CYP4s represent one of the largest human P450 families, only a subset of these enzymes has significant ω-hydroxylase activity toward eicosanoids.
Table 1.
Human CYP4 Enzymes: Tissue Distribution and Fatty Acid ω-Hydroxylase Activities
| Enzyme | Tissue Distribution | Substrates |
|---|---|---|
| CYP4A11 | Liver, kidney | AA, ω3 PUFA, MCFA, LCFA |
| CYP4A22 | Liver, kidney | ? |
| CYP4B1 | Lung, bladder | Inactive in human, SCFA in other species |
| CYP4F2 | Liver, kidney | LTB4, AA, MCFA, LCFA, VLCFA, LX, EET, HETE |
| CYP4F3A | Myeloid tissues | LTB4, ω3 PUFA, MCFA, LCFA, LX, EET, HETE |
| CYP4F3B | Liver, kidney | AA, ω3 PUFA, MCFA, LCFA, VLCFA, LTB4, EET |
| CYP4F8 | Urogenital tissues | PGH1/2 (internal oxidations) |
| CYP4F11 | Liver, kidney | MCFA, LCFA (3-hydroxy analogs), AA, LTB4, LX, HETE |
| CYP4F12 | Liver, gut, kidney | AA, PGH1/2 (internal oxidations) |
| CYP4F22 | Skin | Hepoxilins?, acylceramides? |
| CYP4V2 | Retina | ω3 PUFA, AA, MCFA |
| CYP4X1 | Brain, bronchial airways | No reported ω-hydroxylase activity |
| CYP4Z1 | Mammary tissue | MCFA (internal oxidations), AA? |
AA, arachidonic acid; LTB4, leukotriene B4; PGH1/2, prostaglandin H1/2; SCFA (C7–C9), short-chain fatty acid; MCFA (C10–C16), medium chain fatty acid; LCFA (C17–C21), long-chain fatty acid; VLCFA (C22–C26), very long-chain fatty acid; ω3 PUFA, omega 3 polyunsaturated fatty acid; LX, lipoxin; HETE, hydroxyeicosatrienoic acid; EET, epoxyeicosatrienoic acid.
Information taken from references throughout the text.
The most extensively studied human CYP4 fatty acid ω-hydroxylases are CYP4A11, CYP4F2, CYP4F3A, and CYP4F3B. CYP4A11 (ortholog of rat CYP4A1) is regulated by peroxisome proliferator-activated receptor-α (PPARα), expressed at high levels in liver and kidney, and catalyzes the metabolism of lauric and arachidonic acids with high regioselectivity at the ω-position (Johnson, Palmer, Griffin, & Hsu, 1996). CYP4F2 is also expressed at significant levels in liver and kidney, and although it is not regulated by PPARα, it is induced by some statins that signal through the sterol regulatory element-binding protein (SREBP) (Hsu, Savas, Griffin, & Johnson, 2007b).
CYP4F3 is an unusual human P450 gene in that it undergoes tissue-specific alternative splicing to CYP4F3A and CYP4F3B. The substrate specificities of CYP4F3A and CYP4F3B are determined by the incorporation of either exon 4 or exon 3, respectively, into the translated enzyme (Christmas, Ursino, Fox, & Soberman, 1999). CYP4F3A is expressed in neutrophils and has the highest affinity of all the CYP4F isoforms for LTB4. In contrast, CYP4F3B is primarily expressed in human liver and kidney and has greater activity toward arachidonic acid and omega 3 polyunsaturated fatty acids (ω3 PUFAs) (Christmas et al., 2001; Fer et al., 2008). CYP4F3 enzymes are also reported to be the major ω-hydroxylases of long-chain fatty acid epoxides (Le Quere, Plee-Gautier, Potin, Madec, & Salaun, 2004).
CYP4F8, CYP4F22, and CYP4V2 are all expressed primarily in extrahepatic tissues (Bylund, Hidestrand, Ingelman-Sundberg, & Oliw, 2000; Kelly, Nakano, Rohatgi, Yarov-Yarovoy, & Rettie, 2011), although only CYP4V2 exhibits clear fatty acid ω-hydroxylase activity (Nakano, Kelly, & Rettie, 2009; Nakano, Kelly, Wiek, Hanenberg, & Rettie, 2012). More information is available for CYP4F11 and CYP4F12, both of which are expressed primarily in drug clearance organs, including the liver. Whereas CYP4F12 can metabolize a diverse array of bulky xenobiotics, similar to CYP3A4 (Eksterowicz, Rock, Rock, Wienkers, & Foti, 2014), CYP4F11 is associated with the ω-hydroxylation of endogenous substrates including longer chain fatty acids and vitamin K (Dhar, Sepkovic, Hirani, Magnusson, & Lasker, 2008; Edson et al., 2013).
CYP4X1 and CYP4Z1 are extrahepatic P450s, with sparse information about their biological function. Human CYP4X1 is highly expressed in the brain, skin, and airways, and although it is regulated by PPARα, the enzyme is not a recognized fatty acid ω-hydroxylase, generating only epoxide metabolites of arachidonic acid at low levels (Savas, Hsu, Griffin, Bell, & Johnson, 2005; Stark, Dostalek, & Guengerich, 2008). CYP4Z1 is expressed in mammary tissue and is inducible by glucocorticoids and progesterone (Rieger et al., 2004; Savas et al., 2005). The regioselectivity of CYP4Z1-dependent fatty acid metabolism is strikingly different from other ω-hydroxylases discussed so far in that the recombinant enzyme is a mid-chain hydroxylase of medium-chain fatty acids, adding a hydroxyl group to positions ω-2 to ω-5 (Zollner et al., 2009). Finally, CYP4A22 and CYP4B1 are expressed at very low levels and/or are inactive due to poor enzyme stability (Hsu, Savas, Griffin, & Johnson, 2007a; Wiek et al., 2015).
3 Eicosanoid Pathways for Bioactive Lipid Anabolism and Catabolism
Eicosanoids are lipid mediators derived from arachidonic acid by the actions of cyclooxygenases (COXs), lipoxygenases (LOXs), and CYPs (Fig. 1) that together make up complex signaling networks of over 20 chemical messengers implicated in critical biological actions in practically every tissue, organ, and cell in the body (Funk, 2001).
3.1 COX Pathways
Prostaglandins (PGs) are autocrine and paracrine lipid mediators, synthesized from arachidonic acid by the dual-function cyclooxygenase enzymes, COX-1 and COX-2. COX-1 is constitutively expressed in most cell types, whereas COX-2 expression is generally induced in response to proinflammatory stimuli (Smith, DeWitt, & Garavito, 2000; Vane, Bakhle, & Botting, 1998), though both COX enzymes are involved in homeostatic- and inflammation-induced prostanoid synthesis (Seibert et al., 1997). COXs initially metabolize arachidonic acid, via their COX function, to unstable prostaglandin G2 (PGG2) that is then converted by their peroxidase function to prostaglandin H2 (PGH2). PGH2 serves as a substrate for cell- and tissue-selective prostanoid synthases and isomerases that produce numerous bioactive prostanoids: prostaglandins D2, E2, F2α, I2, and thromboxane A2. Differential expression of these synthases determines the prostanoid profile of a given tissue (Naraba et al., 1998; Smith & Song, 2002). Prostanoids are widely distributed throughout the body and exert diverse physiological functions and responses. Their biosynthesis, mechanism of action, and role in health, inflammation and disease progression have been reviewed in detail elsewhere (Ricciotti & FitzGerald, 2011; Smyth, Grosser, Wang, Yu, & FitzGerald, 2009).
3.2 LOX Pathways
Of the various eicosanoids produced by the LOX pathway of arachidonic acid metabolism, the proinflammatory leukotrienes (LTs) are arguably the most significant in terms of their function in disease pathogenesis. Unlike PGs, which are synthesized by various cell types, LTs are predominantly formed in inflammatory cells such as polymorphonuclear leukocytes (PMNL), macrophages, and mast cells (Funk, 2001). 5-LOX catalyzes the epoxidation of arachidonic acid to leuktotriene A4 (LTA4), via the intermediacy of 5-hydroperoxyeicosatetraenoic acid (5-HPETE). LTA4 then undergoes enzymatic hydrolysis to LTB4 or conjugation with glutathione to form the cysteinyl-leukotrienes: LTC4, LTD4, and LTE4 (Fig. 1). LTB4 is a potent neutrophil chemotactic agent and a key mediator in inflammation, while the cysteinyl-LTs are involved in the contraction of smooth muscle to propagate the inflammatory response (Peters-Golden & Henderson, 2007; Samuelsson, Dahlen, Lindgren, Rouzer, & Serhan, 1987). Aside from LT formation, 5-, 12-, and 15-LOX can also convert arachidonic acid into the proinflammatory 5-, 12-, and 15-HETEs (Powell & Rokach, 2015). Additionally, 15-LOX synthesizes the lipoxins (LXs), arachidonic acid metabolites that act as important mediators in the resolution of inflammation (McMahon, Mitchell, Brady, & Godson, 2001; Serhan, Chiang, Dalli, & Levy, 2015). The biosynthesis, enzymology, and biological action of the LOX-derived arachidonic acid metabolites have been extensively reviewed elsewhere (Haeggstrom & Funk, 2011; Murphy & Gijon, 2007).
3.3 P450 Pathways
The CYP monooxygenases form two series of bioactive oxidized lipids from arachidonic acid (Capdevila & Falck, 2002; Konkel & Schunck, 2011); four regioisomeric epoxyeicosatrienoic acids (EETs), 14,15-, 11,12-, 8,9- and 5,6-EET; and eleven mono-HETEs, 5-, 8-, 9-, 11-, 12-, 15-, 16-, 17-, 18-, 19-, and 20-HETE (Fig. 1). Whereas human CYP2C and CYP2J enzymes are the principal catalysts of EETs formation, the internal HETEs are formed by a wide variety of P450s (Kaspera & Totah, 2009). CYP4 enzymes dominate 20-HETE formation and catabolism of LTB4. The P450-derived eicosanoids exhibit a vast array of biological functions, although much of the research emphasis for the HETEs (and EETs) has been focused on the vascular (Fleming, 2008) and renal systems (Fan, Muroya, & Roman, 2015).
4 ω-Hydroxylases in Inflammation
Eicosanoid metabolites of arachidonic acid, including PGs, LTs, and 20-HETE, are key proinflammatory mediators of the inflammatory cascade (Khanapure, Garvey, Janero, & Letts, 2007). P450 ω-hydroxylases are linked to inflammation, largely through catabolism of the LTs and formation of the proinflammatory metabolite 20-HETE. It is well established that members of the CYP4F family function in the resolution phase of inflammation and in the ω-hydroxylation and deactivation of LTB4 (Kalsotra & Strobel, 2006; Kikuta, Kusunose, & Kusunose, 2000). However, CYP4s may also play a role in the activation and amplification of inflammation through catabolism of other anti-inflammatory eicosanoids, such as the EETs and LXs (Kalsotra & Strobel, 2006; Kikuta, Kusunose, & Kusunose, 2002). In this section, we explore this dual role for P450 ω-hydroxylases in inflammation by reviewing eicosanoid metabolism catalyzed by the CYP4 family and examining what is known about the expression and regulation of CYP4 genes during inflammation to better understand how these genes function in inflammatory diseases.
4.1 LT ω-Hydroxylases
LTB4 and the cysteinyl-LTs, derived from arachidonic acid via the LOX pathway, are important proinflammatory mediators. LTs increase leukocyte tissue infiltration and amplify inflammation through production of cytokines and chemokines. LTB4 is a potent chemotactic agent for neutrophils and other leukocytes and promotes expression of cell adhesion proteins. The cysteinyl-LTs mediate contraction of smooth muscle thereby increasing venopermeability to enhance recruitment of leukocytes. Numerous studies have demonstrated that elevated levels of LTB4 and cysteinyl-LTs are linked to the pathogenesis of several inflammatory diseases, including asthma, Alzheimer's disease, rheumatoid arthritis, psoriasis, diabetes, cardiovascular disease, and cancer, among others (Peters-Golden & Henderson, 2007). Inactivation and elimination of LTs, especially LTB4, is therefore expected to be a crucial part of the resolution of inflammation and return to normal homeostasis. While metabolism of LTB4 is complex involving multiple enzymatic pathways, ω-hydroxylation is established as the primary route of inactivation. Conversion of LTB4 to its 20-hydroxy metabolite (20-OH LTB4) results in a dramatic loss of its chemotactic activity on leukocytes. 20-OH LTB4 is rapidly oxidized to 20-COOH LTB4 and targeted for β-oxidation or conjugation with a glucuronide (Kikuta et al., 1993).
ω-Hydroxylase function in inflammation can be traced back to the discovery of the CYP4F family in the 1980s through a series of studies aimed at identifying a deactivation pathway of LTB4 in human PMNL. Experiments with 18O2 revealed that the terminal hydroxyl group in a newly identified ω-hydroxylated metabolite of LTB4 (20-OH LTB4) was derived from molecular oxygen (Hansson, Lindgren, Dahlen, Hedqvist, & Samuelsson, 1981). Subsequently, it was shown that ω-hydroxylase activity in PMNL was confined to the microsomal fraction and could be inhibited by carbon monoxide (Shak & Goldstein, 1984). Inhibition of this activity by carbon monoxide and by antibodies against P450 reductase indicates catalysis by a P450 enzyme specific for LTB4 ω-hydroxylation. This led to the cloning, expression, and identification of a novel LTB4 ω-hydroxylase, CYP4F3A (Kikuta et al., 1993, 1998).
Human CYP4F2 and CYP4F3B, the major LTB4 ω-hydroxylases expressed in human liver, play a central role in the inactivation and elimination of LTs from systemic circulation during the resolution of inflammation. The eicosanoid substrate selectivity and activity of the CYP4F subfamily as LTB4 ω-hydroxylases varies considerably. Neutrophil-specific CYP4F3A has a much higher affinity for LTB4 (Km = 0.64 µM) than either CYP4F2 (Km = 47 µM) or CYP4F3B (Km = 21 µM) (Christmas et al., 2001; Kikuta et al., 1998). CYP4F11 also possesses LTB4 ω-hydroxylase activity, though much less than that of the other CYP4Fs (Kalsotra, Turman, Kikuta, & Strobel, 2004). Early reports indicated that CYP4A11, purified from human liver microsomes, exhibited negligible ω-hydroxylase activity toward LTB4 (Jin, Koop, Raucy, & Lasker, 1998), although CYP4A11 protein expression correlated with formation of 20-OH LTB4. However, a more recent study demonstrated that CYP4A11 Supersomes™ generated 20-OH LTB4 at a rate of 1.8 nmol/min/nmol P450 (Le Quere et al., 2004), which is ~ 20-fold lower than catalysis by CYP4F3A (Kikuta et al., 1998). Other CYP4F family members also generate 17-, 18-, and 19-OH LTB4 (Berry, Borgeat, Gosselin, Flamand, & Murphy, 2003). Further studies are needed to determine if these latter LT metabolites have any biological activity. It is still not known which P450s ω-hydroxylate and inactivate cysteinyl-LTs, although early work demonstrated ω-oxidation of LTE4 in rat liver microsomes (Orning, 1987), and observed this metabolite in the urine of human subjects (Sala, Voelkel, Maclouf, & Murphy, 1990).
4.2 PG ω-Hydroxylases
PGs are proinflammatory metabolites of arachidonic acid, synthesized de novo at sites of injury by the COX pathway. PGs are largely responsible for the cardinal symptoms of inflammation: rubor (redness), calor (heat), tumor (swelling), and dolor (pain) (Ricciotti & FitzGerald, 2011). While PG metabolism is expected to be a key component of inflammation resolution, human P450 ω-hydroxylases do not appear to contribute significantly to this process. Several studies demonstrated that rabbit and rodent CYP4A isoforms can ω-hydroxylate PGs (Masters et al., 1993; Roman et al., 1993). However, human CYP4A11 does not exhibit PG ω-hydroxylase activity, although it does ω-hydroxylate PGH2 analogs (Oliw, Stark, & Bylund, 2001). CYP4F8, largely expressed in prostate and seminal vesicles, is not a PG ω-hydroxylase but catalyzes the formation of 19-OH PGE2 (Bylund et al., 2000).
4.3 Arachidonic Acid and ω-Hydroxylases
20-HETE plays a role in inflammation by stimulating the production of various proinflammatory mediators: PGE2, cytokine tissue necrosis factor alpha (TNFα), and the chemokines IL-8, IL-12, IL-14 (Ishizuka et al., 2008). CYP4A11, CYP4F2, CYP4F3B, and CYP4F11 all catalyze formation of 20-HETE and are expressed in the liver and kidney, both major sites of 20-HETE synthesis (Christmas et al., 2001; Hsu et al., 2007a; Kalsotra et al., 2004). CYP4F2 is the major catalyst of 20-HETE formation in human liver and kidney microsomes (Lasker et al., 2000; Powell et al., 1998). Production of 20-HETE by the CYP4A and CYP4F enzymes therefore suggests a role for ω-hydroxylases in the activation of inflammation, which may exacerbate 20-HETE's role in tumor progression (see Section 5). In addition to CYP4Fs catalyzing the formation of 20-HETE, these enzymes also ω-hydroxylate and deactivate proinflammatory 5-, 8-, and 12-HETE (Kalsotra & Strobel, 2006; Kikuta et al., 2002), again suggestive of dual roles for the P450 ω-hydroxylases in both the initiation and resolution phases of inflammation.
4.4 EET ω-Hydroxylases
The EETs mediate vasodilation of the vasculature and are endowed with anti-inflammatory properties, including prevention of cytokine-induced leukocyte adhesion to endothelial cells through inhibition of NF-κB, reducing platelet aggregation, and reducing smooth muscle cell migration (Fleming, 2008; Node et al., 1999). Catabolism of EETs proceeds primarily through soluble epoxide hydrolase (sEH) which catalyzes metabolism to the corresponding less active vicinal diol, dihydroxyeicosatrienoic acid (DHET) (Kaspera & Totah, 2009). The ratio of circulating 20-HETE:EETs + DHETs in plasma is usually used as an indicator of the relative rates of hydroxylation versus epoxidation of arachidonic acid, and therefore a gauge for inflammatory status (Theken et al., 2011).
EETs themselves can be ω-hydroxylated by CYP4 enzymes to their respective 20-hydroxyepoxyeicosatrienoic acids (HEETs) (Le Quere et al., 2004). Interestingly, human liver microsomes exhibit the highest ω-hydroxylase activity toward 11,12-EET, whereas 8,9-EET was the best substrate for recombinant CYP4F2, CYP4F3A, and CYP4F3B enzymes. It is not known whether other CYP4Fs or CYP4A11 can catalyze formation of ω-HEETs, although several rat CYP4A isoforms have demonstrated ω-hydroxylation activity toward the various EETs (Cowart et al., 2002), which is consistent with studies showing that ω-HEETs have high affinity toward PPARα and induce peroxisome proliferation in rodents (Gatica et al., 2007). It remains unclear whether ω-hydroxylation of EETs in humans is another deactivation process or a means of synthesizing biologically active compounds, as it is in rodents, as peroxisome proliferation is mediated through suppression of HNF4α and not by PPARα in humans (Hertz, Sheena, Kalderon, Berman, & Bar-Tana, 2001).
4.5 LX ω-Hydroxylases
LXs are anti-inflammatory metabolites of arachidonic acid, whose formation is catalyzed by 15-LOX, although they can also be synthesized from LTA4 through transcellular metabolism (Gronert, Clish, Romano, & Serhan, 1999). LXs play an important role in the resolution phase of inflammation by decreasing leukocyte infiltration and cell adhesion, promoting macrophage clearance of apoptotic cells, and modulating the production of proinfammatory cytokines (Haeggstrom & Funk, 2011; McMahon et al., 2001). While the Km values for LXs are several fold higher than that for LTB4, CYP4F3A quite efficiently catalyzes ω-hydroxylation of both LXA4 and LXB4 but shows much higher affinity toward LXB4 as a substrate (Kikuta et al., 1998). In contrast, CYP4F2 can ω-hydroxylate only LXA4, with similar affinity and rate of formation as LTB4 (Kikuta et al., 2000). The resolution of inflammation is an active process that involves the biosynthesis of proresolving mediators, in addition to catabolism of proinflammatory mediators. Failure of acute inflammation to resolve may lead to excessive tissue damage, chronic inflammation, and propagation of the autoimmune response. The ω-hydroxylation of the LXs by members of the CYP4F family suggests differential roles for these enzymes: promotion of inflammation resolution through deactivation of LTB4 and prolongation of inflammation through LX catabolism.
4.6 ω3 PUFA ω-Hydroxylases
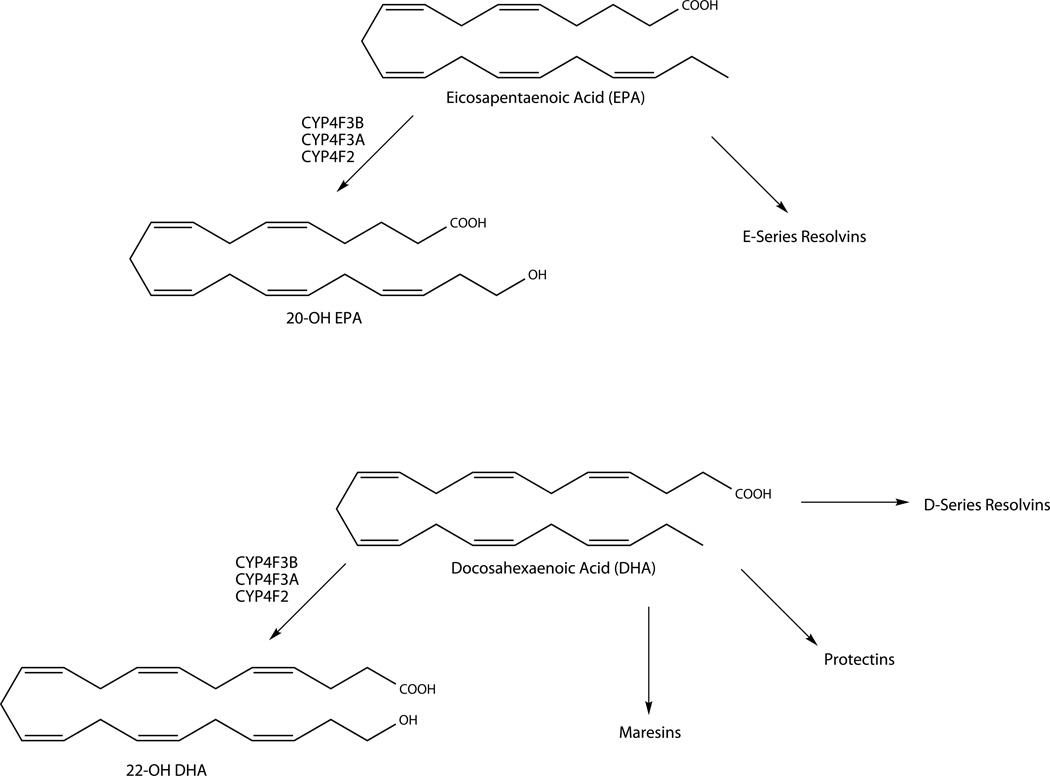
ω3 PUFAs, especially eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), have long been touted as human dietary supplements for their beneficial impact in a wide range of diseases with an inflammatory etiology (Simopoulos, 2002). However, the molecular mechanisms by which they exert their anti-inflammatory and cardioprotective effects are poorly understood. EPA and DHA are precursors for resolvins (Rvs), protectins and maresins (Fig. 3), lipid mediators with potent anti-inflammatory and proresolving properties. Similar to the LXs, Rvs and protectins block leukocytic infiltration, modulate production of cytokines and other proinflammatory mediators, and promote macrophage uptake of apoptotic cells and clearance of cellular debris. Several recent reviews are available detailing the biosynthesis, regulation, and biological actions of this class of compounds (Serhan & Chiang, 2013; Serhan et al., 2015; Serhan & Petasis, 2011).
Figure 3.
Metabolism of ω3 PUFAs by P450 ω-hydroxylases.
Direct effects of the CYP4 enzymes on protectins and Rvs appear to be minimal. There is an isolated report that human neutrophils can ω-hydroxylate RvE1 to 20-OHRvE1 (Arita et al., 2006), but the enzyme that catalyzes the reaction was not identified. It is also not clear if this is a true inactivation pathway, as 20-OHRvE1 retained some of the original RvE1 activity. However, P450 ω-hydroxylases may exert an indirect (proinflammatory) effect through reducing the biosynthesis of Rvs and protectins through ω-hydroxylation of the parent compounds EPA and DHA (Fer et al., 2008). CYP4F3B can ω-hydroxylate EPA and DHA to their 20-hydroxy and 22-hydroxy metabolites, respectively (Harmon et al., 2006). Evidence was presented in the same study that 20-OH EPA and 22-OH DHA may function as lipid mediators rather than degradation products, much like 20-HETE, as these two metabolites were able to bind and activate PPARα in Cos-7 cells. However, the biological actions of these two ω-hydroxylated products are not well characterized.
Addition of EPA or DHA to incubations of arachidonic acid with each of the CYP4F enzymes resulted in decreased 20-HETE formation, suggesting that EPA and DHA compete with arachidonic acid as substrates for ω-oxidation (Fer et al., 2008; Harmon et al., 2006). In rats supplemented with dietary EPA/DHA, substantial replacement of arachidonic acid by EPA and DHA in membrane phospholipids of various organs was observed, providing in vivo evidence that EPA and DHA may compete with arachidonic acid for ω-hydroxylation by P450 (Arnold et al., 2010). In humans, supplementation with DHA/EPA leads to a change in the erythrocyte membrane fatty acid composition with DHA and EPA replacing arachidonic acid in phospholipids (Dawczynski et al., 2013). Conceivably, incorporation of EPA and DHA into membrane phospholipids may prevent conversion of arachidonic acid to the proinflammatory LTs and PGs by serving as alternative substrates for the COX and LOX enzymes to form docosanoids and related products (Fig. 3). Inhibition of arachidonic acid metabolism to proinflammatory mediators is potentially another mechanism by which ω3 PUFAs may exert their beneficial effects.
4.7 Gene Regulation of ω-Hydroxylases in Inflammation
CYP4F2, CYP4F3A, and CYP4F3B enzymes appear to be the primary P450 ω-hydroxylases involved in the inflammatory response, therefore understanding how these CYP4F genes are regulated during inflammation may provide insight into mechanisms involved in the activation and progression of inflammatory diseases. While regulation of the major drug-metabolizing P450s during inflammation and disease has been studied extensively, leading to reduced expression of the majority of P450s (Harvey & Morgan, 2014), little is known regarding the regulation of human CYP4F genes during inflammation. Studies in rat hepatocytes demonstrated induction of CYP4F genes after treatment with proinflammatory cytokines, TNFα, interleukin (IL)-1β, and IL-6, but downregulation of CYP4Fs by an anti-inflammatory cytokine IL-10 (Kalsotra et al., 2007). This is consistent with the view that cytokine-mediated suppression or induction of gene transcription is the primary mechanism for P450 regulation. However, identification of the specific nuclear receptors and transcription factors that mediate these effects has remained elusive, and analogous studies with human CYP4Fs are absent from the literature.
Multiple studies showed that retinoids, specifically 9-cis- and all-trans-retinoic acid (atRA), induce expression of CYP4F2 and 20-OH LTB4 formation in HepG2 cells through the retinoid X receptor (RXR) (Zhang, Chen, & Hardwick, 2000; Zhang & Hardwick, 2000). Strobel and coworkers found that human keratinocytes can ω-hydroxylate LTB4 and that this activity is increased following induction of CYP4F2, CYP4F3A, and CYP4F3B at both the transcript and protein levels by atRA activation of RXR, although the nuclear receptor partner of RXR remains unidentified (Kalsotra et al., 2008). A later study also confirmed that topical administration of retinoids to human skin samples enhanced CYP4F-mediated LTB4 ω-hydroxylation, suggesting one way by which retinoid therapy might exert its anti-inflammatory effects in disorders such as psoriasis and atopic dermatitis (Du, Yin, Morrow, Strobel, & Keeney, 2009). Induction of CYP4F2 has been found to proceed through other pathways in human hepatocytes and HepG2 cells, as well. CYP4F2-specific expression is induced by lovastatin through SREBP (Hsu et al., 2007b) and through an AMP-activated protein kinase pathway by genistein and resveratrol (Hsu, Savas, Lasker, & Johnson, 2011). Understanding how proinflammatory cytokines affect these pathways may help explain how CYP4F2, at least, is regulated during inflammation.
Much of the information regarding CYP4F gene regulation during inflammation has evolved from the use of mouse and animal models administered bacterial endotoxin lipopolysaccharide (LPS) or barium sulfate to induce an acute inflammatory response. However, the use of animal models in understanding human CYP4F gene expression during inflammation is limited by the fact that it is difficult to assign rodent orthologs of human CYP4F isoforms. Moreover, mechanisms of regulation by inflammatory mediators appear to be species specific. For instance, in a murine LPS-induced neuroinflammation model, pretreatment with the peroxisomal proliferator fenofibrate attenuated the inflammatory response and offered neuroprotection through induction of specific mouse Cyp4fs, an effect that was significantly reduced by the CYP4 inhibitor 17-ODYA (17-octadecynoic acid) (Sehgal et al., 2011). In contrast, CYP4F2 expression in HepG2 cells was suppressed by clofibrate, ciprofibrate, and WY14,643 (Zhang et al., 2000). This complements in vivo evidence demonstrating that TNFα and IL-6 increase in response to fibrates in rodents (Rose et al., 1999; Rusyn, Tsukamoto, & Thurman, 1998), while levels of these proinflammatory cytokines decrease in human serum (Madej et al., 1998; Staels et al., 1998).
Additionally, CYP4F enzymes may be differentially regulated by the type of inflammatory insult, as evidenced by studies with murine Cyp4a enzymes. Murine hepatic Cyp4a mRNAs are typically downregulated in response to both LPS and Citrobacter rodentium infection models of inflammation. However, Cyp4a10 and Cyp4a14 knockout mice afforded protection and anti-inflammatory effects, such as reduced proinflammatory cytokine production, only in response to C. rodentium infection but not to LPS injection compared to wild-type mice (Nyagode, Williams, & Morgan, 2014). Similar gene-deletion studies of murine Cyp4f isoforms have not yet been performed. In a rodent model of septic shock, administration of a synthetic 20-HETE mimetic to rats following LPS injection, resulted in protection against inflammation, tachycardia, and hypotension, with attenuation of nitric oxide synthase activity in the heart and kidney being one of the most marked observations (Sari et al., 2014; Tunctan et al., 2013). During inflammation, there is often enhanced release of nitric oxide, which can inhibit 20-HETE formation by CYP ω-hydroxylases (Wang et al., 2003), leading to the resultant hypotension in septic shock. Thus, while CYP4F involvement was not directly investigated in the foregoing examples, study of the tissue-specific regulation of CYP4F enzymes in response to different inflammatory insults appears warranted.
Nonetheless, animal models have proven useful for establishing that individual CYP4F genes are differentially regulated in a tissue- and time-dependent manner. Multiple studies that examined the inflammatory response and CYP4F expression in skin, brain, and lung tissue showed an initial suppression of specific CYP4F isoforms for 12–24 h, then induction, followed by a gradual return to basal levels over a period ranging from hours to days to weeks (Birnie, Morrison, Camara, & Strauss, 2013; Du et al., 2009; Kalsotra et al., 2008; Stoilov et al., 2006; Wang et al., 2008). These changes in CYP4F enzyme expression coincided with an initial increase of LTB4 in the tissues analyzed, consistent with activation and amplification of inflammatory response, followed by a decrease in LTB4 as CYP4F expression increased in the resolution phase. In mice, Cyp4f regulation was found to be tissue- and isoform specific, in addition to being time dependent. P450 transcripts were analyzed in various mouse tissues for up to 48 h following LPS administration. While induction of Cyp4f16 mRNA expression was observed at 24 h in mouse liver, kidney, and heart, Cyp4f13 expression was induced in kidney and heart but suppressed in liver (Theken et al., 2011). Moreover, in mouse liver, the time course of Cyp4f13 expression corresponded to 20-HETE levels, suggesting that Cyp4f13 functions in the initiation phase of inflammation (20-HETE formation), while Cyp4f16 is active in the resolution of inflammation (LTB4 ω-hydroxylation). Downregulation of Cyp2c and Cyp2j transcripts and suppression of EET and DHET formation during the initial 24 h following LPS treatment were observed as well, resulting in an increase in 20-HETE:EET + DHET compared to basal levels. These effects were attenuated at 48 h, indicating a time dependent, anti-inflammatory switch toward resolution of inflammation, suggestive of cross talk in the regulation pathways of epoxygenase and ω-hydroxylase enzymes throughout the inflammatory response. Collectively, these results indicate that CYP4Fs are temporally regulated in an isoform- and tissue-specific manner and have differential roles in the initiation and resolution phases of inflammation.
5 ω-Hydroxylases in Cancer
In contrast to the inflammatory response where ω-hydroxylation of numerous eicosanoids may impinge on the processes of activation and resolution, consequences for eicosanoid ω-hydroxylation in cancer have largely been focused on 20-HETE. This arachidonic acid metabolite initially gained pharmacological interest for its potent vasoconstrictor activity in rat aorta (Schwartzman, Falck, Yadagiri, & Escalante, 1989) and inhibition of sodium transport in renal proximal tubules (Quigley, Baum, Reddy, Griener, & Falck, 2000). In recent years, new attention has been paid to the role of CYP4A/4F-derived arachidonic acid metabolites in cancer (Alexanian & Sorokin, 2013; Panigrahy, Kaipainen, Greene, & Huang, 2010). In this section, we examine the effects of cancer on P450 enyzme activity, CYP4 enzymes as potential cancer biomarkers and review the role for CYP4-mediated formation of 20-HETE in cancer progression.
5.1 P450 Expression and Activity in Cancer
P450-dependent drug elimination is typically reduced in cancer due to an increase in inflammatory cytokines that decrease transcription of the major drug-metabolizing enzyme, CYP3A4, in normal human liver (Harvey & Morgan, 2014; Rivory, Slaviero, & Clarke, 2002). In cancerous tissues, expression of most xenobiotic-metabolizing P450 enzymes, other than CYP1B1 and CYP2J2, is substantially reduced (or even absent) relative to neighboring noncancerous tissue (Forrester et al., 1990; Murray et al., 1997). In contrast, several CYP4 enzymes that are associated with the ω-hydroxylation of endogenous substrates are increased in cancer cells (Alexanian & Sorokin, 2013).
5.2 P450s as Biomarkers in Cancer
A relationship between P450 ω-hydroxylases and cancer can be traced back to the early 1980s when peroxisome proliferators were shown to cause liver cancer in rodents (Reddy, Azarnoff, & Hignite, 1980) with accompanying upregulation of other targets of PPARα, including microsomal CYP4A ω-hydroxylases and mitochondrial and peroxisomal enzymes that regulate fatty acid catabolism (Johnson et al., 1996). Pretreatment of rats with clofibrate facilitated the purification of the then novel “cytochrome P-452” (Gibson, Orton, & Tamburini, 1982), now termed CYP4A1, which preferentially hydroxylates lauric and palmitic acids at their ω-terminus (Aoyama et al., 1990), very similar to the regioselectivity exhibited by human CYP4A11 (Hardwick, 2008). In subsequent years, much effort was devoted to investigating the relationship between target gene induction and genotoxicity, especially given that P450 metabolites may conceivably play a role in the pathophysiology of liver cancer. However, today, we recognize that while peroxisome proliferator-mediated gene induction and liver cancer in rodents is critically dependent on PPAR transactivation (Gonzalez & Shah, 2008), the concomitant upregulation of CYP4A enzymes is merely a biomarker of exposure to these ubiquitous environmental agents. A search in the early 1990s for new CYP4A genes that could be involved in carcinogenesis led to the discovery of the CYP4F subfamily. In rat liver tumors induced by a variety of chemical carcinogens, CYP4A gene expression and associated laurate ω-hydroxylase activities were decreased, but Northern blotting revealed a new gene (CYP4F1) that was expressed at high levels, especially in tumors induced with aflatoxin B1 (Chen & Hardwick, 1993).
In human thyroid, ovarian, breast, and colon cancer tissues, CYP4F2 and CYP4A11 expression is generally upregulated at the mRNA level, although interindividual differences are large (Alexanian, Miller, Roman, & Sorokin, 2012). Higher levels of CYP4F2 protein were confirmed in several tumor samples by Western blotting. Upregulation of mRNA for CYP4A11, CYP4F2, and CYP4F3 has also been reported in pancreatic ductal adenocarcinoma, with the suggestion that these ω-hydroxylases might be used as distinguishing markers in pancreatic pathology (Gandhi et al., 2013).
CYP4Z1 is selectively expressed in mammary tissue and also appears to be upregulated in breast cancer tissue (Rieger et al., 2004). Recently, CYP4Z1 was identified as a potential biomarker of poor prognosis in prostate cancer (Tradonsky et al., 2012). In an extensive series of studies by Murray and colleagues, immunohistochemical analysis of cancer tissue microarrays highlighted CYP4Z1 expression in both breast and ovarian cancer (Downie et al., 2005; Murray, Patimalla, Stewart, Miller, & Heys, 2010), but not colon cancer (Kumarakulasingham et al., 2005). CYP4V2 and CYP4X1 expression has also been correlated with breast tumor grade (Murray et al., 2010). These immunohistochemical studies employed highly selective P450 antibodies against more than 20 P450 enzymes, but unfortunately, the panel did not include antibodies for CYP4A and CYP4F enzymes.
5.3 ω-Hydroxylases and Cancer Progression
Local and distant metastases are the cause of 90% of human mortality to cancer (Weigelt, Peterse, & van 't Veer, 2005). A key event in tumor metastasis is neovascularization and associated angiogenesis (Folkman, 1971). Angiogenesis—the growth of new blood vessels from vascular endothelial cells—is critical to the tumor microenvironment that must provide nutrients and oxygen to support metastatic progression. Angiogenesis results from an imbalance between antiangiogenic and proangiogenic factors such as cytokines and proteases as well as a variety of growth factors, notably vascular endothelial growth factor (VEGF) (Nishida, Yano, Nishida, Kamura, & Kojiro, 2006). Tumor angiogenesis requires binding of VEGF to the VEGF-2 receptor, which activates a proliferative signaling cascade through the mitogen-activated protein kinase (MAPK) pathway (D'Angelo, Struman, Martial, & Weiner, 1995). Over the last decade, 20-HETE has become recognized as a critical modulator of cancer progression, operating together with VEGF (and probably other growth factors) to promote cellular proliferation, neovascularization and angiogenesis, cell migration, tumor growth, and metastasis. The evidence in support of these functions and the role of specific CYP4 ω-hydroxylases is discussed in the following sections.
5.4 Chemical Tools for Dissecting CYP4 Enzymology and Participation of 20-HETE in Cancer Progression
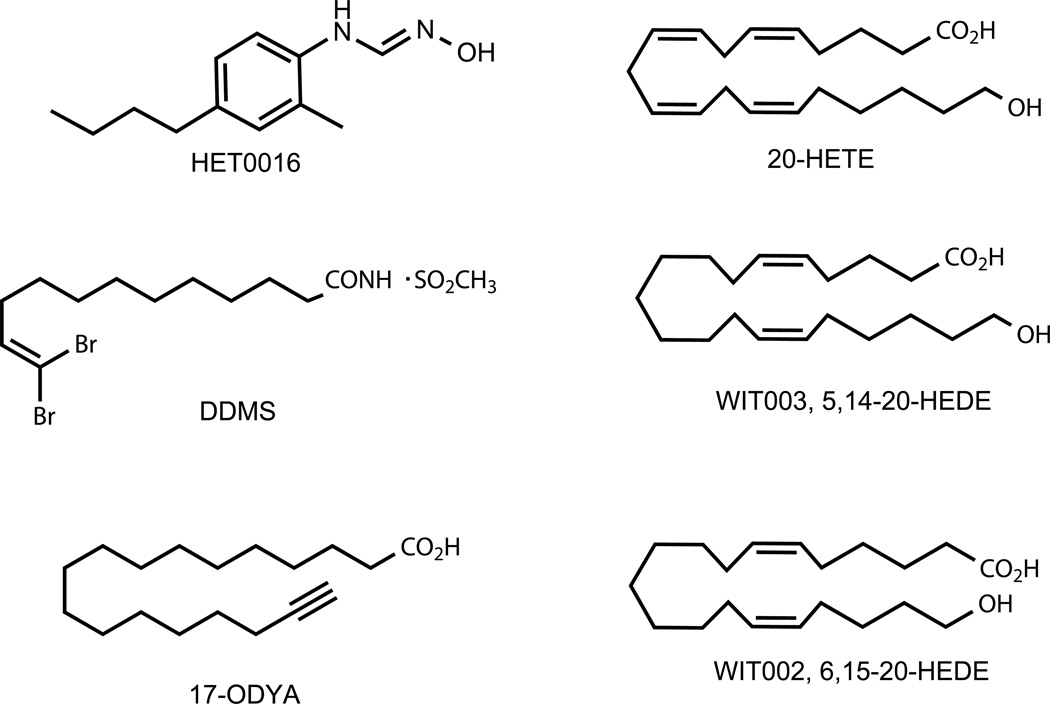
A cellular receptor for 20-HETE has not been identified, although a GPCR that binds 12(S)-HETE with high affinity has been described (Guo, Zhang, et al., 2011). Therefore, the current lexicon of 20-HETE modulators is restricted to chemical inhibitors of CYP ω-hydroxylases and chemical analogs of 20-HETE that act as agonists or antagonists (Fig. 4).
Figure 4.
Structures of chemical tools used widely for dissecting ω-hydroxylase actions in inflammation and cancer.
The availability of chemical inhibitors for the CYP4 ω-hydroxylases has been critical for elucidating the physiological roles of eicosanoids metabolized by these enzymes. The most widely used chemical inhibitors are the mechanism-based inhibitor, 17-ODYA and the tight-binding inhibitor, N-hydroxy-N′-(4-n-butyl-2-methylphenyl)formamidine (HET0016). 17-ODYA is one of a group of terminal acetylenes and halogenated olefins (e.g., DDMS) that destroy ω-hydroxylases upon metabolic turnover to reactive intermediates that bind to the enzyme (Kroetz & Xu, 2005). However, 17-ODYA is not specific for ω-hydroxylases and inhibits renal microsomal 20-HETE and EET formation to an equal extent (Zou et al., 1994).
HET0016 has emerged as both a potent and selective CYP4 chemical inhibitor that has proven exceptionally useful for studying 20-HETE pathways in vitro and in vivo. HET0016 was discovered in a screen to find a selective ω-hydroxylase inhibitor for the treatment of renal hypertension. In rat kidney microsomes, the IC50 for 20-HETE formation was 35 nM, while the IC50 for EET formation in this system was ~ 100 times higher (Miyata et al., 2001; Nakamura et al., 2003; Sato et al., 2001). Although the older literature tends to refer to HET0016 as a selective CYP4A inhibitor, it has the properties of a pan-CYP4 inhibitor, with IC50 values for CYP4A11, CYP4F2, CYP4F3B, CYP4V2, and CYP4B1 all in the 100 nM region or lower (Kehl et al., 2002; Nakano et al., 2009; Parkinson, Kelly, Bezabih, Whittington, & Rettie, 2012). High selectivity for human CYP4 ω-hydroxylases is clearly evident because IC50 values for the other major human liver P450s, CYP3A4, CYP2C9, and CYP2D6, are in the range 4–85 µM (Nakamura et al., 2003). Therefore, HET0016 is a more selective inhibitor of CYP4-dependent ω-hydroxylase activity than is 17-ODYA.
WIT002 and WIT003 are eicosanoid analogs that represent moderately potent (low µM) 20-HETE antagonist and agonists, respectively (Yu et al., 2004). WIT003 and HET0016 are often employed in study designs for evaluating 20-HETE signaling because of their complementary nature; i.e., demonstration of any WIT003-mediated reversal of HET0016’s biological effects provides a powerful argument for the intermediacy of a CYP4 ω-hydroxylase in the pathway under study.
5.5 Cellular Proliferation and Tumor Growth
20-HETE has long been implicated in cellular proliferation (Roman, 2002), often invoking the participation of growth factors, such as VEGF, epidermal growth factor (EGF), fibroblast growth factor (FGF), and platelet-derived growth factor (PDGF). Lin et al. provided an early demonstration that 20-HETE, specifically, was a mitogen in rat proximal tubule cells (Lin, Rios, Falck, Belosludtsev, & Schwartzman, 1995). Notably, this effect was not observed with addition of other ω/ω-1 metabolites, such as 20-carboxy arachidonic acid, 19(S)- or 19(R)-HETE. In the same studies, EGF stimulated both cell growth and endogenous 20-HETE formation, while 17-ODYA attenuated the proliferative effects. In a complementary fashion, HET0016 inhibited the basal and EGF-/PDGF-stimulated growth of 9L rat gliosarcoma cells in culture, while WIT003 increased cellular proliferation (Guo et al., 2006), thereby providing strong evidence for a role in the CYP4:20-HETE pathway. Interestingly, 9L cells did not generate detectable quantities of 20-HETE when incubated with radiolabeled arachidonic acid. Similar effects were seen in vitro with U251 glioma cells (Guo, Roman, Falck, Edwards, & Scicli, 2005). These observations led the authors to question whether HET0016 might have antiproliferative effects unrelated to the compound's ability to inhibit 20-HETE formation—a situation that remains to be resolved.
Modulation of the CYP4:20-HETE pathway has very pronounced effects on tumor size in animal models of brain, kidney, and breast cancer. For example, following implantation into normal rat forebrain of U251 glioma cells that had been transfected with CYP4A1, a 10-fold increase in tumor volume was observed compared with the nontransfected cells (Guo et al., 2008). Similarly, brain implantation of 9L gliosarcoma cells caused rapid tumor growth that caused death in about 17 days. However, chronic treatment with HET0016 increased survival time by 5 days, apparently through a combination of reduced mitosis and increased apoptosis (Guo et al., 2006).
These results on tumor growth are impressive because brain cancers are difficult to treat, as are renal adenocarcinomas, which proliferate and metastasize extensively. Similar to its effects on glioma cells, HET0016 (and WIT002) reduced proliferation of renal adenocarcinoma cells (Alexanian et al., 2009). In the same studies, WIT002 also suppressed the growth of advanced renal carcinoma in a mouse model. Formation of low levels of 20-HETE by renal cancer cells could be demonstrated upon addition of arachidonic acid, and this effect was abolished by HET0016.
Nonsmall cell lung cancer (NSCLC) is another treatment-refractory malignancy. Injection of mice with an NSCLC-derived cell line (A549) transfected with CYP4A11 increased the tumor size and growth rate, both of which were reduced with HET0016 or WIT002 (Yu et al., 2011). Similar results were obtained in a separate lung metastasis model. Collectively, these studies provide strong support for the CYP4:20-HETE pathway as a drug target for combating tumor growth and metastatic potential across a variety of difficult to treat cancers.
5.6 Angiogenesis
The effect of the CYP4:20-HETE system on angiogenesis has also been investigated extensively in recent years. The majority of these efforts evaluated VEGF expression and/or cellular responses to the growth factor because VEGF receptors and their ligands play essential roles in the regulation of angiogenesis (Shibuya, 2013). These studies are of particular interest because they have helped illuminate mechanistic details of the pathway(s) involved for 20-HETE signaling in cancer progression.
Several groups, applying independent experimental approaches, established a role for 20-HETE in angiogenesis in the early 2000s. In one of the first of these studies, angiogenesis induced in skeletal muscle by chronic electrical stimulation was accompanied by a 2.5-fold increase in 20-HETE formation that could be completely blocked by HET0016 (Amaral, Maier, Schippers, Roman, & Greene, 2003). However, immunochemically detectable increases in VEGF in electrically stimulated muscles were not reduced by ω-hydroxylase inhibitors, although a neutralizing VEGF antibody did block electrically induced increases in 20-HETE. These observations led the authors to conclude that 20-HETE acted downstream of VEGF in this pathway for muscle angiogenesis induced by electrical stimulation.
The foregoing study did not directly link angiogenesis involving 20-HETE to CYP4 enzyme expression, but this was subsequently evaluated by Lanaido-Schwartzman and coworkers upon overexpression of rat CYP4A1 in microdissected renal arteries (Jiang et al., 2004). Transduced arteries produced an ~ threefold higher level of 20-HETE in the culture medium and ~ eightfold increase in endothelial cell sprouting. Both effects were blocked by HET0016 whose antiangiogenic activity could be reversed upon addition of a 20-HETE agonist. These ex vivo studies were complemented with an in vivo approach that used sustained release polymer pellets containing growth factors (VEGF, EGF, or FGF), plus or minus HET0016 or DDMS, implanted into rat eyes. Each rat served as its own control for evaluation of the effect of these modulators on corneal neovascularization. In all combinations, the P450 inhibitor completely abrogated the angiogenic response that had been stimulated by the growth factor. Similar results were obtained by these investigators with an experimental design that replaced the growth factors in the implanted pellets with U251 glioma spheroids. Although changes in 20-HETE levels were not confirmed in these experiments, the authors concluded that the similarity in effects caused in vivo by two P450 inhibitors with distinct inhibitory mechanisms (HET0016 and DDMS) argued for a role for the CYP4:20-HETE pathway in growth factor-induced changes in angiogenesis in vivo. More recently, CYP4Z1 overexpression in breast cancer cells has been linked to increased VEGF expression, angiogenesis, cell proliferation, and migration in vitro and increased tumor weight in xenograft models (Yu et al., 2012). These effects were accompanied by increased 20-HETE production and inhibition by HET0016. This is interesting because CYP4Z1 has yet to be directly shown to function as an ω-hydroxylase of arachidonic acid.
In recent years, endothelial progenitor cells (EPCs) have emerged as a model system to investigate the CYP4:20-HETE system in angiogenesis. Circulating EPCs contribute to postnatal vascularization, neovascularization, and tissue damage repair and so have attracted attention as a potential therapeutic target in vascular disease (Kirton & Xu, 2010). EPCs express relatively high levels of CYP4A11 and form 20-HETE and numerous other metabolites when incubated with arachidonic acid (Guo, Janic, et al., 2011). 20-HETE has been shown to regulate EPC angiogenesis both in vitro and in vivo (Chen et al., 2014; Guo, Janic, et al., 2011) and so this appears to be a highly useful cell system for dissecting CYP4 signaling pathways in cancer progression.
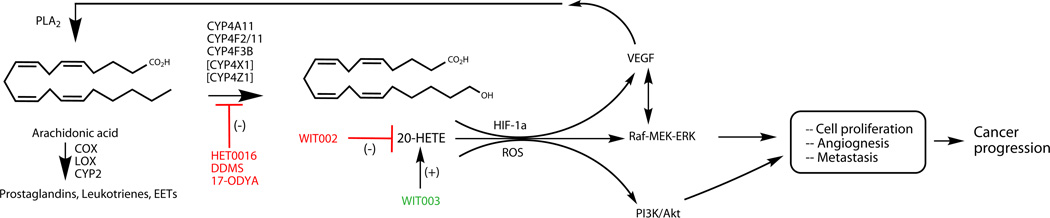
5.7 Signaling Mechanisms in CYP4:20-HETE-Induced Cancer Progression
Many of the studies discussed above highlight the interdependency between 20-HETE and VEGF in promoting critical features of cancer progression, including proliferation, angiogenesis, and metastases. Whereas Amaral et al. concluded that 20-HETE acts downstream of VEGF in skeletal muscle angiogenesis (Amaral et al., 2003), others have provided data that favor the reverse situation (Guo, Janic, et al., 2011).
The MAPK pathway has been invoked in phospholipase A2 activity that could release arachidonic acid, perhaps in concert with VEGF, and thereby amplify 20-HETE formation (Muthalif et al., 1998). Other critical elements of CYP4:20-HETE cancer signaling include activation of PI3K/Akt (Yu et al., 2011), as well as roles for HIF-1α and reactive oxygen species (ROS) (Guo et al., 2007). Very recently, Zeldin and coworkers generated a CYP4F2 transgenic mouse with selective endothelial cell expression (Cheng et al., 2014). Endothelial cells from transgenic animals exhibited twofold increases in levels of 20-HETE, increased growth and tube formation with upregulation of VEGF and the prooxidant enzyme NADPH oxidase, further underlining the importance of the MAPK pathway and ROS in cellular proliferation. Figure 5 attempts to capture essential elements of the complex signaling pathways of CYP4:20-HETE in cancer progression. More detailed mechanistic proposals can be found in two excellent recent reviews (Chen et al., 2014; Hoopes, Garcia, Edin, Schwartzman, & Zeldin, 2015)
Figure 5.
6 Conclusion
Considerable evidence supports the view that P450-dependent ω-hydroxylases play pivotal roles in the progression of cancer and in the resolution of inflammation. However, the CYP4 enzymes that dominate these processes do not act in isolation. Similar to CYP4 enzymes, expression of CYP2J2 is upregulated in tumor cells of human origin compared to adjacent normal cells corresponding to an increase in EET formation (Wang & Dubois, 2010). Interestingly, during cancer progression, EETs generated by CYP2C and CYP2J appear to play rather similar roles to 20-HETE, in that they also stimulate tumor cell proliferation, inhibition of apoptosis, angiogenesis, and metastases in a variety of mouse tumors (Tacconelli & Patrignani, 2014). EETs also appear to participate in signaling pathways that mirror several of those invoked for 20-HETE's protumorigenic effects including VEGF, MAPK, and PI3K/Akt. Administration of synthetic EET analogs or sEH inhibitors recapitulated these effects, whereas EET antagonists or chemical inhibitors reduced the effects on tumor size and metastases (Jiang et al., 2007; Panigrahy et al., 2012). In contrast, during inflammation, EETs appear to oppose 20-HETE effects, suppressing production of proinflammatory cytokines and chemokines and preventing leukocyte cell adhesion, thereby reducing release of LTB4. There is also evidence that EET formation is decreased through downregulation of CYP2C and CYP2J genes during the initiation of acute inflammation, which creates favorable conditions for 20-HETE formation, and thus enhanced production of cytokines and LTB4, resulting in amplification of inflammation (Theken et al., 2011). This suggests a functional balance between ω-hydroxylase and epoxygenase pathways throughout the different phases of the inflammatory response.
In general, it appears that investigators have not fully evaluated the ω-hydroxylase and epoxygenase pathways together in the same cell-based systems or animal models. This may reflect attempts to simplify the complex pharmacology or perhaps circumvent some of the analytical challenges in measuring 20-HETE and EETS in biological samples. Because EETs and 20-HETE are chemically unstable, subject to rapid oxidation, acid sensitive and present in low concentrations in blood and tissue samples, they present a serious analytical challenge. For EET quantification, determining whether downstream DHET analytes derive from the action of sEH or arise as artifacts of the extraction protocol can be quite difficult. Unfortunately, neither 20-HETE nor EETs are among the compounds included on traditional metabolomics or eicosanoid analytical platforms (Kofeler, Fauland, Rechberger, & Trotzmuller, 2012). In the cell, EETs and 20-HETE exist predominantly esterified at the sn-2 position in the phospholipid fraction of the membrane, with less than 5% present as the free acid (Hammond & O'Donnell, 2012). However, many studies only analyze the free EETs and 20-HETE by direct extraction into organic solvents, which will lead to variability in the level of lipids measured and make it difficult to observe changes in concentration following inhibition of CYP4 enzymes or treatment with sEH. There is also variability in analytical tools employed in EET and 20-HETE measurements. While LC–MS/MS is the instrumentation of choice for measuring EETs and 20-HETE owing to its sensitivity and rapid quantification of several lipids simultaneously, recent ELISA assays have emerged for 20-HETE and specific DHETs (Kroot et al., 2010). These varied methodologies render it even more difficult to interpret altered levels of lipid mediators in healthy versus cancerous tissue from different studies and determine how their concentrations change in cancer progression and metastases.
Another analytical challenge for helping understand the interplay between hydroxylation and epoxidation pathways is the quantitation of CYP4 and CYP2 enzymes in the cell type or tissue under investigation. However, recent advances in mass spectrometry-based quantitation of P450 enzymes in whole and broken cell systems make this a realizable goal that does not rely on the availability of monospecific antibodies or the presumption that transcript profiling will always translate directly to protein levels (Aebersold, Burlingame, & Bradshaw, 2013). A wider array of P450 transgenic and knockout animal models can also be expected to provide valuable new tools for unveiling ω-hydroxylase biology in vivo. A few Cyp4a and Cyp4f knockout mice have been generated (Bardowell, Duan, Manor, Swanson, & Parker, 2012; Holla et al., 2001), but their utility for answering complex questions about disease in humans is limited by the expansion of these P450 subfamilies in mice relative to humans. In the absence of a clear identification of the mouse homolog for, say CYP4F2, either the entire Cyp4 locus would need to be deleted or human CYP4F2 would need to be knocked in at high enough levels in a tissue-specific manner to facilitate data interpretation. As noted earlier, Zeldin and coworkers have generated a CYP4F2 transgenic mouse with selective endothelial cell expression (Cheng et al., 2014). Future development of animal models for other ω-hydroxylases will further expand the toolbox required for fully evaluating the biological roles of these enzymes in cancer and inflammation.
Finally, the development of drugs that selectively inhibit arachidonic acid pathways has been a seductive goal for many years, with clear therapeutic successes in aspirin, NSAIDs and zileuton, COX and LOX inhibitors, respectively, to draw on for inspiration. The CYP4F ω-hydroxylases also provide a potential drug target in the attenuation of the inflammatory response, as induction of these enzymes increases formation of 20-OH LTB4. However, greater understanding of the differential tissue expression and regulation of the various CYP4Fs is needed before this becomes a truly viable option. Elucidating mechanisms by which EETs and ω3 PUFAs exert their anti-inflammatory benefits, and the extent to which ω-hydroxylases are involved, will also be an important focus in future development of novel anti-inflammatory therapies. sEH inhibitors have been developed and advanced into clinical trials with the intent of increasing in vivo EET levels for the potential treatment of hypertension, nociceptive pain, and diabetes (Kodani & Hammock, 2015). However, this approach has the potential downside that the undesirable protumorigenic features of the EETs might be unmasked in certain patients. Therefore, selective modulation of the 20-HETE pathway to combat cancer progression may be more tractable. Indeed, the tumor-shrinking properties of HET0016 are striking, although several studies have failed to link HET0016-mediated CYP4 inhibition to reductions in tumor levels of 20-HETE, raising the possibility that HET0016 might be acting through a different mechanism. Alternatively, extremely low 20-HETE levels, measuring only free 20-HETE or upregulated secondary metabolic pathways might explain these results. Continuing improvements in the sensitivity of mass spectrometry-based analysis of arachidonate metabolites (Tsikas & Zoerner, 2014) combined with a more thorough understanding of the metabolic fate of 20-HETE and EETs can be expected to further our understanding of the critical role these compounds play in the complex lipid signaling involved in inflammation and cancer.
Acknowledgments
CYP4 research in the Rettie laboratory is currently supported by NIH Grant R01 GM109743. CYP2J2 and EET research in the Totah laboratory is supported by NIH Grant RHL096706.
Abbreviations
- 17-ODYA
17-octadecynoic acid
- Akt
protein kinase B
- AMP
5′ adenosine monophosphate
- COX
cyclooxygenase
- CYP
cytochrome P450
- DDMS
dibromo-dodecenyl-methylsulfimide
- DHA
docosahexaenoic acid
- DHET
dihydroxyeicosatrienoic acid
- EET
epoxyeicosatrienoic acid
- EGF
epidermal growth factor
- EPA
eicosapentaenoic acid
- EPC
endothelial progenitor cells
- FGF
fibroblast growth factor
- HEET
hydroxyepoxyeicosatrienoic acid
- HET0016
N-hydroxy-N′-(4-n-butyl-2-methylphenyl)formamidine
- HETE
hydroxyeicosatetraenoic acid
- HIF-1α
hypoxia-inducible factor 1-alpha
- HNF4α
hepatocyte nuclear factor 4-alpha
- HPETE
hydroperoxyeicosatetraenoic acid
- IL
interleukin
- LOX
lipoxygenase
- LPS
lipopolysaccharide
- LT
leukotriene
- LX
lipoxin
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor kappa-B
- NSCLC
nonsmall cell lung cancer
- PDGF
platelet-derived growth factor
- PG
prostaglandin
- PI3K
phosphoinositide 3-kinase
- PMNL
polymorphonuclear leukocytes
- PPAR
peroxisome proliferator-activated receptor
- PUFA
polyunsaturated fatty acid
- ROS
reactive oxygen species
- Rv
resolvin
- sEH
soluble epoxide hydrolase
- SREBP
sterol regulatory element-binding protein
- TNFα
tissue necrosis factor alpha
- VEGF
vascular endothelial growth factor
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
REFERENCES
- Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: Time to turn the tables? Molecular & Cellular Proteomics. 2013;12:2381–2382. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexanian A, Miller B, Roman RJ, Sorokin A. 20-HETE-producing enzymes are up-regulated in human cancers. Cancer Genomics Proteomics. 2012;9:163–169. [PMC free article] [PubMed] [Google Scholar]
- Alexanian A, Rufanova VA, Miller B, Flasch A, Roman RJ, Sorokin A. Down-regulation of 20-HETE synthesis and signaling inhibits renal adenocarcinoma cell proliferation and tumor growth. Anticancer Research. 2009;29:3819–3824. [PMC free article] [PubMed] [Google Scholar]
- Alexanian A, Sorokin A. Targeting 20-HETE producing enzymes in cancer— Rationale, pharmacology, and clinical potential. Onco Targets and therapy. 2013;6:243–255. doi: 10.2147/OTT.S31586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral SL, Maier KG, Schippers DN, Roman RJ, Greene AS. CYP4A metabolites of arachidonic acid and VEGF are mediators of skeletal muscle angiogenesis. American Journal of Physiology Heart and Circulatory Physiology. 2003;284:H1528–H1535. doi: 10.1152/ajpheart.00406.2002. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Hardwick JP, Imaoka S, Funae Y, Gelboin HV, Gonzalez FJ. Clofibrate-inducible rat hepatic P450s IVA1 and IVA3 catalyze the omega- and (omega-1)-hydroxylation of fatty acids and the omega-hydroxylation of prostaglandins E1 and F2 alpha. Journal of Lipid Research. 1990;31:1477–1482. [PubMed] [Google Scholar]
- Arita M, Oh SF, Chonan T, Hong S, Elangovan S, Sun YP, et al. Metabolic inactivation of resolvin E1 and stabilization of its anti-inflammatory actions. The Journal of Biological Chemistry. 2006;281:22847–22854. doi: 10.1074/jbc.M603766200. [DOI] [PubMed] [Google Scholar]
- Arnold C, Markovic M, Blossey K, Wallukat G, Fischer R, Dechend R, et al. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of {omega}-3 fatty acids. The Journal of Biological Chemistry. 2010;285:32720–32733. doi: 10.1074/jbc.M110.118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardowell SA, Duan F, Manor D, Swanson JE, Parker RS. Disruption of mouse cytochrome p450 4f14 (Cyp4f14 gene) causes severe perturbations in vitamin E metabolism. The Journal of Biological Chemistry. 2012;287:26077–26086. doi: 10.1074/jbc.M112.373597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar MM, Schwartzman M, Ibraham NG, McGiff JC, Mullane KM. Conversion of arachidonic acid to two novel products by a cytochrome P450-dependent mixed-function oxidase in polymorphonuclear leukocytes. Biochemical and Biophysical Research Communications. 1984;123:581–588. doi: 10.1016/0006-291x(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Berry KA, Borgeat P, Gosselin J, Flamand L, Murphy RC. Urinary metabolites of leukotriene B4 in the human subject. The Journal of Biological Chemistry. 2003;278:24449–24460. doi: 10.1074/jbc.M300856200. [DOI] [PubMed] [Google Scholar]
- Birnie M, Morrison R, Camara R, Strauss KI. Temporal changes of cytochrome P450 (Cyp) and eicosanoid-related gene expression in the rat brain after traumatic brain injury. BMC Genomics. 2013;14:303. doi: 10.1186/1471-2164-14-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund J, Hidestrand M, Ingelman-Sundberg M, Oliw EH. Identification of CYP4F8 in human seminal vesicles as a prominent 19-hydroxylase of prostaglandin endoperoxides. The Journal of Biological Chemistry. 2000;275:21844–21849. doi: 10.1074/jbc.M001712200. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Chacos N, Werringloer J, Prough RA, Estabrook RW. Liver microsomal cytochrome P-450 and the oxidative metabolism of arachidonic acid. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:5362–5366. doi: 10.1073/pnas.78.9.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila JH, Falck JR. Biochemical and molecular properties of the cytochrome P450 arachidonic acid monooxygenases. Prostaglandins & Other Lipid Mediators. 2002:68–69. 325–344. doi: 10.1016/s0090-6980(02)00038-2. [DOI] [PubMed] [Google Scholar]
- Chen L, Ackerman R, Saleh M, Gotlinger KH, Kessler M, Mendelowitz LG, et al. 20-HETE regulates the angiogenic functions of human endothelial progenitor cells and contributes to angiogenesis in vivo. The Journal of Pharmacology and Experimental Therapeutics. 2014;348:442–451. doi: 10.1124/jpet.113.210120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hardwick JP. Identification of a new P450 subfamily, CYP4F1, expressed in rat hepatic tumors. Archives of Biochemistry and Biophysics. 1993;300:18–23. doi: 10.1006/abbi.1993.1003. [DOI] [PubMed] [Google Scholar]
- Cheng J, Edin ML, Hoopes SL, Li H, Bradbury JA, Graves JP, et al. Vascular characterization of mice with endothelial expression of cytochrome P450 4F2. The FASEB Journal. 2014;28:2915–2931. doi: 10.1096/fj.13-241927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas P, Jones JP, Patten CJ, Rock DA, Zheng Y, Cheng SM, et al. Alternative splicing determines the function of CYP4F3 by switching substrate specificity. The Journal of Biological Chemistry. 2001;276:38166–38172. doi: 10.1074/jbc.M104818200. [DOI] [PubMed] [Google Scholar]
- Christmas P, Ursino SR, Fox JW, Soberman RJ. Expression of the CYP4F3 gene. Tissue-specific splicing and alternative promoters generate high and low K(m) forms of leukotriene B(4) omega-hydroxylase. The Journal of Biological Chemistry. 1999;274:21191–21199. doi: 10.1074/jbc.274.30.21191. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Wei S, Hsu MH, Johnson EF, Krishna MU, Falck JR, et al. The CYP4A isoforms hydroxylate epoxyeicosatrienoic acids to form high affinity peroxisome proliferator-activated receptor ligands. The Journal of Biological Chemistry. 2002;277:35105–35112. doi: 10.1074/jbc.M201575200. [DOI] [PubMed] [Google Scholar]
- D'Angelo G, Struman I, Martial J, Weiner RI. Activation of mitogen-activated protein kinases by vascular endothelial growth factor and basic fibroblast growth factor in capillary endothelial cells is inhibited by the antiangiogenic factor 16-kDa N- terminal fragment of prolactin. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6374–6378. doi: 10.1073/pnas.92.14.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawczynski C, Massey KA, Ness C, Kiehntopf M, Stepanow S, Platzer M, et al. Randomized placebo-controlled intervention with n-3 LC-PUFA-supplemented yoghurt: Effects on circulating eicosanoids and cardiovascular risk factors. Clinical Nutrition. 2013;32:686–696. doi: 10.1016/j.clnu.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Dhar M, Sepkovic DW, Hirani V, Magnusson RP, Lasker JM. Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. Journal of Lipid Research. 2008;49:612–624. doi: 10.1194/jlr.M700450-JLR200. [DOI] [PubMed] [Google Scholar]
- Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID, et al. Profiling cytochrome P450 expression in ovarian cancer: Identification of prognostic markers. Clinical Cancer Research. 2005;11:7369–7375. doi: 10.1158/1078-0432.CCR-05-0466. [DOI] [PubMed] [Google Scholar]
- Draye JP, Vamecq J. The gluconeogenicity of fatty acids in mammals. Trends in Biochemical Sciences. 1989;14:478–479. doi: 10.1016/0968-0004(89)90176-x. [DOI] [PubMed] [Google Scholar]
- Du L, Yin H, Morrow JD, Strobel HW, Keeney DS. 20-Hydroxylation is the CYP-dependent and retinoid-inducible leukotriene B4 inactivation pathway in human and mouse skin cells. Archives of Biochemistry and Biophysics. 2009;484:80–86. doi: 10.1016/j.abb.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson KZ, Prasad B, Unadkat JD, Suhara Y, Okano T, Guengerich FP, et al. Cytochrome P450-dependent catabolism of vitamin K: Omega-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry. 2013;52:8276–8285. doi: 10.1021/bi401208m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eksterowicz J, Rock DA, Rock BM, Wienkers LC, Foti RS. Characterization of the active site properties of CYP4F12. Drug Metabolism and Disposition. 2014;42:1698–1707. doi: 10.1124/dmd.114.059626. [DOI] [PubMed] [Google Scholar]
- Fan F, Muroya Y, Roman RJ. Cytochrome P450 eicosanoids in hypertension and renal disease. Current Opinion in Nephrology and Hypertension. 2015;24:37–46. doi: 10.1097/MNH.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fer M, Corcos L, Dreano Y, Plee-Gautier E, Salaun JP, Berthou F, et al. Cytochromes P450 from family 4 are the main omega hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. Journal of Lipid Research. 2008;49:2379–2389. doi: 10.1194/jlr.M800199-JLR200. [DOI] [PubMed] [Google Scholar]
- Fleming I. Vascular cytochrome p450 enzymes: Physiology and pathophysiology. Trends in Cardiovascular Medicine. 2008;18:20–25. doi: 10.1016/j.tcm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Folkman J. Tumor angiogenesis: Therapeutic implications. The New England Journal of Medicine. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- Forrester LM, Hayes JD, Millis R, Barnes D, Harris AL, Schlager JJ, et al. Expression of glutathione S-transferases and cytochrome P450 in normal and tumor breast tissue. Carcinogenesis. 1990;11:2163–2170. doi: 10.1093/carcin/11.12.2163. [DOI] [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Gandhi AV, Saxena S, Relles D, Sarosiek K, Kang CY, Chipitsyna G, et al. Differential expression of cytochrome P450 omega-hydroxylase isoforms and their association with clinicopathological features in pancreatic ductal adenocarcinoma. Annals of Surgical Oncology. 2013;20(Suppl 3):S636–S643. doi: 10.1245/s10434-013-3128-x. [DOI] [PubMed] [Google Scholar]
- Gatica A, Aguilera MC, Contador D, Loyola G, Pinto CO, Amigo L, et al. P450 CYP2C epoxygenase and CYP4A omega-hydroxylase mediate ciprofibrate- induced PPARalpha-dependent peroxisomal proliferation. Journal of Lipid Research. 2007;48:924–934. doi: 10.1194/jlr.M700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Gibson GG, Orton TC, Tamburini PP. Cytochrome P-450 induction by clofibrate. Purification and properties of a hepatic cytochrome P-450 relatively specific for the 12- and 11-hydroxylation of dodecanoic acid (lauric acid) The Biochemical Journal. 1982;203:161–168. doi: 10.1042/bj2030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YM. PPARalpha: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Gronert K, Clish CB, Romano M, Serhan CN. Transcellular regulation of eicosanoid biosynthesis. Methods in Molecular Biology. 1999;120:119–144. doi: 10.1385/1-59259-263-5:119. [DOI] [PubMed] [Google Scholar]
- Guo AM, Arbab AS, Falck JR, Chen P, Edwards PA, Roman RJ, et al. Activation of vascular endothelial growth factor through reactive oxygen species mediates 20-hydroxyeicosatetraenoic acid-induced endothelial cell proliferation. The Journal of Pharmacology and Experimental Therapeutics. 2007;321:18–27. doi: 10.1124/jpet.106.115360. [DOI] [PubMed] [Google Scholar]
- Guo AM, Janic B, Sheng J, Falck JR, Roman RJ, Edwards PA, et al. The cytochrome P450 4A/F-20-hydroxyeicosatetraenoic acid system: A regulator of endothelial precursor cells derived from human umbilical cord blood. The Journal of Pharmacology and Experimental Therapeutics. 2011;338:421–429. doi: 10.1124/jpet.111.179036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Roman RJ, Falck JR, Edwards PA, Scicli AG. Human U251 glioma cell proliferation is suppressed by HET0016 [N-hydroxy-N'-(4-butyl-2-methylphenyl)formamidine], a selective inhibitor of CYP4A. The Journal of Pharmacology and Experimental Therapeutics. 2005;315:526–533. doi: 10.1124/jpet.105.088567. [DOI] [PubMed] [Google Scholar]
- Guo M, Roman RJ, Fenstermacher JD, Brown SL, Falck JR, Arbab AS, et al. 9L gliosarcoma cell proliferation and tumor growth in rats are suppressed by N-hydroxy-N'-(4-butyl-2-methylphenol) formamidine (HET0016), a selective inhibitor of CYP4A. The Journal of Pharmacology and Experimental Therapeutics. 2006;317:97–108. doi: 10.1124/jpet.105.097782. [DOI] [PubMed] [Google Scholar]
- Guo AM, Sheng J, Scicli GM, Arbab AS, Lehman NL, Edwards PA, et al. Expression of CYP4A1 in U251 human glioma cell induces hyperproliferative phenotype in vitro and rapidly growing tumors in vivo. The Journal of Pharmacology and Experimental Therapeutics. 2008;327:10–19. doi: 10.1124/jpet.108.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhang W, Giroux C, Cai Y, Ekambaram P, Dilly AK, et al. Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)- hydroxyeicosatetraenoic acid. The Journal of Biological Chemistry. 2011;286:33832–33840. doi: 10.1074/jbc.M110.216564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeggstrom JZ, Funk CD. Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chemical Reviews. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- Hammond VJ, O'Donnell VB. Esterified eicosanoids: Generation, characterization and function. Biochimica et Biophysica Acta. 2012;1818:2403–2412. doi: 10.1016/j.bbamem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson G, Lindgren JA, Dahlen SE, Hedqvist P, Samuelsson B. Identification and biological activity of novel omega-oxidized metabolites of leukotriene B4 from human leukocytes. FEBS Letters. 1981;130:107–112. doi: 10.1016/0014-5793(81)80676-x. [DOI] [PubMed] [Google Scholar]
- Hardwick JP. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochemical Pharmacology. 2008;75:2263–2275. doi: 10.1016/j.bcp.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Harmon SD, Fang X, Kaduce TL, Hu S, Raj Gopal V, Falck JR, et al. Oxygenation of omega-3 fatty acids by human cytochrome P450 4F3B: Effect on 20- hydroxyeicosatetraenoic acid production. Prostaglandins, Leukotrienes, and Essential Fatty Acids. 2006;75:169–177. doi: 10.1016/j.plefa.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Harvey RD, Morgan ET. Cancer, inflammation, and therapy: Effects on cytochrome p450-mediated drug metabolism and implications for novel immunotherapeutic agents. Clinical Pharmacology and Therapeutics. 2014;96:449–457. doi: 10.1038/clpt.2014.143. [DOI] [PubMed] [Google Scholar]
- Hashizume T, Imaoka S, Mise M, Terauchi Y, Fujii T, Miyazaki H, et al. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. The Journal of Pharmacology and Experimental Therapeutics. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- He X, Cryle MJ, De Voss JJ, de Montellano PR. Calibration of the channel that determines the omega-hydroxylation regiospecificity of cytochrome P4504A1: Catalytic oxidation of 12-HALODOdecanoic acids. The Journal of Biological Chemistry. 2005;280:22697–22705. doi: 10.1074/jbc.M502632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz R, Sheena V, Kalderon B, Berman I, Bar-Tana J. Suppression of hepatocyte nuclear factor-4alpha by acyl-CoA thioesters of hypolipidemic peroxisome proliferators. Biochemical Pharmacology. 2001;61:1057–1062. doi: 10.1016/s0006-2952(01)00578-0. [DOI] [PubMed] [Google Scholar]
- Holla VR, Adas F, Imig JD, Zhao X, Price E, Jr, Olsen N, et al. Alterations in the regulation of androgen-sensitive Cyp 4a monooxygenases cause hypertension. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5211–5216. doi: 10.1073/pnas.081627898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes SL, Garcia V, Edin ML, Schwartzman ML, Zeldin DC. Vascular actions of 20-HETE. Prostaglandins & Other Lipid Mediators. 2015 doi: 10.1016/j.prostaglandins.2015.03.002. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Griffin KJ, Johnson EF. Human cytochrome p450 family 4 enzymes: Function, genetic variation and regulation. Drug Metabolism Reviews. 2007a;39:515–538. doi: 10.1080/03602530701468573. [DOI] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Griffin KJ, Johnson EF. Regulation of human cytochrome P450 4F2 expression by sterol regulatory element-binding protein and lovastatin. The Journal of Biological Chemistry. 2007b;282:5225–5236. doi: 10.1074/jbc.M608176200. [DOI] [PubMed] [Google Scholar]
- Hsu MH, Savas U, Lasker JM, Johnson EF. Genistein, resveratrol, and 5- aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside induce cytochrome P450 4F2 expression through an AMP-activated protein kinase-dependent pathway. The Journal of Pharmacology and Experimental Therapeutics. 2011;337:125–136. doi: 10.1124/jpet.110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Cheng J, Singh H, Vitto MD, Manthati VL, Falck JR, et al. 20-Hydroxyeicosatetraenoic acid stimulates nuclear factor-kappaB activation and the production of inflammatory cytokines in human endothelial cells. The Journal of Pharmacology and Experimental Therapeutics. 2008;324:103–110. doi: 10.1124/jpet.107.130336. [DOI] [PubMed] [Google Scholar]
- Jiang M, Mezentsev A, Kemp R, Byun K, Falck JR, Miano JM, et al. Smooth muscle-specific expression of CYP4A1 induces endothelial sprouting in renal arterial microvessels. Circulation Research. 2004;94:167–174. doi: 10.1161/01.RES.0000111523.12842.FC. [DOI] [PubMed] [Google Scholar]
- Jiang JG, Ning YG, Chen C, Ma D, Liu ZJ, Yang S, et al. Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Research. 2007;67:6665–6674. doi: 10.1158/0008-5472.CAN-06-3643. [DOI] [PubMed] [Google Scholar]
- Jin R, Koop DR, Raucy JL, Lasker JM. Role of human CYP4F2 in hepatic catabolism of the proinflammatory agent leukotriene B4. Archives of Biochemistry and Biophysics. 1998;359:89–98. doi: 10.1006/abbi.1998.0880. [DOI] [PubMed] [Google Scholar]
- Johnson EF, Palmer CN, Griffin KJ, Hsu MH. Role of the peroxisome proliferator-activated receptor in cytochrome P450 4A gene regulation. The FASEB Journal. 1996;10:1241–1248. doi: 10.1096/fasebj.10.11.8836037. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Anakk S, Brommer CL, Kikuta Y, Morgan ET, Strobel HW. Catalytic characterization and cytokine mediated regulation of cytochrome P450 4Fs in rat hepatocytes. Archives of Biochemistry and Biophysics. 2007;461:104–112. doi: 10.1016/j.abb.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Du L, Wang Y, Ladd PA, Kikuta Y, Duvic M, et al. Inflammation resolved by retinoid X receptor-mediated inactivation of leukotriene signaling pathways. The FASEB Journal. 2008;22:538–547. doi: 10.1096/fj.07-9244com. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Strobel HW. Cytochrome P450 4F subfamily: At the crossroads of eicosanoid and drug metabolism. Pharmacology & Therapeutics. 2006;112:589–611. doi: 10.1016/j.pharmthera.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Turman CM, Kikuta Y, Strobel HW. Expression and characterization of human cytochrome P450 4F11: Putative role in the metabolism of therapeutic drugs and eicosanoids. Toxicology and Applied Pharmacology. 2004;199:295–304. doi: 10.1016/j.taap.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Kaspera R, Totah RA. Epoxyeicosatrienoic acids: Formation, metabolism and potential role in tissue physiology and pathophysiology. Expert Opinion on Drug Metabolism & Toxicology. 2009;5:757–771. doi: 10.1517/17425250902932923. [DOI] [PubMed] [Google Scholar]
- Kehl F, Cambj-Sapunar L, Maier KG, Miyata N, Kametani S, Okamoto H, et al. 20-HETE contributes to the acute fall in cerebral blood flow after subarachnoid hemorrhage in the rat. American Journal of Physiology Heart and Circulatory Physiology. 2002;282:H1556–H1565. doi: 10.1152/ajpheart.00924.2001. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Nakano M, Rohatgi P, Yarov-Yarovoy V, Rettie AE. Finding homes for orphan cytochrome P450s: CYP4V2 and CYP4F22 in disease states. Molecular Interventions. 2011;11:124–132. doi: 10.1124/mi.11.2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanapure SP, Garvey DS, Janero DR, Letts LG. Eicosanoids in inflammation: Biosynthesis, pharmacology, and therapeutic frontiers. Current Topics in Medicinal Chemistry. 2007;7:311–340. doi: 10.2174/156802607779941314. [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Kusunose E, Endo K, Yamamoto S, Sogawa K, Fujii-Kuriyama Y, et al. A novel form of cytochrome P-450 family 4 in human polymorphonuclear leukocytes. cDNA cloning and expression of leukotriene B4 omega-hydroxylase. The Journal of Biological Chemistry. 1993;268:9376–9380. [PubMed] [Google Scholar]
- Kikuta Y, Kusunose E, Kusunose M. Characterization of human liver leukotriene B(4) omega-hydroxylase P450 (CYP4F2) Journal of Biochemistry. 2000;127:1047–1052. doi: 10.1093/oxfordjournals.jbchem.a022696. [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Kusunose E, Kusunose M. Prostaglandin and leukotriene omega- hydroxylases. Prostaglandins & Other Lipid Mediators. 2002:68–69. 345–362. doi: 10.1016/s0090-6980(02)00039-4. [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Kusunose E, Sumimoto H, Mizukami Y, Takeshige K, Sakaki T, et al. Purification and characterization of recombinant human neutrophil leukotriene B4 omega-hydroxylase (cytochrome P450 4F3) Archives of Biochemistry and Biophysics. 1998;355:201–205. doi: 10.1006/abbi.1998.0724. [DOI] [PubMed] [Google Scholar]
- Kirton JP, Xu Q. Endothelial precursors in vascular repair. Microvascular Research. 2010;79:193–199. doi: 10.1016/j.mvr.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Kodani SD, Hammock BD. The 2014 bernard B. Brodie award lecture-epoxide hydrolases: Drug metabolism to therapeutics for chronic pain. Drug Metabolism and Disposition. 2015;43:788–802. doi: 10.1124/dmd.115.063339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofeler HC, Fauland A, Rechberger GN, Trotzmuller M. Mass spectrometry based lipidomics: An overview of technological platforms. Metabolites. 2012;2:19–38. doi: 10.3390/metabo2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel A, Schunck WH. Role of cytochrome P450 enzymes in the bioactivation of polyunsaturated fatty acids. Biochimica et Biophysica Acta. 2011;1814:210–222. doi: 10.1016/j.bbapap.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Kovarik JM, Dole K, Riviere GJ, Pommier F, Maton S, Jin Y, et al. Ketoconazole increases fingolimod blood levels in a drug interaction via CYP4F2 inhibition. Journal of Clinical Pharmacology. 2009;49:212–218. doi: 10.1177/0091270008329553. [DOI] [PubMed] [Google Scholar]
- Kroetz DL, Xu F. Regulation and inhibition of arachidonic acid omega- hydroxylases and 20-HETE formation. Annual Review of Pharmacology and Toxicology. 2005;45:413–438. doi: 10.1146/annurev.pharmtox.45.120403.100045. [DOI] [PubMed] [Google Scholar]
- Kroot JJ, Laarakkers CM, Geurts-Moespot AJ, Grebenchtchikov N, Pickkers P, van Ede AE, et al. Immunochemical and mass-spectrometry-based serum hepcidin assays for iron metabolism disorders. Clinical Chemistry. 2010;56:1570–1579. doi: 10.1373/clinchem.2010.149187. [DOI] [PubMed] [Google Scholar]
- Kumarakulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, et al. Cytochrome p450 profile of colorectal cancer: Identification of markers of prognosis. Clinical Cancer Research. 2005;11:3758–3765. doi: 10.1158/1078-0432.CCR-04-1848. [DOI] [PubMed] [Google Scholar]
- Lasker JM, Chen WB, Wolf I, Bloswick BP, Wilson PD, Powell PK. Formation of 20-hydroxyeicosatetraenoic acid, a vasoactive and natriuretic eicosanoid, in human kidney. Role of Cyp4F2 and Cyp4A11. The Journal of Biological Chemistry. 2000;275:4118–4126. doi: 10.1074/jbc.275.6.4118. [DOI] [PubMed] [Google Scholar]
- Le Quere V, Plee-Gautier E, Potin P, Madec S, Salaun JP. Human CYP4F3s are the main catalysts in the oxidation of fatty acid epoxides. Journal of Lipid Research. 2004;45:1446–1458. doi: 10.1194/jlr.M300463-JLR200. [DOI] [PubMed] [Google Scholar]
- Lewis DF, Lake BG. Molecular modelling of CYP4A subfamily members based on sequence homology with CYP102. Xenobiotica; The Fate of Foreign Compounds in Biological Systems. 1999;29:763–781. doi: 10.1080/004982599238227. [DOI] [PubMed] [Google Scholar]
- Lin F, Rios A, Falck JR, Belosludtsev Y, Schwartzman ML. 20- Hydroxyeicosatetraenoic acid is formed in response to EGF and is a mitogen in rat proximal tubule. The American Journal of Physiology. 1995;269:F806–F816. doi: 10.1152/ajprenal.1995.269.6.F806. [DOI] [PubMed] [Google Scholar]
- Lu AY, Coon MJ. Role of hemoprotein P-450 in fatty acid omega-hydroxylation in a soluble enzyme system from liver microsomes. The Journal of Biological Chemistry. 1968;243:1331–1332. [PubMed] [Google Scholar]
- Madej A, Okopien B, Kowalski J, Zielinski M, Wysocki J, Szygula B, et al. Effects of fenofibrate on plasma cytokine concentrations in patients with atherosclerosis and hyperlipoproteinemia IIb. International Journal of Clinical Pharmacology and Therapeutics. 1998;36:345–349. [PubMed] [Google Scholar]
- Masters BS, Clark JE, Roman LJ, Nishimoto M, McCabe TJ, Ortiz de Montellano PR, et al. Functional aspects of eicosanoid hydroxylation by lung and kidney cytochromes P450. Expression of cDNAs in mammalian cells and E. coli. Journal of Lipid Mediators. 1993;6:353–360. [PubMed] [Google Scholar]
- McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Molecular Pharmacology. 2009;75:1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon B, Mitchell S, Brady HR, Godson C. Lipoxins: Revelations on resolution. Trends in Pharmacological Sciences. 2001;22:391–395. doi: 10.1016/s0165-6147(00)01771-5. [DOI] [PubMed] [Google Scholar]
- Miyata N, Taniguchi K, Seki T, Ishimoto T, Sato-Watanabe M, Yasuda Y, et al. HET0016, a potent and selective inhibitor of 20-HETE synthesizing enzyme. British Journal of Pharmacology. 2001;133:325–329. doi: 10.1038/sj.bjp.0704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RC, Gijon MA. Biosynthesis and metabolism of leukotrienes. The Biochemical Journal. 2007;405:379–395. doi: 10.1042/BJ20070289. [DOI] [PubMed] [Google Scholar]
- Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. Profiling the expression of cytochrome P450 in breast cancer. Histopathology. 2010;57:202–211. doi: 10.1111/j.1365-2559.2010.03606.x. [DOI] [PubMed] [Google Scholar]
- Murray GI, Taylor MC, McFadyen MC, McKay JA, Greenlee WF, Burke MD, et al. Tumor-specific expression of cytochrome P450 CYP1B1. Cancer Research. 1997;57:3026–3031. [PubMed] [Google Scholar]
- Muthalif MM, Benter IF, Karzoun N, Fatima S, Harper J, Uddin MR, et al. 20-Hydroxyeicosatetraenoic acid mediates calcium/calmodulin-dependent protein kinase II-induced mitogen-activated protein kinase activation in vascular smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12701–12706. doi: 10.1073/pnas.95.21.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sato M, Kakinuma H, Miyata N, Taniguchi K, Bando K, et al. Pyrazole and isoxazole derivatives as new, potent, and selective 20-hydroxy-5,8,11,14- eicosatetraenoic acid synthase inhibitors. Journal of Medicinal Chemistry. 2003;46:5416–5427. doi: 10.1021/jm020557k. [DOI] [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Rettie AE. Expression and characterization of CYP4V2 as a fatty acid omega-hydroxylase. Drug Metabolism and Disposition. 2009;37:2119–2122. doi: 10.1124/dmd.109.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kelly EJ, Wiek C, Hanenberg H, Rettie AE. CYP4V2 in Bietti's crystalline dystrophy: Ocular localization, metabolism of omega-3-polyunsaturated fatty acids, and functional deficit of the p.H331P variant. Molecular Pharmacology. 2012;82:679–686. doi: 10.1124/mol.112.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naraba H, Murakami M, Matsumoto H, Shimbara S, Ueno A, Kudo I, et al. Segregated coupling of phospholipases A2, cyclooxygenases, and terminal prostanoid synthases in different phases of prostanoid biosynthesis in rat peritoneal macrophages. Journal of Immunology. 1998;160:2974–2982. [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vascular Health and Risk Management. 2006;2:213–219. doi: 10.2147/vhrm.2006.2.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, et al. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyagode BA, Williams IR, Morgan ET. Altered inflammatory responses to Citrobacter rodentium infection, but not bacterial lipopolysaccharide, in mice lacking the Cyp4a10 or Cyp4a14 genes. Inflammation. 2014;37:893–907. doi: 10.1007/s10753-013-9809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliw EH, Stark K, Bylund J. Oxidation of prostaglandin H(2) and prostaglandin H(2) analogues by human cytochromes P450: Analysis of omega-side chain hydroxy metabolites and four steroisomers of 5-hydroxyprostaglandin I(1) by mass spectrometry. Biochemical Pharmacology. 2001;62:407–415. doi: 10.1016/s0006-2952(01)00683-9. [DOI] [PubMed] [Google Scholar]
- Orning L. Omega-oxidation of cysteine-containing leukotrienes by rat-liver microsomes. Isolation and characterization of omega-hydroxy and omega-carboxy metabolites of leukotriene E4 and N-acetylleukotriene E4. European Journal of Biochemistry. 1987;170:77–85. doi: 10.1111/j.1432-1033.1987.tb13669.x. [DOI] [PubMed] [Google Scholar]
- Panigrahy D, Edin ML, Lee CR, Huang S, Bielenberg DR, Butterfield CE, et al. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. The Journal of Clinical Investigation. 2012;122:178–191. doi: 10.1172/JCI58128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahy D, Kaipainen A, Greene ER, Huang S. Cytochrome P450-derived eicosanoids: The neglected pathway in cancer. Cancer Metastasis Reviews. 2010;29:723–735. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker RS, Sontag TJ, Swanson JE, McCormick CC. Discovery, characterization, and significance of the cytochrome P450 omega-hydroxylase pathway of vitamin E catabolism. Annals of the New York Academy of Sciences. 2004;1031:13–21. doi: 10.1196/annals.1331.002. [DOI] [PubMed] [Google Scholar]
- Parkinson OT, Kelly EJ, Bezabih E, Whittington D, Rettie AE. Bioactivation of 4-Ipomeanol by a CYP4B enzyme in bovine lung and inhibition by HET0016. Journal of Veterinary Pharmacology and Therapeutics. 2012;35:402–405. doi: 10.1111/j.1365-2885.2011.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Golden M, Henderson WR., Jr Leukotrienes. The New England Journal of Medicine. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- Powell WS, Rokach J. Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochimica et Biophysica Acta. 2015;1851:340–355. doi: 10.1016/j.bbalip.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell PK, Wolf I, Jin R, Lasker JM. Metabolism of arachidonic acid to 20- hydroxy-5,8,11, 14-eicosatetraenoic acid by P450 enzymes in human liver: Involvement of CYP4F2 and CYP4A11. The Journal of Pharmacology and Experimental Therapeutics. 1998;285:1327–1336. [PubMed] [Google Scholar]
- Preiss B, Bloch K. Omega-oxidation of long chain fatty acids in rat liver. The Journal of Biological Chemistry. 1964;239:85–88. [PubMed] [Google Scholar]
- Quigley R, Baum M, Reddy KM, Griener JC, Falck JR. Effects of 20- HETE and 19(S)-HETE on rabbit proximal straight tubule volume transport. American Journal of Physiology Renal Physiology. 2000;278:F949–F953. doi: 10.1152/ajprenal.2000.278.6.F949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JK, Azarnoff DL, Hignite CE. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature. 1980;283:397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger MA, Ebner R, Bell DR, Kiessling A, Rohayem J, Schmitz M, et al. Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Research. 2004;64:2357–2364. doi: 10.1158/0008-5472.can-03-0849. [DOI] [PubMed] [Google Scholar]
- Rivory LP, Slaviero KA, Clarke SJ. Hepatic cytochrome P450 3A drug metabolism is reduced in cancer patients who have an acute-phase response. British Journal of Cancer. 2002;87:277–280. doi: 10.1038/sj.bjc.6600448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiological Reviews. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- Roman LJ, Palmer CN, Clark JE, Muerhoff AS, Griffin KJ, Johnson EF, et al. Expression of rabbit cytochromes P4504A which catalyze the omega- hydroxylation of arachidonic acid, fatty acids, and prostaglandins. Archives of Biochemistry and Biophysics. 1993;307:57–65. doi: 10.1006/abbi.1993.1560. [DOI] [PubMed] [Google Scholar]
- Rose ML, Rivera CA, Bradford BU, Graves LM, Cattley RC, Schoonhoven R, et al. Kupffer cell oxidant production is central to the mechanism of peroxisome proliferators. Carcinogenesis. 1999;20:27–33. doi: 10.1093/carcin/20.1.27. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Tsukamoto H, Thurman RG. WY-14 643 rapidly activates nuclear factor kappaB in Kupffer cells before hepatocytes. Carcinogenesis. 1998;19:1217–1222. doi: 10.1093/carcin/19.7.1217. [DOI] [PubMed] [Google Scholar]
- Sala A, Voelkel N, Maclouf J, Murphy RC. Leukotriene E4 elimination and metabolism in normal human subjects. The Journal of Biological Chemistry. 1990;265:21771–21778. [PubMed] [Google Scholar]
- Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and lipoxins: Structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- Sari AN, Korkmaz B, Serin MS, Kacan M, Unsal D, Buharalioglu CK, et al. Effects of 5,14-HEDGE, a 20-HETE mimetic, on lipopolysaccharide- induced changes in MyD88/TAK1/IKKbeta/IkappaB-alpha/NF-kappaB pathway and circulating miR-150, miR-223, and miR-297 levels in a rat model of septic shock. Inflammation Research: Official Journal of the European Histamine Research Society … [et al]***. 2014;63:741–756. doi: 10.1007/s00011-014-0747-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Ishii T, Kobayashi-Matsunaga Y, Amada H, Taniguchi K, Miyata N, et al. Discovery of a N'-hydroxyphenylformamidine derivative HET0016 as a potent and selective 20-HETE synthase inhibitor. Bioorganic & Medicinal Chemistry Letters. 2001;11:2993–2995. doi: 10.1016/s0960-894x(01)00614-x. [DOI] [PubMed] [Google Scholar]
- Savas U, Hsu MH, Griffin KJ, Bell DR, Johnson EF. Conditional regulation of the human CYP4X1 and CYP4Z1 genes. Archives of Biochemistry and Biophysics. 2005;436:377–385. doi: 10.1016/j.abb.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Schwartzman ML, Falck JR, Yadagiri P, Escalante B. Metabolism of 20-hydroxyeicosatetraenoic acid by cyclooxygenase. Formation and identification of novel endothelium-depende t vasoconstrictor metabolites. The Journal of Biological Chemistry. 1989;264:11658–11662. [PubMed] [Google Scholar]
- Sehgal N, Agarwal V, Valli RK, Joshi SD, Antonovic L, Strobel HW, et al. Cytochrome P4504f, a potential therapeutic target limiting neuroinflammation. Biochemical Pharmacology. 2011;82:53–64. doi: 10.1016/j.bcp.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Isakson P. Distribution of COX-1 and COX-2 in normal and inflamed tissues. Advances in Experimental Medicine and Biology. 1997;400A:167–170. doi: 10.1007/978-1-4615-5325-0_24. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Chiang N. Resolution phase lipid mediators of inflammation: Agonists of resolution. Current Opinion in Pharmacology. 2013;13:632–640. doi: 10.1016/j.coph.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Chiang N, Dalli J, Levy BD. Lipid mediators in the resolution of inflammation. Cold Spring Harbor Perspectives in Biology. 2015;7:a016311. doi: 10.1101/cshperspect.a016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chemical Reviews. 2011;111:5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shak S, Goldstein IM. Omega-oxidation is the major pathway for the catabolism of leukotriene B4 in human polymorphonuclear leukocytes. The Journal of Biological Chemistry. 1984;259:10181–10187. [PubMed] [Google Scholar]
- Shibuya M. Vascular endothelial growth factor and its receptor system: Physiological functions in angiogenesis and pathological roles in various diseases. Journal of Biochemistry. 2013;153:13–19. doi: 10.1093/jb/mvs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. Journal of the American College of Nutrition. 2002;21:495–505. doi: 10.1080/07315724.2002.10719248. [DOI] [PubMed] [Google Scholar]
- Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: Structural, cellular, and molecular biology. Annual Review of Biochemistry. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- Smith WL, Song I. The enzymology of prostaglandin endoperoxide H synthases-1 and-2. Prostaglandins & Other Lipid Mediators. 2002:68–69. 115–128. doi: 10.1016/s0090-6980(02)00025-4. [DOI] [PubMed] [Google Scholar]
- Smyth EM, Grosser T, Wang M, Yu Y, FitzGerald GA. Prostanoids in health and disease. Journal of Lipid Research. 2009;50(Suppl.):S423–S428. doi: 10.1194/jlr.R800094-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, et al. Activation of human aortic smooth-muscle cells is inhibited by PPARalpha but not by PPARgamma activators. Nature. 1998;393:790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- Stark K, Dostalek M, Guengerich FP. Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. The FEBS Journal. 2008;275:3706–3717. doi: 10.1111/j.1742-4658.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov I, Krueger W, Mankowski D, Guernsey L, Kaur A, Glynn J, et al. The cytochromes P450 (CYP) response to allergic inflammation of the lung. Archives of Biochemistry and Biophysics. 2006;456:30–38. doi: 10.1016/j.abb.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Tacconelli S, Patrignani P. Inside epoxyeicosatrienoic acids and cardiovascular disease. Frontiers in Pharmacology. 2014;5:239. doi: 10.3389/fphar.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theken KN, Deng Y, Kannon MA, Miller TM, Poloyac SM, Lee CR. Activation of the acute inflammatory response alters cytochrome P450 expression and eicosanoid metabolism. Drug Metabolism and Disposition. 2011;39:22–29. doi: 10.1124/dmd.110.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradonsky A, Rubin T, Beck R, Ring B, Seitz R, Mair S. A search for reliable molecular markers of prognosis in prostate cancer: A study of 240 cases. American Journal of Clinical Pathology. 2012;137:918–930. doi: 10.1309/AJCPF3QWIG8FWXIH. [DOI] [PubMed] [Google Scholar]
- Tsikas D, Zoerner AA. Analysis of eicosanoids by LC-MS/MS and GC-MS/MS: A historical retrospect and a discussion. Journal of Chromatography B, Analytical Technologies in the Biomedical and Life Sciences. 2014;964:79–88. doi: 10.1016/j.jchromb.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Tunctan B, Korkmaz B, Sari AN, Kacan M, Unsal D, Serin MS, et al. Contribution of iNOS/sGC/PKG pathway, COX-2, CYP4A1, and gp91(phox) to the protective effect of 5,14-HEDGE, a 20-HETE mimetic, against vasodilation, hypotension, tachycardia, and inflammation in a rat model of septic shock. Nitric Oxide: Biology and Chemistry/Official Journal of the Nitric Oxide Society. 2013;33:18–41. doi: 10.1016/j.niox.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane JR, Bakhle YS, Botting RM. Cyclooxygenases 1 and 2. Annual Review of Pharmacology and Toxicology. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- Verkade PE, Elzas M, van der Lee J, de Wolff HH, Verkade-Sandbergen A, van der Sande D. Untersuchungen über den Fettstoffwechsel I. Hoppe-Seyler's Zeitschrift für physiologische Chemie. 1933;215:225–257. [Google Scholar]
- Wakabayashi K, Shimazono N. Studies on omega-oxidation of fatty acids in vitroIOverall reaction and intermediate. Biochimica et Biophysica Acta. 1963;70:132–142. doi: 10.1016/0006-3002(63)90733-9. [DOI] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nature Reviews Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MZ, Saulter JY, Usuki E, Cheung YL, Hall M, Bridges AS, et al. CYP4F enzymes are the major enzymes in human liver microsomes that catalyze the O- demethylation of the antiparasitic prodrug DB289 [2,5-bis(4-amidinophenyl)furan-bis- O-methylamidoxime] Drug Metabolism and Disposition: The Biological Fate of Chemicals. 2006;34:1985–1994. doi: 10.1124/dmd.106.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MH, Wang J, Chang HH, Zand BA, Jiang M, Nasjletti A, et al. Regulation of renal CYP4A expression and 20-HETE synthesis by nitric oxide in pregnant rats. American Journal of Physiology Renal Physiology. 2003;285:F295–F302. doi: 10.1152/ajprenal.00065.2003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao J, Kalsotra A, Turman CM, Grill RJ, Dash PK, et al. CYP4Fs expression in rat brain correlates with changes in LTB4 levels after traumatic brain injury. Journal of Neurotrauma. 2008;25:1187–1194. doi: 10.1089/neu.2008.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: Markers and models. Nature Reviews Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Wiek C, Schmidt EM, Roellecke K, Freund M, Nakano M, Kelly EJ, et al. Identification of amino acid determinants in CYP4B1 for optimal catalytic processing of 4-ipomeanol. The Biochemical Journal. 2015;465:103–114. doi: 10.1042/BJ20140813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Cambj-Sapunar L, Kehl F, Maier KG, Takeuchi K, Miyata N, et al. Effects of a 20-HETE antagonist and agonists on cerebral vascular tone. European Journal of Pharmacology. 2004;486:297–306. doi: 10.1016/j.ejphar.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Yu W, Chai H, Li Y, Zhao H, Xie X, Zheng H, et al. Increased expression of CYP4Z1 promotes tumor angiogenesis and growth in human breast cancer. Toxicology and Applied Pharmacology. 2012;264:73–83. doi: 10.1016/j.taap.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Chen L, Yang YQ, Falck JR, Guo AM, Li Y, et al. Cytochrome P450 omega-hydroxylase promotes angiogenesis and metastasis by upregulation of VEGF and MMP-9 in non-small cell lung cancer. Cancer Chemotherapy and Pharmacology. 2011;68:619–629. doi: 10.1007/s00280-010-1521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen L, Hardwick JP. Promoter activity and regulation of the CYP4F2 leukotriene B(4) omega-hydroxylase gene by peroxisomal proliferators and retinoic acid in HepG2 cells. Archives of Biochemistry and Biophysics. 2000;378:364–376. doi: 10.1006/abbi.2000.1836. [DOI] [PubMed] [Google Scholar]
- Zhang X, Hardwick JP. Regulation of CYP4F2 leukotriene B4 omega-hydroxylase by retinoic acids in HepG2 cells. Biochemical and Biophysical Research Communications. 2000;279:864–871. doi: 10.1006/bbrc.2000.4020. [DOI] [PubMed] [Google Scholar]
- Zollner A, Dragan CA, Pistorius D, Muller R, Bode HB, Peters FT, et al. Human CYP4Z1 catalyzes the in-chain hydroxylation of lauric acid and myristic acid. Biological Chemistry. 2009;390:313–317. doi: 10.1515/BC.2009.030. [DOI] [PubMed] [Google Scholar]
- Zou AP, Ma YH, Sui ZH, Ortiz de Montellano PR, Clark JE, Masters BS, et al. Effects of 17-octadecynoic acid, a suicide-substrate inhibitor of cytochrome P450 fatty acid omega-hydroxylase, on renal function in rats. The Journal of Pharmacology and Experimental Therapeutics. 1994;268:474–481. [PubMed] [Google Scholar]