SUMMARY
Near-infrared fluorescent proteins (NIR FPs) engineered from bacterial phytochromes (BphPs) are the probes of choice for deep-tissue imaging. Detection of several processes requires spectrally distinct NIR FPs. We developed an NIR FP, BphP1-FP, which has the most blue-shifted spectra and the highest fluorescence quantum yield among BphP-derived FPs. We found that these properties result from the binding of the biliverdin chromophore to a cysteine residue in the GAF domain, unlike natural BphPs and other BphP-based FPs. To elucidate the molecular basis of the spectral shift, we applied biochemical, structural and mass spectrometry analyses and revealed the formation of unique chromophore species. Mutagenesis of NIR FPs of different origins indicated that the mechanism of the spectral shift is general and can be used to design multicolor NIR FPs from other BphPs. We applied pairs of spectrally distinct point cysteine mutants to multicolor cell labeling and demonstrated that they perform well in model deep-tissue imaging.
INTRODUCTION
Bacterial phytochrome photoreceptors (BphPs) are of great interest as protein templates for the development of genetically encoded near-infrared (NIR) fluorescent probes for in vivo imaging (Piatkevich et al., 2013a; Shcherbakova et al., 2015). In an NIR optical window (650–900 nm), mammalian tissue is relatively transparent to light because the combined absorption of water, hemoglobin, and melanin is minimal, and autofluorescence is low (Weissleder, 2001). Therefore, NIR FPs are the probes of choice for truly (up to 20 mm) deep-tissue imaging in neuroscience, stem cell biology, developmental biology, and cancer research (Jiguet-Jiglaire et al., 2014; Sanders et al., 2013; Zhu et al., 2013). Non-invasive imaging of several biological processes in vivo requires spectrally distinct NIR FPs, which all have excitation and emission near or above 650 nm. Understanding of the chromophore-protein interactions responsible for spectral properties will allow us to rationally design such probes from any NIR FP type.
BphPs belongs to a family of phytochrome photoreceptors found in plants, algae, fungi, bacteria, and cyanobacteria, which use linear tetrapyrroles, also known as bilins, as a chromophore (Auldridge and Forest, 2011; Giraud and Vermeglio, 2008; Karniol et al., 2005; Rockwell and Lagarias, 2010). Importantly, BphPs incorporate the most near-infrared-absorbing bilin, biliverdin IXα (BV) (Bhoo et al., 2001), unlike plant and cyanobacterial phytochromes, which utilize phytochromobilin (PΦB) and phycocyanobilin (PCB) (Rockwell and Lagarias, 2010). BV happens to be naturally present in cells and tissues of higher animals, including flies, fishes, and mammals, as a product of enzymatic heme degradation (Kapitulnik and Maines, 2012; Shu et al., 2009; Tran et al., 2014).
In all phytochromes, the bilin chromophore is positioned in a pocket of the GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) domain (Burgie et al., 2014; Essen et al., 2008; Wagner et al., 2005; Yang et al., 2009). The GAF and the N-terminal PAS (Per-ARNT-Sim) domains are minimally required for covalent BV binding (Wagner et al., 2005). BV is covalently attached to a conserved Cys in the N-terminal extension of the PAS domain (Lamparter et al., 2003, 2004). The N-terminal extension containing binding Cys passes through a knot loop structure formed by residues of the GAF domain (Wagner et al., 2005). Unlike BphPs, plant and cyanobacterial phytochromes are not able to bind BV. Instead, they bind PΦB and PCB to conserved Cys residue in the GAF domain (Rockwell and Lagarias, 2010).
The PAS and GAF domains are followed by the PHY (phytochrome-specific) domain, which is important for chromophore photoconversion and light-driven signal transduction (Takala et al., 2014; Wu and Lagarias, 2000; Yang et al., 2008). Light absorption induces a photo-isomerization of bilin at its 15/16 double bond. Two states corresponding to distinct BV conformations are Pr (red absorbing, cis 15/16 double bond) and Pfr (far-red absorbing, trans 15/16 double bond). Typical absorbance maxima of BphPs are 680–710 nm for the Pr state and 750–770 nm for the Pfr state (Giraud and Vermeglio, 2008). In addition to the main absorbance peaks of the Pr and Pfr states (Q band), phytochromes also absorb at ~400 nm (Soret band), which does not depend on covalent binding of BV and is a common band for tetrapyrrole compounds (Franzen and Boxer, 1997).
Weak fluorescence of natural phytochromes has been known for years (quantum yields are ~1%–4%) (Auldridge et al., 2012; Toh et al., 2011), although the utility of phytochromes as templates was first explored a decade ago by Fischer and Lagarias (2004). To engineer NIR FPs, BphPs need to be truncated to PAS-GAF domains, and mutations stabilizing the Pr state should be introduced to prevent non-radiative energy dissipation (Auldridge et al., 2012; Toh et al., 2011). In recent years, a number of NIR FPs were engineered from different BphPs. Deinococcus radiodurans DrBphP was engineered into IFP1.4 (Shu et al., 2009), IFP2.0 (Yu et al., 2014), IFP1.4rev (Bhattacharya et al., 2014), and Wi-Phy (Auldridge et al., 2012), and Bradyrhizobium BrBphP was engineered into mIFP (Yu et al., 2015). Several proteins of an iRFP series were developed based on Rhodopseudomonas palustris RpBphP2 and RpBphP6 (Filonov et al., 2011; Filonov and Verkhusha, 2013; Shcherbakova and Verkhusha, 2013). The iRFP series was engineered to specifically incorporate endogenous BV, resulting in their high fluorescence in mammalian cells.
Imaging several cell populations, tissues, and organs in animals requires spectrally distinct NIR FPs. Attempts to stabilize the Pfr state and make it fluorescent have been unsuccessful so far. Recently, we found that by mutating residues located close to the chromophore it was possible to shift the spectra of NIR FPs (Shcherbakova and Verkhusha, 2013). Using this strategy, we developed iRFP670, iRFP682, iRFP702, iRFP713, and iRFP720, starting from two BphPs, RpBphP2 and RpBphP6. Interestingly, blue-shifted iRFP670 and iRFP682 have higher quantum yields than their respective red-shifted NIR FP variants.
Here we developed a novel NIR FP, named BphP1-FP, from wild-type RpBphP1. BphP1-FP exhibits the most blue-shifted absorbance and fluorescence emission and the highest quantum yield among the available BphP-derived NIR FPs. We performed biochemical, structural, and mass spectrometry analyses of BphP1-FP and its mutants to characterize the binding of BV chromophore. We found that the BphP1-FP mutant forms two unique BV adducts. Based on this novel phenomenon, we propose a mechanism for the chromophore blue shift and a rational design strategy to tune color and brightness of NIR FPs.
RESULTS
Development of a Blue-Shifted Near-Infrared Protein BphP1-FP
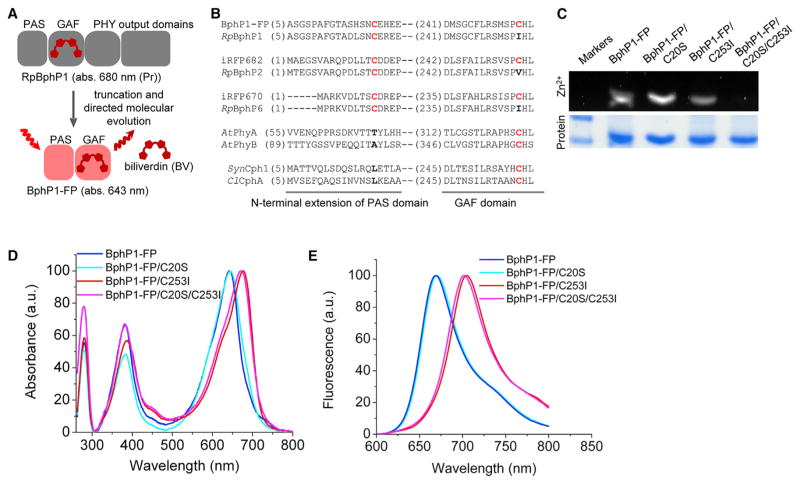
We chose RpBphP1 as a starting template for directed molecular evolution. The Pr state of this BphP absorbs at 680 nm, which is 20 and 30 nm blue-shifted compared with the Pr states of RpBphP6 and RpBphP2 (Giraud and Vermeglio, 2008). The crystal structure is available for this protein, although at a low resolution (2.9 Å) (Bellini and Papiz, 2012).
First, we truncated RpBphP1 to the chromophore-binding PAS-GAF domain. This protein was weakly fluorescent with a quantum yield of 3.3% (Table S1). Then we randomly mutated the key residue Asp201 and adjacent Ile202 in a conserved PXSDIP motif and screened the library of mutants for brightness in bacteria producing BV. These mutations were shown to stabilize the chromophore in the Pr state and increase the fluorescence quantum yield (Auldridge et al., 2012; Shcherbakova and Verkhusha, 2013). A mixture of brightest mutants was subjected to random mutagenesis in bacteria. Using flow cytometry followed by screening of bacterial colonies on Petri dishes, we specifically selected mutants with the blue-shifted fluorescence spectra. Following three rounds of random mutagenesis, we obtained the protein named BphP1-FP. This FP had an excitation peak at 639 nm and an emission peak at 669 nm. The absorbance peak was at 643 nm, which is 36 nm shorter than the absorbance of RpBphP1 in the Pr state (Figure 1A). Interestingly, the fluorescence quantum yield of BphP1-FP was 13%, which is the highest quantum yield observed in BphP-derived FPs.
Figure 1. Chromophore-Binding Cysteine in the GAF Domain Is Responsible for the Hypsochromic Shift in BphP1-FP.
(A) Schematic representation of a directed molecular evolution used for development of BphP1-FP. Absorbance peaks for BphP1-FP and parental RpBphP1 in the Pr form are indicated.
(B) Sequence alignment of the blue-shifted NIR FPs with their BphP templates and representative plant and cyanobacterial phytochromes in the regions of PAS and GAF domains containing bilin binding Cys (in red). BphPs (Rhodopseudomonas palustris RpBphP1, RpBphP2, and RpBphP6) covalently bind BV via Cys in PAS domain, whereas plant phytochromes (Arabidopsis thaliana AtPhyA and AtPhyB) and cyanobacterial phytochromes (Synechocystis Cph1 and Calothrix CphA) covalently bind PΦB and PCB via Cys in GAF domain.
(C) Assay of covalent BV binding. Proteins in SDS-PAGE were incubated with ZnCl2 for zinc-induced fluorescence of bound bilin (Zn2+) and then stained with Coomassie Blue (Protein).
(D) Overlay of absorbance spectra of BphP1-FP, BphP1-FP/C20S, BphP1-FP/C253I, and BphP1-FP/C20S/C253I.
(E) Overlay of emission spectra of BphP1-FP, BphP1-FP/C20S, BphP1-FP/C253I, and BphP1-FP/C20S/C253I. See also Figure S1, Table S1, and Supplemental Data File.
Spectral Blue Shift is Caused by a Cysteine in the GAF Domain
We noticed that, in addition to conserved Cys at the N-extension of the PAS domain (C20), BphP1-FP contains Cys in the conserved -SPXH- motif in the GAF domain (C253). Two Cys residues at the same positions were also observed in previously reported blue-shifted iRFP670 and iRFP682, developed from RpBphP6 and RpBphP2, respectively (Figure 1B and Supplemental Data File). Cys253 is located at the same position as a chromophore binding Cys in plant and cyanobacterial phytochromes.
We hypothesized that Cys253 in the GAF domain may covalently bind BV causing the observed spectral blue shift. To test this, we obtained mutants with a single Cys in either the PAS (BphP1-FP/C253I) or the GAF (BphP1-FP/C20S) domains, and a mutant without both cysteine residues. The proteins were expressed and purified from bacteria producing BV. We found that both Cys20 and Cys253 covalently bind BV, whereas the BphP1-FP/C20S/C253I double mutant does not bind BV covalently (Figure 1C). Next, we characterized the spectral properties of the mutants. The C253I mutation in BphP1-FP resulted in a red shift. The absorbance shifted to 677 nm and the emission to 704 nm, respectively (Figures 1D and 1E). The C20S mutation resulted in a protein with absorbance and fluorescence spectra very similar to those of BphP1-FP (Figures 1D and 1E). This suggests that in the protein with two Cys residues BV is preferably bound to Cys in the GAF domain. The double mutant BphP1-FP/C20S/C253I had red-shifted spectra similar to BphP1-FP/C253I, which is consistent with previous observations (Borucki et al., 2009; Wagner et al., 2007) (Figures 1D and 1E; Table 1). BphP1-FP/C20S had a quantum yield of 13.9% (Table 1). Quantum yields of both red-shifted BphP1-FP/C253I and BphP1-FP/C20S/C253I were considerably lower and did not exceed 4%. Interested by the effect of Cys253, we tested the properties of the wild-type RpBphP1 (PAS-GAF domains) with C20S/I253C mutations. We found that this mutant with the chromophore-binding Cys in the GAF domain was blue-shifted relative to the wild-type protein, with an absorbance peak at 650 nm and an emission peak at 671 nm (Table S1 and Figures S1A–S1C). The quantum yield of this mutant (4.1%) was higher than that of wild-type RpBphP1 (3.3%).
Table 1.
Properties of BphP1-FP and its Mutants Expressed in Bacteria with Either BV or PΦB
| NIR FP | Absorbance Maximum (nm) | Excitation Maximum (nm) | Emission Maximum (nm) | Extinction Coefficient (M−1 cm−1) | Quantum Yield (%) | Relative Molecular Brightness (%) |
|---|---|---|---|---|---|---|
| BphP1-FP + BV | 643 | 639 | 669 | 60,000 | 13.0 | 100 |
| BphP1-FP/C20S + BV | 642 | 639 | 670 | 82,000 | 13.9 | 146 |
| BphP1-FP/C253I + BV | 677 | 675 | 704 | 70,000 | 4.0 | 36 |
| BphP1-FP/C20S/C253I + BV | 673 | 671 | 702 | 59,000 | 3.8 | 29 |
| BphP1-FP/C20S + PΦB | 641 | 639 | 669 | 105,000 | 15.4 | 207 |
| BphP1-FP/C253I + PΦB | 677 | 675 | 703 | 59,000 | NA | NA |
NA, not available. See also Table S5.
To check for possible heterogeneity of the chromophore and the presence of protoporphyrin IX (PPIX) in BphP1-FP/C20S, we recorded emission spectra at different excitation wavelengths. We found no significant heterogeneity and no presence of PPIX (manifested by emission peaks with maxima at 622–626 nm according to Wagner et al., 2008) (Figures S1D–S1G). We concluded that covalent BV adducts bound to the Cys253 in the GAF domain are responsible for the high brightness and the blue-shifted spectra of BphP1-FP and BphP1-FP/C20S.
Structure of BV Chromophore Bound to the Cysteine in the GAF Domain
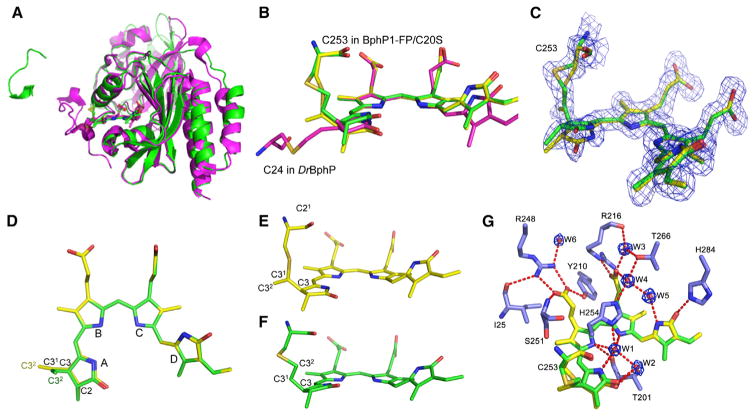
To study the nature of the BV adduct bound to Cys in the GAF domain, we crystallized BphP1-FP/C20S. The structure was determined at 1.64-Å resolution (Tables S2 and S3).
Overall, BphP1-FP/C20S has a fold typical of the chromophore-binding domains of BphPs (Figures 2A and S2). In contrast to other BphP structures, the N-terminal extension of BphP1-FP/C20S is mainly disordered, as there is no stabilizing covalent bond between residue 20 and BV. The figure-eight knot common to all BphPs is preserved (Figure S2C). The chromophore species in BphP1-FP/C20S are linked to Cys253 in the GAF domain (Figure 2B).
Figure 2. Structure of BphP1-FP/C20S.
(A) Superposition of the structures of BphP1-FP/C20S (PDB: 4XTQ, in green) and Deinococcus radiodurans DrBphP PAS-GAF domains (PDB: 2O9C, in magenta).
(B) Superimposed structures as in (A) showing the chromophores and binding cysteine residues. Biliverdin adducts are green and yellow in BphP1-FP/C20S and magenta in DrBphP PAS-GAF.
(B–E) Carbon atoms of the molecules connected to C253 via C31 and C32 atoms are shown in yellow and green, respectively. Nitrogen atoms are in blue, oxygen in red, sulfur in dark yellow. (C) Biliverdin adducts in FEM electron density mapping countered at 1.5σ. (D) Top view of the two overlaid biliverdin adducts. Pyrrole rings and carbon atoms in ring A are labeled. (E and F) Side views of the two biliverdin adducts connected via C31 (E) or C32 (F) atoms.
(G) The nearest chromophore environment with a detailed view of the water molecules in the FEM electron density mapping countered at 2.5σ. See also Figure S2; Tables S2 and S3.
Electron density maps of the chromophore revealed two chromophore species that form thioether covalent bonds with Cys253 in the GAF domain. These species are linked to Cys253 via C31 or C32 carbon atoms of the A ring (Figure 2C). The presence of two chromophores was confirmed by both composite OMIT Fo-Fc and conventional 2Fo-Fc maps. We also confirmed it by calculated feature-enhanced electron density mapping (Afonine et al., 2015) (see also Experimental Procedures). Similarly to other BphPs in the Pr state (Auldridge et al., 2012; Wagner et al., 2007; Yu et al., 2014), both chromophores adopt a 5Zsyn,10Zsyn,15Zanti conformation for the methylene linkers connecting the four pyrrole rings (Figures 2C and 2D). However, there is a remarkable difference between the A rings of chromophores in BphP1-FP/C20S and BV adducts observed in other BphP-derived proteins. In both chromophores linked to Cys253, C2, and C3, carbon atoms assume tetrahedral geometry indicative of sp3 hybridization similarly to A rings in reduced PCB and PΦB bilins (Figure 2C). The methyl group of C21 atom points away from the GAF cysteine residue. The two chromophore species differ by the orientations of the A rings. Whereas the chromophore linked to the Cys253 via C31 atom has ring A in a plane with rings B and C, the chromophore linked to the Cys253 via the C32 atom has ring A rotated by 11° out of this plane (Figures 2E and 2F).
The nearest chromophore environment in BphP1-FP/C20S in general is similar to that of other BphP-based FPs with some unique characteristics (Figure 2G). In addition to a well-ordered “pyrrole water” (W1), there is an additional water molecule W2, which is located within hydrogen bond distance to W1 and forms a hydrogen bond with the carbonyl oxygen of the chromophore A ring (Figures 2G and S2D). Another unique feature is the orientation of the propionate group of the C ring, which is rotated toward the side chain of Arg216 and is stabilized by a hydrogen bond with this residue.
BV and PΦB Bound to the Cysteine in the GAF Domain Exhibit Similar Spectral Properties
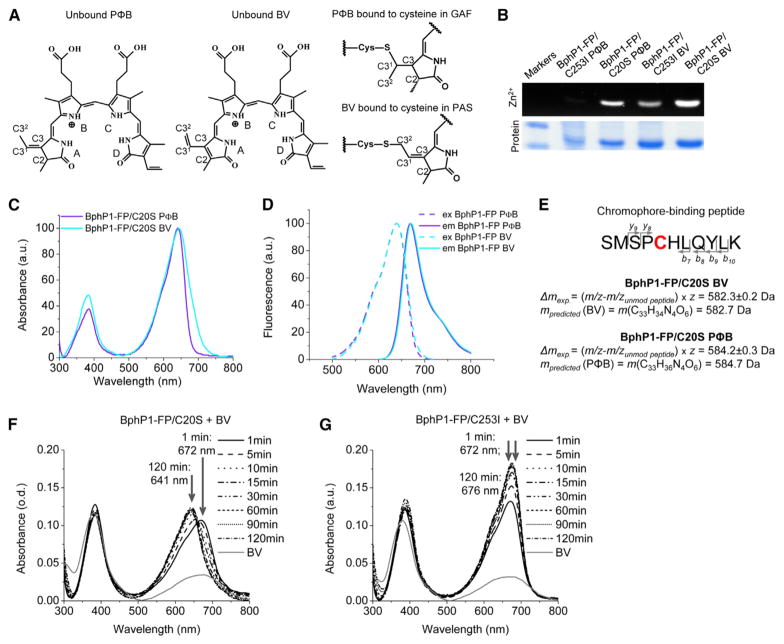
As the C3 atom in the A ring does not form a double bond, the BV adducts in BphP1-FP/C20S should have the same number of conjugated double bonds as bound PΦB (Figure 3A). PΦB is enzymatically produced from BV by ferredoxin-dependent bilin reductase (Rockwell and Lagarias, 2010). Importantly, in plant phytochromes, PΦB is covalently bound to Cys in the GAF domain.
Figure 3. Spectral Properties and Chromophore Binding in BphP1-FP Cysteine Mutants Assembled with P.
ΦB and BV
(A) Chemical structures of unbound PΦB (left) and BV (middle), and rings A of PΦB bound to cysteine in the GAF domain and BV bound to cysteine in the PAS domain (right). Pyrrole rings and carbon atoms in ring A are labeled. Bound PΦB has no double bond linked to the C3 atom, similar to BV adducts observed in BphP1-FP/C20S.
(B) Assay of covalent bilin binding for BphP1-FP/C20S and BphP1-FP/C253I assembled with either BV or PΦB. Proteins in SDS-PAGE were incubated with ZnCl2 for zinc-induced fluorescence of bound bilin (Zn2+) and then stained with Coomassie Blue (Protein).
(C) Overlay of absorbance spectra of BphP1-FP/C20S assembled with BV and PΦB.
(D) Overlay of excitation and emission spectra of BphP1-FP/C20S assembled with BV and PΦB.
(E) Mass spectrometry analysis of BphP1-FP/C20S assembled with BV or PΦB. Chromophore-binding peptide, its fragments used for analysis, calculated masses of the chromophores, and their predicted masses with corresponding molecular formulas are shown. Chromophore masses were calculated as differences between mass-to-charge ratios (m/z) of peptide fragments with and without modification, multiplied by charge (z).
(F and G) Assembly of purified BphP1-FP/C20S and BphP1-FP/C253I apoproteins with BV in vitro. Free BV in PBS buffer containing 1 mM DTT was mixed with the 1.5-fold molar excess of the purified apoproteins BphP1-FP/C20S (F) and BphP1-FP/C253I (G) and absorbance spectra were monitored for 2 hr. See also Figures S3 and S4, and Table S4.
We tested chromophore binding and measured the spectral properties of the BphP-FP mutants expressed in bacteria producing PΦB. BphP1-FP/C20S readily incorporated PΦB and bound it covalently, in contrast to control BphP1-FP/C253I with Cys in the PAS domain (Figure 3B). The absorbance spectrum of BphP1-FP/C20S with PΦB was similar to that with BV but narrower (Figure 3C and Table 1). The excitation and emission spectra of BphP1-FP/C20S expressed either with PΦB or with BV were nearly identical (Figure 3D). Interestingly, BphP1-FP/C20S assembled with PΦB exhibited a high quantum yield of 15.4% (Table 1). Control BphP1-FP/C253I incorporated PΦB very weakly, with excitation and emission spectra similar to the spectra of BphP1-FP/C253I expressed with BV (Figures S3A and S3B). Since the chemical structures of non-covalently bound PΦB and BV covalently bound to the Cys in the PAS domain are similar (Figure 3A), we can conclude that weak fluorescence of BphP1-FP/C253I assembled with PΦB originates from non-covalently bound PΦB.
The similarity of the fluorescence spectra of BphP1-FP/C20S expressed with BV and with PΦB suggests that at least one of the chromophore species in BphP1-FP/C20S may be reduced and structurally identical to PΦB. To test this, we analyzed BphP1-FP/C20S assembled with BV and PΦB using mass spectroscopy. We found that the mass of the chromophore bound in BphP1-FP/C20S assembled with BV corresponds to BV, whereas the mass of the chromophore bound to the protein assembled with PΦB is ~2 Da larger and corresponds to PΦB (Figures 3E and S4; Table S3). Thus, the BV chromophore is not reduced when assembled with BphP1-FP/C20S.
Holoprotein with BV Bound to the Cysteine in the GAF Domain Assembles Autocatalytically
Next, we tested whether a holoprotein can be assembled without any additional enzymes and cofactors. We purified the BphP1-FP/C20S and control BphP1-FP/C253I apoproteins and assembled them with BV in vitro as described previously (Quest and Gartner, 2004).
When BphP1-FP/C20S was mixed with BV, the absorbance maximum at the Q band gradually shifted toward shorter wavelengths (Figure 3F). Starting from 672 nm, which corresponds to non-covalently bound BV (Table 1, see BphP1-FP/C20S/C253I), the absorbance maximum reached 641 nm and this matches the value observed for BphP1-FP/C20S assembled in bacteria. Initially, a similar maximum of 672 nm was observed for BphP1-FP/C253I. The absorbance then shifted to 676 nm, which corresponds to the maximum observed for this protein expressed with BV in bacteria (Figures 3G and S3C, and S3D with an overlay of fluorescence spectra). Thus, the blue-shifted chromophore bound to Cys in the GAF domain forms autocatalytically.
The Nature of the Hypsochromic Spectral Shift
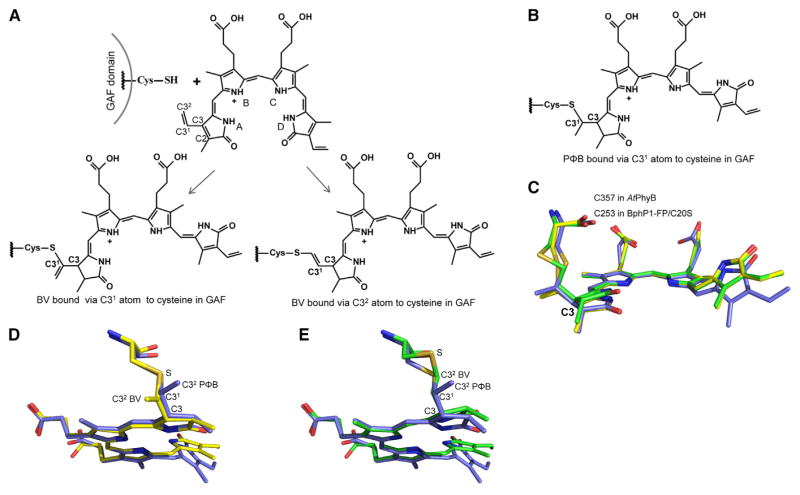
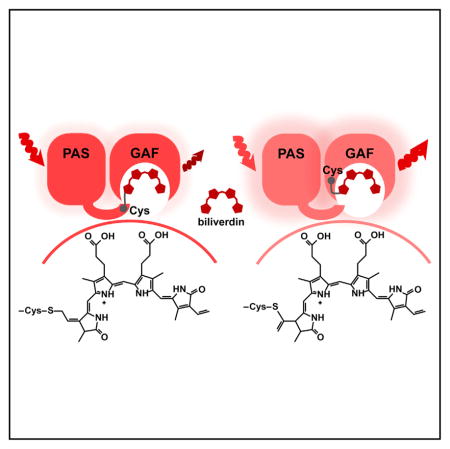
The structural and biochemical analyses indicate that both BV adducts bound to Cys253 via either C31 or C32 atoms have no double bonds formed by the C3 atom in ring A, and that they are not chemically reduced and form without additional enzymes and cofactors.
These data suggest that, upon binding, BV adducts autocatalytically isomerize to change the initial position of the double bond between C3 and C2 or C3 and C31 atoms to the double bond between C31 and C32 atoms (Figure 4). The re-arrangement of initially bound intermediates (Figure S5) with the double bond between C3 and C2 or C3 and C31 atoms should reduce the steric constraint. In contrast to the chromophore-binding Cys in the PAS domain lying almost in the same plane with BV, the Cys in the GAF domain is located above the BV chromophore plane (Figure 2B). The latter Cys location causes a steric constraint if the C3 atom forms a double bond with C2 or C31 atoms and has a planar geometry.
Figure 4. Chemical Structures of BV Chromophores Bound to the Cysteine in the GAF Domain.
(A) The BV chromophores in BphP1-FP/C20S bound to cysteine in the GAF domain. Pyrrole rings and carbon atoms in ring A are labeled.
(B) PΦB chromophore bound to the cysteine in the GAF domain.
(C) Superposition of the BV chromophore species in crystal structures of BphP1-FP/C20S (PDB: 4XTQ, the chromophore bound via C31 is shown in yellow, the chromophore bound via C32 is shown in green) and AtPhyB with bound PΦB (PDB: 4OUR, in blue). Note the tetrahedral geometry of C3 atoms in both structures.
(D and E) Overlays of PΦB chromophore and BV chromophores bound via the C31 atom (D) and the C32 atom (E) are shown separately in the orientation, highlighting the tetrahedral geometry of the C31 atom in the PΦB adduct, and the planar geometry of C31 atoms in the BV adducts. See also Figure S5.
The proposed BV adducts explain the significant blue shift in absorbance and fluorescence observed in the NIR FPs with BV bound to Cys in the GAF domain. The nearly identical spectra of BphP1-FP/C20S assembled with BV and assembled with PΦB (Figures 3C and 3D) correlate with the similarity between the π-conjugated systems of the bound chromophores (Figures 4A and 4B).
In both BV and PΦB chromophores bound to the Cys in the GAF domain, C3 atoms in ring A do not form double bonds and have tetrahedral geometry (sp3 hybridization) that can be seen from the overlay of chromophores in BphP1-FP/C20S and plant phytochrome from Arabidopsis thaliana AtPhyB (Burgie et al., 2014) (Figure 4C). According to the chemical structures of the BV adducts (Figure 4A), the C31 atom in ring A forms a double bond with the C32 atom and thus adopts planar geometry (sp2 hybridization). This is not the case for the PΦB chromophore, in which the C31 atom is tetrahedral (sp3 hybridization) (Figure 4B). The overlays of each of the BV chromophores with the PΦB chromophore indicate the presence of a double bond between C31 and C32 atoms in BV adducts (Figures 4D and 4E).
Engineering of Spectrally Distinct NIR FPs for Multicolor Imaging
To test whether the proposed mechanism of the blue shift in BphP-based FPs is broadly applicable, we tested it on two previously reported blue-shifted NIR FPs, iRFP670 and iRFP682, which both acquired Cys in the -SPXH- motif of GAF domains in the process of molecular evolution (Shcherbakova and Verkhusha, 2013). Whereas BphP1-FP was developed from RpBphP1, iRFP670 and iRFP682 were engineered from RpBphP6 and RpBphP2, respectively. We obtained iRFP670 and iRFP682 mutants containing a single Cys either in the PAS or in the GAF domains. The spectral properties of these mutants corresponded to those of the respective mutants of BphP1-FP (Table S4). The spectra of iRFP670 and iRFP682 mutants with Cys in the PAS domain demonstrated a 35- to 40-nm red shift, while the mutants with Cys in the GAF domain had spectra similar to the original proteins with two Cys residues.
The proposed mechanism of the blue shift in BphPs (Figure 4) can be applied to engineer spectrally distinct NIR FPs for multicolor in vivo imaging. We suggest that each BphP can be developed into two NIR FPs with a spectral difference of up to 40 nm, which have chromophore-binding Cys either in the PAS or in the GAF domains. First, the blue-shifted NIR FPs can be designed by introduction of chromophore-binding Cys in the GAF domain of a BphP template at the early stages of molecular evolution. Then the red-shifted NIR FPs can be obtained from the respective blue-shifted NIR FPs by removing Cys from the GAF domain, and both obtained variants can be improved by subsequent random mutagenesis.
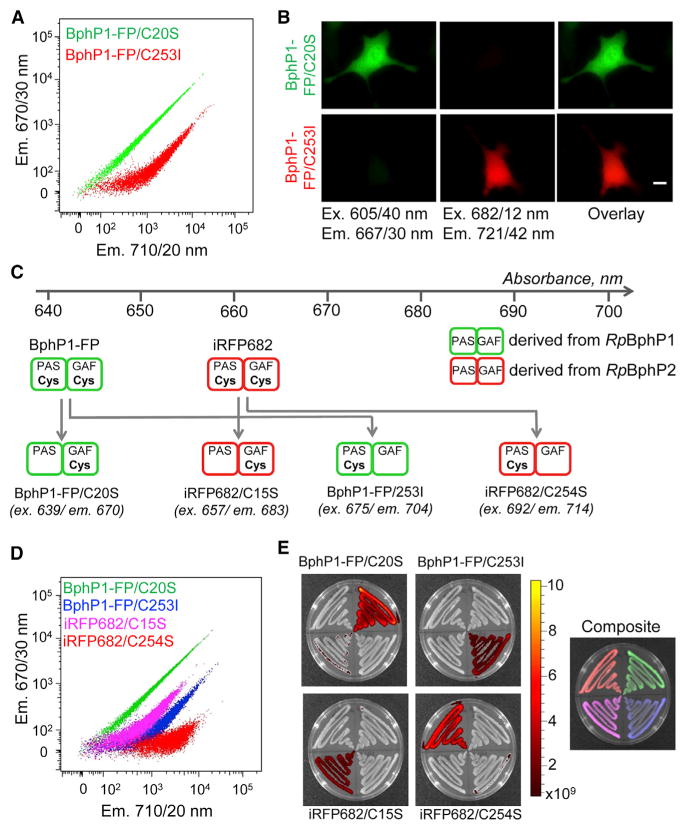
We tested whether Cys point mutants of BphP1-FP can be directly applied for multicolor labeling in mammalian cells. The BphP1-FP/C20S- and BphP1-FP/C253I-expressing cells were successfully spectrally resolved using standard flow cytometry and epifluorescence microscopy (Figures 5A and 5B). Similar two-color detection was also possible for the Cys point mutants of iRFP670 and iRFP682 (Figure S6). All tested NIR FP variants efficiently bound endogenous biliverdin, which is abundant in eukaryotes.
Figure 5. Multicolor Labeling Using Cysteine Point Mutants of NIR FPs Derived from Different BphP Templates.
(A) Flow cytometry analysis of live HeLa cells expressing either BphP1/C20S or BphP1-FP/C253I.
(B) Representative images of HeLa cells expressing BphP1/C20S and BphP1-FP/C253I. The images in the 605/40 nm excitation and 667/30 nm emission channel are shown in a green pseudo-color, and images in the 682/12 nm excitation and 721/42 nm emission channel are shown in a red pseudocolor. Scale bar represents 10 μm.
(C) Cys point mutations of BphP1-FP and iRFP682 produce four spectrally resolvable NIR FP variants.
(D) Flow cytometry analysis of live HeLa cells expressing BphP1/C20S, BphP1-FP/C253I, iRFP682/C15S, and iRFP682/C254S.
(E) Multicolor spectral imaging of bacterial streaks expressing the same four NIR FP variants as in (D). The color bar indicates the fluorescent radiant efficiency in [photon/s/cm2/steradian]/[μW/cm2]. See also Figure S6.
The use of several different BphPs will allow engineering of more than two multicolor NIR FPs. The Pr state absorption maxima of known BphPs vary from 670 to 710 nm (Giraud and Vermeglio, 2008), providing an additional 40-nm range to the ~40-nm spectral difference between NIR FPs developed from a single BphP. Thus, it should be possible to develop several pairs of spectrally distinct NIR FPs, of which the most blue-shifted and red-shifted ones will differ by ~80 nm.
To demonstrate how the pairs of NIR FPs obtained from different BphP templates can be applied for multicolor imaging, we tested the Cys point mutants of BphP1-FP and iRFP682, which are all spectrally distinct (Figure 5C). All four NIR FPs were spectrally resolved using flow cytometry of live mammalian cells (Figure 5D). Moreover, these proteins were also spectrally separated using a commercial multispectral imaging system (Figure 5E).
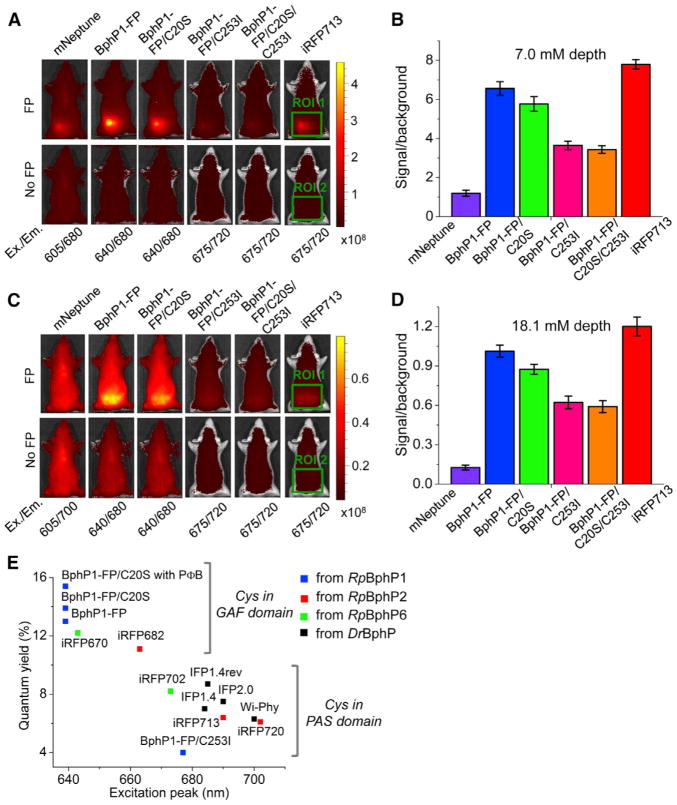
To test the performance of bright blue-shifted NIR FPs in deep-tissue imaging, we quantitatively compared BphP1-FP and its mutants with a widely used red-shifted NIR FP, iRFP713 (Filonov et al., 2011), and one of the brightest far-red GFP-like FPs, mNeptune (Lin et al., 2009). Equal amounts of purified proteins were placed inside a fluorescent mouse phantom engineered to have absorbance, light scattering, and autofluorescence matching those of mouse tissues. The fluorescence signals were imaged at two depths (7.0 and 18.1 mm) in different filter channels (Figures 6A and 6C), and signal-to-autofluorescence background ratios were quantified. One can see that blue-shifted BphP1-FP and BphP1-FP/C20S exhibit signal-to-background ratios comparable with that of iRFP713 and ~6–8 higher than that of mNeptune (Figures 6B and 6D).
Figure 6. Brightness of Blue-Shifted NIR FPs Compared with Other NIR FPs.
(A–D) Performance of blue-shifted NIR FP BphP1-FP and its mutants in deep-tissue imaging. Equal amounts of purified FPs (top row) or buffer (bottom row) were imaged in epifluorescence mode inside the fluorescent mouse phantom at 7.0 mm (A) or 18.1 mm (C) depth using different filter channels. Images with the highest fluorescence signal-to-autofluorescence background ratio among the channels are shown for each FP. The color bar indicates the total fluorescence radiant efficiency [photon/s/cm2/steradian]/[μW/cm2]. (B and D) The quantification of the signal-to-background ratios for the images shown in (A) and (C), respectively. Total radiant efficiencies of the indicated area with (ROI1) and without the FP (ROI2) were measured, and signal-to-background ratios (ROI1 – ROI2)/ROI2 were calculated. Error bars, SD (n = 3). Ex./Em., excitation maximum/emission maxima.
(E) Blue-shifted NIR FPs with chromophore-binding cysteine in the GAF domain have higher quantum yields than NIR FPs with BV bound to cysteine in the PAS domain. Positions of solid squares correspond to the excitation peaks and reported quantum yields for each NIR FP. They are colored according to the parental BphP for each protein: RpBphP1 is in blue, RpBphP2 is in red, RpBphP6 is in green, and DrBphP is in black. Characteristics of iRFP670 and iRFP682 were re-measured after expression and purification similarly to the BphP1-FP variants.
BphP1-FP and its mutant with Cys in the GAF domain exhibit a higher quantum yield than the mutant containing Cys only in the PAS domain. Intrigued by this, we analyzed the quantum yields of all known BphP-based FPs engineered from the PAS-GAF domains (Auldridge et al., 2012; Bhattacharya et al., 2014; Filonov et al., 2011; Shcherbakova and Verkhusha, 2013; Shu et al., 2009; Yu et al., 2014). We found that all NIR FPs containing Cys in the GAF domain exhibit a higher quantum yield than the FPs with Cys in the PAS domain (Figure 6E). Possibly, the higher quantum yield in NIR FPs with Cys in the GAF domain is caused by tightening of the interaction between BV and the protein. It is likely that the bilin bound to the structurally restrained Cys in the GAF domain has a higher structural rigidity than the bilin bound to the structurally loose Cys in the PAS domain and, thus, radiationless decay processes are partially inhibited.
DISCUSSION
We engineered the RpBphP1 phytochrome into the near-infrared BphP1-FP, which exhibits the most blue-shifted spectra and the highest quantum yield of 13% among the BphP-derived FPs. We found that a cysteine residue introduced in the -SPXH- motif of the GAF domain is the key residue responsible for these changes. This cysteine covalently binds BV via either C31 or C32 atoms of ring A. Upon binding, both BV chromophores autocatalytically isomerize, possibly to reduce the structural steric constraints. This results in a spectral blue shift because the isomerized double bond between C31 and C32 is no longer conjugated with the rest of the π-electron system of the chromophore.
We found that spectral shifts can be engineered in NIR FPs derived from different BphP templates. Evidently, the BV isomerization phenomenon is not limited to BphP-derived NIR FPs, but can also occur in the PAS-GAF domains of BphPs as we demonstrated for the PAS-GAF domain of RpBphP1 (Table S1). Previously, it was shown that DrBphP/C24A/M259C truncated to the PAS-GAF domains binds BV via Cys259 in the GAF (Wagner et al., 2007). An absorbance spectrum of this mutant contained two peaks, a major one at 698 nm and a minor one at 673 nm. We hypothesize that the major peak corresponded to non-covalent incorporation of BV, because (1) its absorbance maximum is identical to the maximum of the mutant DrBphP/C24A with no chromophore-binding Cys residues, and (2) DrBphP/C24A/M259C lacks Cys in the PAS and is thus unable to form a covalent bond with BV. We suggest that the minor peak corresponded to the same covalently bound BV adduct bound to the Cys in the GAF domain that is observed in BphP1-FP/C20S. Interestingly, when the chromophore-binding Cys was introduced into the GAF domains of full-length BV-binding Calothrix CphB (Quest and Gartner, 2004) and Agrobacterium tumefaciens Agp1 (Borucki et al., 2009), these BphPs started to covalently incorporate PCB. However, BV did not bind covalently, and no spectral shift was observed. Possibly, the photosensory module (PAS-GAF-PHY domains) in full-length BphPs restricted the covalent binding of BV to Cys in the GAF domain.
Our findings provide a rational design strategy to engineer spectrally distinct NIR FPs. To demonstrate this, we applied the Cys point mutants in multicolor imaging in cells (Figure 5). We found that the blue-shifted NIR FPs with Cys in the GAF domain have higher fluorescence quantum yields than NIR FPs with Cys in the PAS domain. We also showed that bright blue-shifted NIR FPs perform as well as the red-shifted NIR FPs, and considerably better than far-red GFP-like FPs in deep-tissue imaging (Figure 6).
The chemical structures of the blue-shifted BV adducts suggest that it is possible to further increase brightness of BphP-based FPs with Cys in the GAF domain by shifting a ratio between two BV adducts. The structural similarity of the BV adduct linked via the C31 atom with a PΦB adduct (Figures 4A and 4B) suggests that the protein species with a BV chromophore bound via C31 have higher quantum yield than protein molecules with BV bound via C32. We hypothesize that by mutating residues close to Cys in the GAF domain it should be possible to enrich the BV adduct linked via the C31 atom. This rational design, combined with the directed molecular evolution, should result in enhanced NIR FPs with substantially higher quantum yield than currently observed in BphP-derived FPs. Twice higher fluorescence brightness will allow non-invasive tracking of four-fold smaller objects and cell populations, imaging of four-fold deeper organs and tissues, and detection of developmental, cancer, and infection processes at substantially earlier stages.
The possibility to engineer spectrally shifted NIR FPs based on any BphP template demonstrated here can be applied to NIR FPs of other phenotypes. For example, by introducing Cys point mutations, it will be possible to create spectrally distinct bimolecular fluorescence complementation reporters based on split NIR FPs, such as iSplit (Filonov and Verkhusha, 2013), and spectrally resolvable monomeric NIR FP for protein tagging. The multicolor NIR FPs will enable simultaneous labeling and tracking of two and more cellular proteins, protein-protein interactions, cell populations, tissues, and organs in live animals in normal and pathogenic conditions.
SIGNIFICANCE
NIR FPs engineered from bacterial phytochromes enable non-invasive in vivo imaging. The lack of understanding of chromophore-protein interactions that influence the spectral properties of these FPs has hindered their development. We engineered the most blue-shifted NIR FP with the highest fluorescence quantum yield, and uncovered the molecular basis of its properties. Biochemical, structural, and mass spectrometry analyses revealed unique BV chromophore adducts. We showed that the formation of these BV adducts is generally applicable to NIR FPs of different origins and that they can be used for the rational design of spectrally distinct NIR FPs and biosensors for multicolor imaging in whole animals. We demonstrated that pairs of cysteine mutants allow multicolor imaging in live cells and that they perform well in deep-tissue imaging modeling.
EXPERIMENTAL PROCEDURES
Mutagenesis and Directed Molecular Evolution
The DNA fragment corresponding to PAS-GAF domains (first 315 amino acids) of RpBphP1 was amplified by PCR and inserted as the BglII/EcoRI fragment into a pBAD/His-B vector (Invitrogen). Site-specific mutagenesis was performed using a Quick Change mutagenesis kit (Stratagene). Random mutagenesis was performed with a GeneMorph II random mutagenesis kit (Stratagene), using conditions that resulted in a mutation frequency of up to 16 mutations per 103 base pairs. After mutagenesis, a mixture of mutated genes was electroporated into LMG194 bacterial cells (Invitrogen) containing the pWA23h plasmid encoding heme oxygenase (hmuO) from Bradyrhizobium ORS278 under the rhamnose promoter (Piatkevich et al., 2013b; Shcherbakova and Verkhusha, 2013). Typical mutant libraries consisted of more than 106 independent clones. The LMG194 cells were grown overnight at 37°C in RM minimal medium supplemented with ampicillin and kanamycin. Protein expression in the libraries was induced with 0.002% arabinose and 0.02% rhamnose. The cells were grown for 6–8 hr at 37°C and then at 18°C for 24 hr. For flow cytometry screening, bacterial cells were washed with PBS and diluted with PBS to an optical density of 0.03 nm at 600 nm. The libraries were screened using a MoFlo XDP (Beckman Coulter) fluorescence-activated cell sorter using a 592-nm laser for excitation and a 680-nm LP emission filter for positive selection. The 200–1,000 brightest infrared bacterial cells collected were rescued in SOC medium at 37°C for 1 hr, grown on LB/ampicillin/kanamycin Petri dishes supplemented with 0.02% arabinose and 0.2% rhamnose overnight at 37°C, and then incubated at 18°C. Screening for spectrally blue-shifted mutants on Petri dishes was performed with a Leica MZ16F fluorescence stereomicroscope equipped with 605/40 and 650/45 nm excitation filters and 640 and 690 nm LP emission filters (Chroma). Spectral properties of 10–20 selected mutants were tested, and their DNA was sequenced. A mixture of several selected mutants was then used as a template for the next round of mutagenesis.
Protein Expression and Purification
For expression of apoproteins, the genes encoding the proteins in the pBAD/His-B vector (Invitrogen) were expressed in TOP10 bacterial cells (Invitrogen). Bacterial cells were grown in RM medium supplemented with ampicillin. To initiate protein expression, 0.002% arabinose was added and bacterial culture was incubated for 12 hr at 37°C, followed by 24 hr at 18°C. Proteins were purified using Ni-NTA agarose (Qiagen).
For expression of proteins in bacteria in the presence of BV, the genes coding for the proteins in pBAD/His-B vector (Invitrogen) were expressed either in LMG194 or TOP10 bacterial cells (Invitrogen) bearing the pWA23h plasmid (Piatkevich et al., 2013b; Shcherbakova and Verkhusha, 2013). Bacterial cells were grown in RM medium supplemented with ampicillin and kanamycin. First, heme oxygenase was expressed with addition of 0.02% rhamnose for 5 hr at 37°C. Then 0.002% arabinose was added and the bacterial culture was incubated for an additional 12 hr at 37°C, followed by 24 hr at 18°C. Proteins were purified using Ni-NTA agarose (Qiagen). In the elution buffer, imidazole was substituted for 100 mM EDTA. The elution buffer was replaced with PBS buffer using PD-10 desalting columns (GE Healthcare).
For expression of proteins in bacteria in the presence of PΦB, the genes coding for the proteins in pBAD/His-B vector (Invitrogen) were expressed in TOP10 bacterial cells (Invitrogen) bearing the pPL-PΦB (Fischer et al., 2005; Gambetta and Lagarias, 2001) according to the protocol described by Burgie et al. (2014). In brief, cells were grown in terrific broth containing 0.4% glycerol and 1 mM MgCl2 at 37°C until the cells reach an optical density of 0.4–0.6 at 600 nm. The temperature was then decreased to 18°C, and 100 μM δ-amino-levulinic acid was added. After 1 hr, isopropyl β-D-1-thiogalactopyranoside was added to 0.4 mM, followed by addition of 0.02% arabinose after an additional 1 hr. The cells were then cultured for 16 hr. Proteins were purified using Ni-NTA agarose as described earlier.
Spectral and Biochemical Characterization
Fluorescence spectra were recorded using a FluoroMax-3 spectrofluorometer (Jobin Yvon). A Hitachi U-2000 spectrophotometer was used for absorbance measurements. If not specified in the figure legends, excitation spectra were recorded using 750 nm emission; and emission spectra were recorded using 590 nm excitation. The extinction coefficient was calculated from a comparison of absorbance values at the main peak at the Q band with the absorbance value at Soret band, assuming that the latter had an extinction coefficient of free BV of 39,900 M−1 cm−1 (Filonov et al., 2011; Shu et al., 2009). To determine quantum yield, we compared the fluorescence of a purified FP with that of an equally absorbing Nile blue dye (quantum yield is 0.27 in acidic ethanol (Sens and Drexhage, 1981)) using several dilutions.
For studies of BV assembly, purified apoproteins (15 μM) were mixed with 10 μM BV in PBS in the presence of 1 mM DTT. Absorbance spectra were monitored immediately after mixing (in about 1 min) and then at the indicated time points until the absorbance reached a steady state.
For multicolor spectral imaging of bacterial cells, Petri dishes with bacterial streaks of LMG194 strain expressing each of four NIR FPs were prepared. For linear spectral unmixing, a set of images in 19 filter channels was collected on the IVIS Spectrum. Pure fluorescence signals of each NIR FP were identified and spectrally unmixed using Living Image v.4.3.1 software (PerkinElmer).
Fluorescence Imaging in Phantom Mouse
Far-red GFP-like FP mNeptune was expressed in LMG194 bacterial cells and then purified using Ni-NTA agarose (Qiagen). All purified FPs were diluted to equal concentrations of 8 μM, calculated from the extinction coefficients at the chromophore absorbance maxima. A 5-μl volume of each FP was placed into available bores in an XFM-2 phantom mouse (PerkinElmer) at a depth of 7.0 or 18.1 mm. Images were taken in 19 different combinations of the farred and infrared excitation and emission channels using an IVIS Spectrum (PerkinElmer). A fluorescence signal-to-autofluorescence background ratio was calculated for each wavelength combination for each FP; the phantom mouse without FP was used as a background reference. All quantitative measurements of the fluorescence signal were performed using the Living Image v.4.3.1 software (PerkinElmer). To remove cross-bleed from the excitation light, we performed an adaptive background subtraction.
Mass Spectrometry Analysis
Aliquots of 10 μg of purified BphP1-FP/C20S proteins with bound BV or with bound PΦB were added in 100 μl of 50 mM ammonium bicarbonate buffer. Tryptic digestion was performed at 37°C for 16 hr using 20 μg/ml trypsin (Promega) contained in 50 mM ammonium bicarbonate buffer. The resulting digest was then diluted with 2% acetonitrile/0.2% trifluoroacetic acid. The volume of the digest solutions were reduced to 50 μl in a speedvac, and stored at −80°C until liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Nanospray LC-MS/MS was performed on an LTQ linear ion trap mass spectrometer (LTQ, Thermo-Fisher) interfaced with a Rapid Separation LC3000 system (Dionex). Thirty-five microliters of the sample was loaded on an Acclaim PepMap C18 Nanotrap column (5 μm, 100 Å/100 μm, i.d. × 2 cm) from the autosampler, with a 50-μl sample loop with the loading buffer (2% acetonitrile/water +0.1% trifluoroacetic acid) at a flow rate of 8 μl/min. After 15 min, the trap column was switched in line with the Acclaim PepMap RSLC C18 column (2 μm, 100 Å, 75 μm, i.d. × 25 cm) (Dionex). The trap and analytical columns were kept at a constant temperature of 40°C. The peptides were eluted with gradient separation using mobile phase A (2% acetonitrile/water with 0.1% formic acid) and mobile phase B (80% acetonitrile/water + 0.1% formic acid). Solvent B was increased from 2% to 32% over 40 min, increased to 90% over a 5-min period and held at 90% for 10 min at a flow rate of 350 nl/min. The ten ions with the highest intensity with a charge state from +2 to +4 determined from an initial survey scan after LC/MS from 300 to 1600 m/z were selected for fragmentation (MS/MS). MS/MS was performed using an isolation width of 2 m/z, normalized collision energy of 35%, activation time of 30 ms and a minimum signal intensity of 2,500 counts. The dynamic exclusion option was enabled. Once a certain ion was selected once for MS/MS in 7 s, this ion was excluded from being selected again for a period of 30 s.
Crystallization, Data Acquisition, Structure Solution, and Refinement
For crystallization, the PCR-amplified BglII/EcoRI fragment encoding BphP1-FP/C20S was cloned into a pBAD/His-B vector (Invitrogen), which was modified by shortening the N-terminal polyhistidine tag to the MGSHHHHHHGRS-amino acid sequence. Expressed and purified as above, the protein was transferred to a buffer containing 20 mM Tris-HCl, 200 mM NaCl (pH 8.0), and concentrated to 20 mg/ml using an Amicon Ultra centrifugal filter (Millipore). An initial search for crystallization conditions was carried out with Mosquito Robotic Crystallization System (TTP LabTech). The successful hits were further optimized manually. Large-scale crystallization was set up using the hanging drop vapor diffusion method. Typically, 2 μl of protein stock solution was mixed with 2 μl of the well solution and incubated against 500 μl of the same reservoir at 20°C for 2 weeks. The best crystals of BphP1-FP/C20S were obtained from 3.2 M NaCl, 0.08 M Tris-HCl (pH 8.0) buffer.
X-Ray diffraction data were collected at the Advanced Photon Source, SER-CAT beamline 22-BM (Argonne National Laboratory). Diffraction intensities were registered on a MAR225 CCD detector (Rayonix). Prior to data acquisition, a single crystal was dipped into cryoprotecting solution comprised of 20% (v/v) glycerol and 80% (v/v) of the corresponding well solution, and flash cooled in a 100-K nitrogen stream. Cryogenic temperature was maintained throughout the diffraction experiment with an Oxford Cryostream cooling device (Oxford Cryosystems). Diffraction images were indexed, integrated, and scaled with HKL2000 (Otwinowski and Minor, 1997). The statistics of data processing are shown in Table S2.
The structure of BphP1-FP/C20S was solved by a molecular replacement method with MOLREP (Vagin and Teplyakov, 2010), using the PAS and GAF domains of RpBphP1 (PDB: 4GW9 (Bellini and Papiz, 2012)) as a search model. Maximum likelihood refinement and real space model correction were performed with REFMAC5 (Murshudov et al., 2011), PHENIX.REFINE (Adams et al., 2002) and COOT (Emsley and Cowtan, 2004). Ordered water molecules were added to the appropriate difference electron density peaks with COOT and PHENIX.REFINE. Improved noiseless feature-enhanced maps (FEMs) were calculated with PHENIX.FEM (Afonine et al., 2015). Structure quality was validated with PROCHECK (Laskowski et al., 1993) and COOT. The refinement statistics are summarized in Table S3. Structure figures were generated with PyMOL (Delano Scientific).
The presence of unusual chromophore species observed in the structure was confirmed by feature-enhanced electron density mapping (Afonine et al., 2015). The FEM procedure modifies the 2mFobs-DFcalc σA-weighted map to reduce noise and model bias, retaining and enhancing existing features. The FEM algorithm computes a large ensemble of slightly perturbed maps and combines them into one map, exploiting the fact that map artifacts are more sensitive to randomization than the signal. It also performs signal equalization, making strong and weak signals similar in strength, and applies an OMIT map filter to reduce model bias.
Expression in Live Mammalian Cells
To construct mammalian expression plasmids, the respective genes of FPs were PCR-amplified as AgeI-NotI fragments and swapped with a gene encoding EGFP in the pEGFP-N1 plasmid (Clontech).
HeLa cells were grown in DMEM medium supplemented with 10% FBS, 0.5% penicillin-streptomycin and 2 mM glutamine (Invitrogen). For microscopy, cells were cultured in 35-mm glass-bottom Petri dishes with no. 1 coverglass (MatTek). Plasmid transfections were performed using an Effectene reagent (Qiagen).
Flow Cytometry and Fluorescence Microscopy
Flow cytometry analysis was performed using a BD LSRII flow cytometer equipped with 488 and 640 nm lasers and 530/40, 670/30, and 710/20 nm emission filters. A total of 20,000 events for each cell type were analyzed. The obtained dot plots were superimposed.
Epifluorescence microscopy of live HeLa cells was performed 48 hr after the transfection. HeLa cells were imaged using an Olympus IX81 inverted epifluorescence microscope equipped with a 200-W Me-Ha arc lamp (Lumen220Pro, Prior), 100× 1.4 NA oil immersion objective lens (UPlanSApo, Olympus), and two filter sets (605/40 nm exciter and 667/30 nm emitter, and 682/10 nm exciter and 721/42 nm emitter) (Chroma). SlideBook v.4.1 software (Intelligent Imaging Innovations) was used to operate the microscope.
Supplementary Material
Highlights.
Biliverdin binds to cysteine in the GAF domain of blue-shifted bacterial phytochromes
Two unique chromophores bound to cysteine in the GAF domain cause the spectral shift
Near-infrared fluorescent proteins with these chromophores have a high quantum yield
Pairs of point cysteine mutants of fluorescent proteins allow multicolor imaging
Acknowledgments
We thank Eric Giraud (Institute for Research and Development, Montpellier, France) for the RpBphP1 gene, Clark Lagarias (University of California at Davis) and Richard Vierstra (University of Wisconsin at Madison) for the plasmids for production of BV and PΦB in bacteria. We are grateful to Clark Lagarias for useful comments on the manuscript. We thank Jinghang Zhang for assistance with flow cytometry. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This work was supported in part with US Federal funds from the National Cancer Institute of the NIH under contract HHSN261200800001E, the Intramural Research Program of the NIH, by the NIH grants CA164468, GM073913 and GM108579 (all to V.V.V.) and ERC-2013-ADG-340233 (to V.V.V.) grant from the EU FP7 program.
Footnotes
ACCESSION NUMBERS
The coordinates and structure factors for BphP1-FP/C20S have been deposited in the PDB under the accession code PDB: 4XTQ.
Supplemental Information includes a Supplemental Data File, six figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2015.10.007.
AUTHOR CONTRIBUTIONS
D.M.S. and M.B. developed the protein and its mutants and characterized them in vitro. M.B. characterized the protein variants in mammalian cells. S.P., V.N.M., and Z.D. crystallized the protein, and S.P. performed its structural analysis. H.X. performed the mass spectrometry analysis. V.V.V. planned and directed the project and together with D.M.S. designed the experiments, analyzed the data, and wrote the manuscript.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Afonine PV, Moriarty NW, Mustyakimov M, Sobolev OV, Terwilliger TC, Turk D, Urzhumtsev A, Adams PD. FEM: feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Satyshur KA, Anstrom DM, Forest KT. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem. 2012;287:7000–7009. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini D, Papiz MZ. Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure. 2012;20:1436–1446. doi: 10.1016/j.str.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Auldridge ME, Lehtivuori H, Ihalainen JA, Forest KT. Origins of fluorescence in evolved bacteriophytochromes. J Biol Chem. 2014;289:32144–32152. doi: 10.1074/jbc.M114.589739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- Borucki B, Seibeck S, Heyn MP, Lamparter T. Characterization of the covalent and noncovalent adducts of Agp1 phytochrome assembled with biliverdin and phycocyanobilin by circular dichroism and flash photolysis. Biochemistry. 2009;48:6305–6317. doi: 10.1021/bi900436v. [DOI] [PubMed] [Google Scholar]
- Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci USA. 2014;111:10179–10184. doi: 10.1073/pnas.1403096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol. 2013;20:1078–1086. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Lagarias JC. Harnessing phytochrome’s glowing potential. Proc Natl Acad Sci USA. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, Duan Y, Lei H, Lagarias JC. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry. 2005;44:15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen S, Boxer SG. On the origin of heme absorption band shifts and associated protein structural relaxation in myoglobin following flash photolysis. J Biol Chem. 1997;272:9655–9660. doi: 10.1074/jbc.272.15.9655. [DOI] [PubMed] [Google Scholar]
- Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98:10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Vermeglio A. Bacteriophytochromes in anoxygenic photosynthetic bacteria. Photosynth Res. 2008;97:141–153. doi: 10.1007/s11120-008-9323-0. [DOI] [PubMed] [Google Scholar]
- Jiguet-Jiglaire C, Cayol M, Mathieu S, Jeanneau C, Bouvier-Labit C, Ouafik L, El-Battari A. Noninvasive near-infrared fluorescent protein-based imaging of tumor progression and metastases in deep organs and intraosseous tissues. J Biomed Opt. 2014;19:16019. doi: 10.1117/1.JBO.19.1.016019. [DOI] [PubMed] [Google Scholar]
- Kapitulnik J, Maines MD. The role of bile pigments in health and disease: effects on cell signaling, cytotoxicity, and cytoprotection. Front Pharmacol. 2012;3:136. doi: 10.3389/fphar.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparter T, Michael N, Caspani O, Miyata T, Shirai K, Inomata K. Biliverdin binds covalently to agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J Biol Chem. 2003;278:33786–33792. doi: 10.1074/jbc.M305563200. [DOI] [PubMed] [Google Scholar]
- Lamparter T, Carrascal M, Michael N, Martinez E, Rottwinkel G, Abian J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry. 2004;43:3659–3669. doi: 10.1021/bi035693l. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alber T, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski W, Minor F. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev. 2013a;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat Commun. 2013b;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quest B, Gartner W. Chromophore selectivity in bacterial phytochromes: dissecting the process of chromophore attachment. Eur J Biochem. 2004;271:1117–1126. doi: 10.1111/j.1432-1033.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–632. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens R, Drexhage KH. Fluorescence quantum yield of oxazine and carbazine laser dyes. J Lumin. 1981;24:709–712. [Google Scholar]
- Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Baloban M, Verkhusha VV. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol. 2015;27:52–63. doi: 10.1016/j.cbpa.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H, Bjorling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KC, Stojkovic EA, van Stokkum IH, Moffat K, Kennis JT. Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs. Phys Chem Chem Phys. 2011;13:11985–11997. doi: 10.1039/c1cp00050k. [DOI] [PubMed] [Google Scholar]
- Tran MT, Tanaka J, Hamada M, Sugiyama Y, Sakaguchi S, Nakamura M, Takahashi S, Miwa Y. In vivo image analysis using iRFP transgenic mice. Exp Anim. 2014;63:311–319. doi: 10.1538/expanim.63.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, von Stetten D, Gunther M, Murgida DH, Mroginski MA, Walker JM, Forest KT, Hildebrandt P, Vierstra RD. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J Biol Chem. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Wu SH, Lagarias JC. Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochemistry. 2000;39:13487–13495. doi: 10.1021/bi001123z. [DOI] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci USA. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc Natl Acad Sci USA. 2009;106:15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP, Thayer DA, Huang H, Kornberg TB, Royant A, et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Wu G, Robinson H, Wilganowski N, Hall MA, Ghosh SC, Pinkston KL, Azhdarinia A, Harvey BR, Sevick-Muraca EM. Tumor margin detection using quantitative NIRF molecular imaging targeting EpCAM validated by far red gene reporter iRFP. Mol Imaging Biol. 2013;15:560–568. doi: 10.1007/s11307-013-0637-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.