Abstract
Background
Changes in tumor DNA mutation status during chemotherapy can provide insights into tumor biology and drug resistance. The purpose of this study is to analyse the presence or absence of mutations in cancer-related genes using baseline breast tumor samples and those obtained after exposure to one cycle of chemotherapy to determine if any differences exist, and to correlate these differences with clinical and pathological features.
Methods
Paired breast tumor core biopsies obtained pre- and post-first cycle doxorubicin (n = 18) or docetaxel (n = 22) in treatment-naïve breast cancer patients were analysed for 238 mutations in 19 cancer-related genes by the Sequenom Oncocarta assay.
Results
Median age of patients was 48 years (range 32–64); 55% had estrogen receptor-positive tumors, and 60% had tumor reduction ≥25% after cycle 1. Mutations were detected in 10/40 (25%) pre-treatment and 11/40 (28%) post-treatment samples. Four mutation pattern categories were identified based on tumor mutation status pre- → post-treatment: wildtype (WT)→WT, n = 24; mutant (MT)→MT, n = 5; MT→WT, n = 5; WT→MT, n = 6. Overall, the majority of tumors were WT at baseline (30/40, 75%), of which 6/30 (20%) acquired new mutations after chemotherapy. Pre-treatment mutations were predominantly in PIK3CA (8/10, 80%), while post-treatment mutations were distributed in PIK3CA, EGFR, PDGFRA, ABL1 and MET. All 6 WT→MT cases were treated with docetaxel. Higher mutant allele frequency in baseline MT tumors (n = 10; PIK3CA mutations n = 8) correlated with less tumor reduction after cycle 1 chemotherapy (R = -0.667, p = 0.035). No other associations were observed between mutation pattern category with treatment, clinicopathological features, and tumor response or survival.
Conclusion
Tumor mutational profiles can change as quickly as after one cycle of chemotherapy in breast cancer. Understanding of these changes can provide insights on potential therapeutic options in residual resistant tumors.
Trial Registration
ClinicalTrials.gov NCT00212082
Introduction
Carcinogenesis is a multi-step process characterized by acquisition of molecular alterations in various signaling pathways involved in growth and development. Activation of oncogenes leading to cancer occurs via mechanisms such as gene amplification, chromosomal translocations, and point mutations that enhance the function of the "oncoprotein"[1]. Aberrations in oncogenes may be important in natural selection during tumorigenesis or upon treatment, and knowledge of clinically relevant mutations can aid tumor classification as well as facilitate patient stratification for deployment of targeted therapeutics.
Neoadjuvant chemotherapy for breast tumors provides a unique opportunity for acquisition of early information on in vivo tumor response and disease biology, and also lends an ideal model to evaluate biomarkers as the primary tumor may be sampled readily and repeatedly.
Chemotherapy-induced gene expression changes in breast tumors after one cycle or even as early as 24 hours after chemotherapy have been well described [2,3,4], but studies evaluating mutation shifts during neoadjuvant chemotherapy are at present very limited. A recent study using exome sequencing revealed that loss of PIK3CA and TP53 mutations in breast tumors after 3–6 cycles of neoadjuvant chemotherapy translated to improved clinical responses and survival outcomes [5].
In order to gain an improved perspective into earlier molecular changes following chemotherapy, we sought to characterize the mutation profile of treatment-naïve primary breast tumors pre-treatment and 3 weeks after first exposure to doxorubicin or docetaxel chemotherapy using a high-throughput mutation analysis assay interrogating 238 mutations in 19 cancer-related genes [6]. We found distinct changes in mutation patterns pre- and post-treatment which allowed us to categorize our samples into 4 subgroups according to mutation profile variations with treatment. The mutation patterns were also assessed for their correlation with clinical parameters including chemotherapy type, estrogen receptor (ER) status, disease extent, tumor response, progression-free and overall survival.
Materials and Methods
Patients and tissues
Breast tumor samples were prospectively collected as part of a phase II randomized study that recruited Southeast Asian patients with newly diagnosed histologically or cytologically proven clinical stage II-IV breast cancer with measurable primary tumor in order to discover gene expression profiles that predicted for chemosensitivity (ClinicalTrials.gov ID: NCT00212082; S1 Text) [4]. This study was approved by the National Healthcare Group Domain Specific Review Board institutional ethics review committee, and all participants provided written informed consent. Patients were recruited from our institution and randomised at a ratio of 1:1 to one of two alternating sequences of doxorubicin (A) and docetaxel (T), starting with either doxorubicin 75mg/m2 or docetaxel 75mg/m2 every 3 weeks for 6 cycles (Arm A: A→T→A→T→A→T; Arm B: T→A→T→A→T→A) as primary systemic therapy. One to two core biopsies from the primary breast tumor were taken each at baseline and approximately 3 weeks after the first cycle of chemotherapy, snap frozen in liquid nitrogen and stored at -80°C until analysis. Only RNA was extracted from the first tumor core for gene expression analysis, the data of which has been reported previously [4,7], while both DNA and RNA was extracted from the second tumor core if available, for DNA mutational analysis and other RNA expression work. Bidimensional breast tumor assessments were performed at every cycle. Tumors were deemed to be intrinsically sensitive or resistant to cycle 1 chemotherapy if they demonstrated ≥25% or < 25% reduction in tumor dimensions respectively after cycle 1. After 6 cycles, the overall response rates (ORR) were classified as complete response, partial response, stable disease, or progressive disease in accordance with World Health Organisation criteria [8]. The study was approved by the institutional ethics review board and all patients provided written informed consent.
Mutational analysis of invasive tumor
Tumor DNA was extracted from frozen tissues using Allprep DNA/RNA/Protein mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The quality and quantity of DNA was assessed by spectrophotometer on the Nanodrop 2000c (Wilmington, DE) but no microdissection was done. DNA mutation analysis was performed using the pre-designed OncoCarta™ mutation panel (Sequenom, San Diego, CA; S1 Table) on the MassARRAY Analyzer Compact (Sequenom) according to the manufacturer’s protocols. Each analysis consisted of 251 assays involving 24 pools of primer pairs and extension primers, and interrogated 238 non-synonymous mutations in 19 cancer-related genes [6]. Five-hundred nanograms of DNA from each patient sample were used for the assay. Mutations calls for each sample were determined using MassArray Typer Analyzer software 4.0.4.20 (Sequenom), and were identified by comparing ratios of the spectral wild type peaks to that of all suspected mutants. The RKO cell line containing the mutations BRAF V600E and PIK3CA H1047R was used as a positive control and included in every run. Associations were evaluated in relation to clinicopathological features such as type of chemotherapy exposure, age, race, presence or absence of metastases, ER status, post-cycle 1 tumor response and overall treatment responses. HER2/neu testing was only carried out in 4 samples and hence was not used in the analysis. We also analysed mutant allele frequency in baseline mutant tumors in relation to tumor responses post-cycle 1 chemotherapy and overall response after 6 cycles of chemotherapy.
Statistical Analysis
For analysis of association with clinicopathological features, cases were divided into two groups consisting of those with no mutations in pre- and post-chemotherapy samples (WT→WT), and a group consisting of all other cases, including those with mutations in both pre- and post-chemotherapy samples (MT→MT), those with mutations in pre-chemotherapy samples but no mutations in post-chemotherapy samples (MT→WT), and those with no mutations in pre-chemotherapy samples but with mutations in post-chemotherapy samples (WT→MT). Associations with categorical variables were examined using Fisher’s exact test. Associations with age as a continuous variable were analyzed using the two-sample t-test.
Logistic regression was carried out to determine the association between mutational transition (MT→WT or WT→MT) and type of chemotherapy exposure (treatment arm A vs arm B) while adjusting for pre-chemotherapy mutation profile, presence of distant metastases and ER status.
Pearson's chi-squared test was used to evaluate the association between the types of mutations (PIK3CA vs others) in the 10 baseline mutant samples with respect to tumour response post-cycle 1 and after 6 cycles of chemotherapy (overall response). Pearson's chi-squared test was also used to determine the relation between high versus low mutant allele frequency (≥25% vs <25%) in the 10 baseline mutant samples and tumor response post-cycle 1 and overall response. A Pearson correlation coefficient test was carried out between the absolute tumour reduction (or increase) after one cycle of chemotherapy and baseline sample mutation allele frequency in baseline mutant tumors.
Progression-free survival (PFS) and overall survival (OS) of the WT→WT subgroup was compared with the other subgroups (MT→MT, MT→WT or WT→MT) using the Kaplan Meier method and logrank test. Multivariate Cox regression analyses were performed to test the effect of mutation subgroup type (WT→WT versus the rest) on PFS and OS while adjusting for T stage (T3 versus T4), presence of metastases (Yes versus No), ER and progesterone receptor (PgR) status (positive versus negative). All statistical analyses were two-sided, and performed using IBM SPSS Statistics Desktop 20 software (IBM, Armonk, NY). A p-value less than 0.05 was considered statistically significant.
Results
Clinical Characteristics
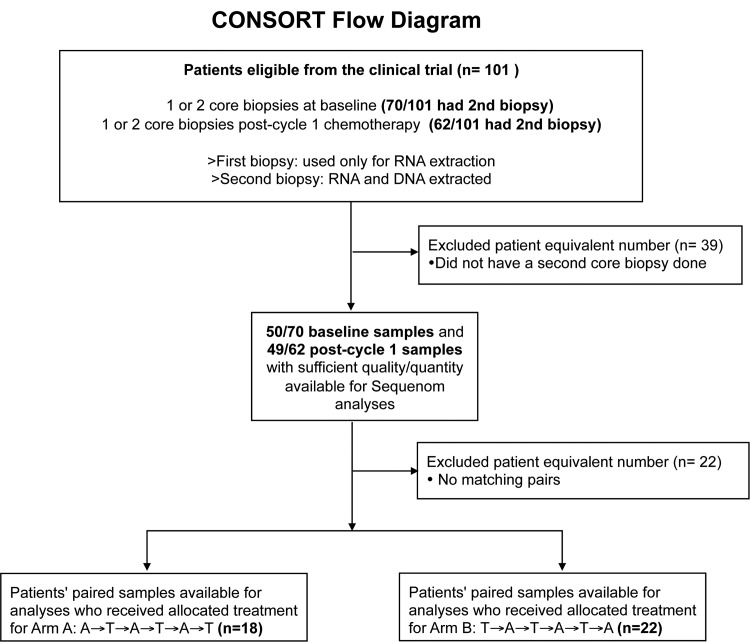
Matched pre- and post-chemotherapy tumor DNA samples from 40 patients who were randomised between 26th April 2002 and 3rd June 2005 were available for mutation analyses (Arm A: n = 18, Arm B: n = 22). Sixty-one patients’ paired samples were not feasible for analysis either due to unavailability of samples (n = 39) or inadequate DNA content (n = 22; Fig 1). An accompanying paraffin-embedded sample was taken at each time point, and the average cellularity of the baseline samples was 63% (range 10–95%), and that for the post-cycle 1 chemotherapy samples was 42% (range 10–90%).
Fig 1. CONSORT flow chart in which the number of paired samples with data available for Sequenom analysis are shown.
None of the participants were lost to follow-up with the last follow-up date being 18th February 2014.
The median age of the cohort was 48 years (range 32–64); 75% were Chinese (n = 30), 23% were Malay (n = 9) and one was Indian. Twenty-five percent (10/40) of the patients had metastatic disease at diagnosis, 28% (11/40) and 72% (29/40) had clinical T3 and T4 disease respectively, and 55% (22/40) had ER-positive tumors. Sixty percent of patients (24/40) achieved at least 25% tumor reduction after cycle 1 chemotherapy. After 6 cycles of chemotherapy, the overall objective clinical responses were as follows: complete clinical response (4/40; 10%); partial response (28/40; 70%); stable disease (7/40, 18%); one was non-evaluable. At the time of analysis, median follow-up was 74.5 months. Median PFS and OS of the entire cohort was 30 months and 75 months respectively.
Mutation Analysis
Mutations were detected in 25% (10/40) of pre-treatment samples, with the majority being in PIK3CA (n = 8: H1047R, n = 3; H1047L, n = 2; E542K, n = 1; E545K, n = 1; N345K, n = 1), and the others being in EGFR S768I (n = 1) and KIT Y503-F504insAY (n = 1) (Fig 2, Table 1). A total of 28% (11/40) of post-treatment samples had mutations, the majority also being in PIK3CA (n = 6: H1047R n = 3; E542K n = 1; E545K n = 1; N345K n = 1), followed by EGFR (n = 2: H773-V774insH and N771-P772>SVDNR), PDGFRA I843-S847>T (n = 1), ABL Y253H (n = 1), and MET Y1230C (n = 1).
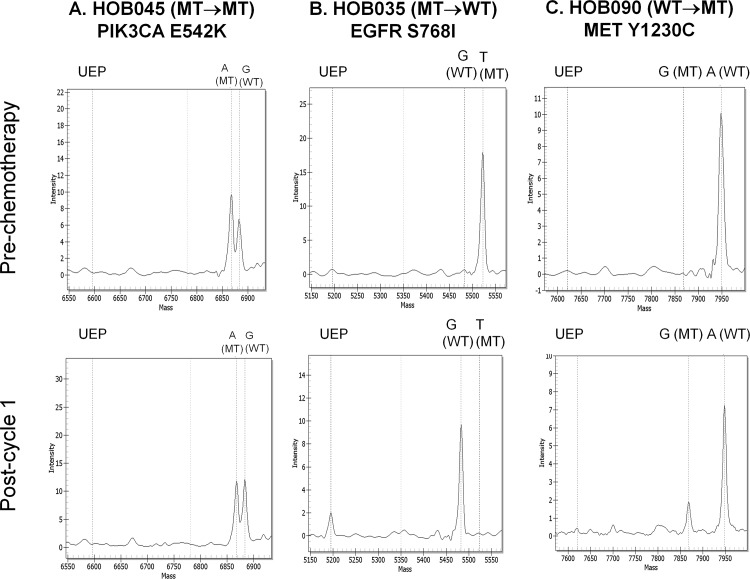
Fig 2. Oncogenic mutations detected pre- and post-chemotherapy.
Representative chromatograms of (A) PIK3CA E542K in a MT→MT case HOB045, (B) EGFR S768I in a MT→WT case HOB035, and (C) MET Y1230C detected in a WT→MT case HOB090. The expected positions for the unextended primer (UEP) and the nucleotide and mutation status (mutant [MT] or wildtype [WT]) based on the size of the extension products are indicated above the gray vertical dashed lines.
Table 1. Study cases according to mutation status in pre- and post-chemotherapy samples and their clinicopathological and tumor response characteristics (n = 40 pairs).
| Category/Case | Mutation | Pre a /mutant allele frequency (if applicable) | Post b /mutant allele frequency (if applicable) | Arm c | Age d | Race | Baseline T stage | Met | ER | PgR | C1 | ORR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT→WT cases | ||||||||||||
| HOB002 | none | WT | WT | A | 47 | Malay | T4 | - | + | + | + | PR |
| HOB005 | none | WT | WT | B | 55 | Chinese | T4 | - | - | - | + | NE |
| HOB007 | none | WT | WT | B | 63 | Chinese | T4 | - | - | - | + | PR |
| HOB014 | none | WT | WT | A | 47 | Chinese | T4 | - | + | + | + | PR |
| HOB015 | none | WT | WT | A | 51 | Chinese | T4 | - | - | - | + | PR |
| HOB019 | none | WT | WT | B | 64 | Malay | T4 | + | + | - | - | SD |
| HOB027 | none | WT | WT | B | 45 | Chinese | T3 | - | - | - | - | SD |
| HOB028 | none | WT | WT | B | 40 | Malay | T4 | - | - | - | + | PR |
| HOB031 | none | WT | WT | A | 48 | Chinese | T4 | - | + | - | + | PR |
| HOB036 | none | WT | WT | B | 46 | Chinese | T4 | - | + | - | - | SD |
| HOB042 | none | WT | WT | B | 59 | Indian | T4 | + | - | - | - | SD |
| HOB043 | none | WT | WT | A | 44 | Chinese | T4 | - | - | - | + | PR |
| HOB046 | none | WT | WT | B | 44 | Chinese | T4 | + | + | - | + | PR |
| HOB054 | none | WT | WT | A | 43 | Chinese | T3 | - | + | + | + | CR |
| HOB055 | none | WT | WT | B | 43 | Chinese | T4 | - | + | + | + | PR |
| HOB060 | none | WT | WT | A | 36 | Chinese | T3 | - | - | - | + | PR |
| HOB064 | none | WT | WT | A | 38 | Chinese | T3 | - | - | - | - | SD |
| HOB069 | none | WT | WT | A | 37 | Chinese | T3 | - | + | - | - | PR |
| HOB071 | none | WT | WT | A | 56 | Malay | T4 | + | - | + | - | CR |
| HOB073 | none | WT | WT | B | 45 | Malay | T4 | - | + | + | + | PR |
| HOB084 | none | WT | WT | A | 45 | Malay | T4 | - | - | + | - | PR |
| HOB089 | none | WT | WT | B | 49 | Chinese | T4 | + | + | + | - | PR |
| HOB091 | none | WT | WT | A | 63 | Chinese | T4 | - | + | + | - | PR |
| HOB099 | none | WT | WT | B | 32 | Chinese | T3 | - | + | + | + | PR |
| MT→MT cases | ||||||||||||
| HOB062 | PIK3CA H1047R | MT/18% | MT/24% | A | 64 | Chinese | T4 | - | + | + | + | PR |
| HOB085 | PIK3CA H1047R | MT/26% | MT/19% | A | 64 | Chinese | T4 | - | + | + | - | PR |
| HOB045 | PIK3CA E542K | MT/60% | MT/50% | B | 40 | Chinese | T3 | - | + | + | + | PR |
| HOB056 | PIK3CA E545K | MT/41% | MT/100% | A | 44 | Chinese | T4 | + | + | + | + | CR |
| HOB063 | PIK3CA N345K | MT/50% | MT/78% | A | 56 | Chinese | T4 | - | - | + | - | PR |
| MT→WT cases | ||||||||||||
| HOB076 | PIK3CA H1047R | MT/9% | WT | A | 34 | Chinese | T4 | - | + | + | + | CR |
| HOB026 | PIK3CA H1047L | MT/12% | WT | A | 54 | Chinese | T4 | - | + | + | + | PR |
| HOB044 | PIK3CA H1047L | MT/66% | WT | B | 48 | Malay | T3 | - | - | - | - | PR |
| HOB035 | EGFR S768I | MT/98% | WT | B | 56 | Malay | T4 | + | - | - | - | SD |
| HOB086 | KIT Y503-F504insAY | MT/20% | WT | B | 51 | Chinese | T3 | - | - | - | + | PR |
| WT→MT cases | ||||||||||||
| HOB096 | ABL Y253H | WT | MT/24% | B | 56 | Chinese | T3 | - | - | - | - | PR |
| HOB088 | EGFR H773-V774insH | WT | MT/51% | B | 45 | Chinese | T4 | + | - | - | + | PR |
| HOB077 | EGFR N771-P772>SVDNR | WT | MT/100% | B | 63 | Chinese | T4 | + | + | - | - | SD |
| HOB090 | MET Y1230C | WT | MT/20% | B | 61 | Chinese | T4 | - | + | - | + | PR |
| HOB052 | PDGFRA I843-S847>T | WT | MT/13% | B | 54 | Chinese | T4 | + | - | - | + | PR |
| HOB080 | PIK3CA H1047R | WT | MT/26% | B | 58 | Malay | T3 | - | + | - | + | PR |
Abbreviations: C1, + = tumor response ≥25% after cycle 1 chemotherapy, C1,— = tumour response <25% after cycle 1 chemotherapy; CR, complete response; ER, estrogen receptor status; Met, Presence of metastasis; MT, mutant; NE, non evaluable (patient had already started on another line of treatment); ORR, overall response rate; PgR, progesterone receptor; PR, partial response; SD, stable disease; T3, >50mm in greatest dimension; T4, tumor of any size with direct extension to the chest wall and/or to the skin (ulceration or skin nodules); WT, wildtype; -, negative/no; +, positive/yes
a Status of respective mutation in pre-chemotherapy sample
b Status of respective mutation in post-chemotherapy sample
c Arm A, randomized to receive doxorubicin in first cycle; Arm B, randomized to receive docetaxel in first cycle
d Age in years
Based on tumor mutation status before and after one cycle of chemotherapy, patient samples were divided into 4 categories: (1) WT→WT (24/40, 60%), (2) MT→MT (5/40, 13%), (3) MT→WT (5/40, 13%), (4) WT→MT (6/40, 15%). All samples with detectable mutations harboured only a single mutation. Of the 5 tumor pairs that had mutations before and after chemotherapy (MT→MT subgroup), the pre and post-chemotherapy mutations were identical in each tumor, and all mutations were in PIK3CA (H1047R n = 2; E542K n = 1; E545K n = 1; N345K n = 1). There were 5 MT→WT cases, consisting of 3 cases that originally carried PIK3CA mutations (H1047R n = 1; H1047L n = 2), and 2 with an EGFR S768I and KIT Y503-F504insAY mutations respectively. A WT→MT pattern occurred in 6 cases; with the post-treatment mutations being ABL1 Y253H, EGFR H773-V774insH, EGFR N771-P772>SVDNR, MET Y1230C, PDGFRA I843-S847>T, and PIK3CA H1047R, respectively. The observed mutant allele frequencies are listed in Table 1. The mean mutant allele frequency in the 10 pre-treatment MT tumor samples was 40% (range 9–98%), while the mutant allele frequency in the 11 post-treatment MT tumor samples was 47.9% (range 13–100%).
Association with Clinical Features
The distribution of exposure to either first-cycle doxorubicin or docetaxel were well balanced in the WT→WT (doxorubicin: 12 patients; docetaxel: 12 patients), and MT→WT groups (doxorubicin: 2 patients; docetaxel: 3 patients). However, 4 out of 5 patients were exposed to doxorubicin in the MT→MT subgroup, while all 6 patients in the WT→MT subgroup were treated initially with docetaxel. ER-status (positive versus negative) was distributed relatively evenly in all groups except in the MT→MT subgroup in which all but one tumor pair with PIK3CA mutations were ER-positive. The tumor sample harbouring a PIK3CA mutation that was ER-negative was progesterone receptor (PgR)-positive. In the MT→WT category, all the pre-treatment samples with PIK3CA mutations were ER-positive except one that was ER- and PgR-negative. Thus, 7 out of 9 paired samples with PIK3CA mutations in pre- and/or post-treatment samples were ER-positive.
No significant difference was observed between WT→WT and the combination of the other 3 subgroups (MT→MT, MT→WT, WT→MT) with respect to type of chemotherapy exposure, age, race, presence or absence of metastases, ER status, post-cycle 1 tumor responses and overall treatment responses (Table 2).
Table 2. Association of pre- and post-chemotherapy mutation patterns with clinicopathological features.
| Feature | Wildtype (WT)→WT Cases | Other Cases a | P value b |
|---|---|---|---|
| Total | N = 24 | N = 16 | |
| Treatment Arm | |||
| A | 12 (50%) | 6 (38%) | 0.526 |
| B | 12 (50%) | 10 (63%) | |
| Age in Years | |||
| Mean±Standard Deviation | 47.5±8.7 | 53.0±8.8 | 0.059 c |
| Race | |||
| Chinese | 17 (71%) | 13 (81%) | 0.711 |
| Non-Chinese | 7 (29%) | 3 (19%) | |
| Metastases | |||
| Yes | 5 (21%) | 5 (31%) | 0.482 |
| No | 19 (79%) | 11(69%) | |
| Estrogen Receptor Status | |||
| Positive | 13 (54%) | 9 (56%) | 1.000 |
| Negative | 11 (46%) | 7 (44%) | |
| Cycle 1 Response ≥25% | |||
| Yes | 14 (58%) | 10 (63%) | 0.528 |
| No | 10 (42%) | 6 (38%) | |
| Overall Response Rate d | |||
| Complete Response (CR) | 2 (8%) | 2 (13%) | |
| Partial Response (PR) | 16 (67%) | 12 (75%) | 0.678 e |
| Stable Disease (SD) | 5 (21%) | 2 (13%) |
a Mutant→Mutant, Mutant→Wildtype, Wildtype→Mutant cases
b Fisher’s exact test, two-sided P value
c Two-sample t-test, two-sided P value
d One WT→WT was not evaluable for overall response
e P value from analysis of CR/PR vs SD
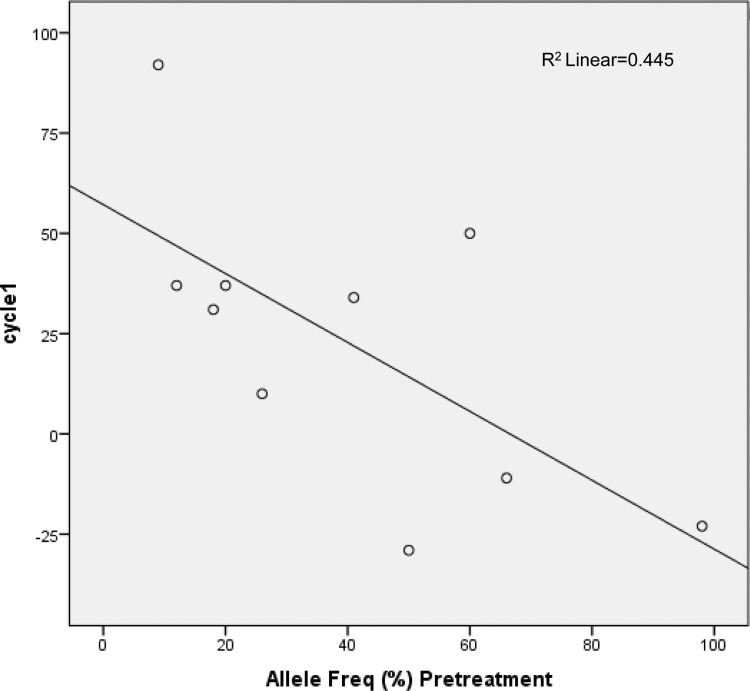
Cases from treatment arm B were significantly more likely to have change in mutation profile (MT→WT or WT→MT) than arm A, after adjustment for pre-chemotherapy mutation profile, presence of distant metastasis and ER status (adjusted odds ratio 12.00 [95% CI 1.11–129.87]). There was no significant association between baseline mutant samples harboring PIK3CA mutations (n = 8) versus those with other mutations (n = 2) in terms of tumor responses post-cycle 1 (p = 0.747) and overall tumor response (p = 0.098). However, baseline mutant tumors with <25% mutant allele frequency were more likely to achieve tumor reduction of ≥25% after cycle 1 chemotherapy (66.7% vs 33.3%, p = 0.035) than those with ≥25% mutant allele frequency. The same analysis (baseline tumors ≥25% vs <25% mutant allele frequency) for overall clinical response did not detect a significant difference between the mutation groups (P-value = 0.679). Upon Pearson correlation coefficient analysis, an inverse relationship was found between percentage mutant allele in the baseline tumour samples and post-cycle 1 chemotherapy absolute tumour reduction (R = -0.667, p = 0.035; Fig 3), suggesting that tumor containing a mutant allele was less likely to respond if it contained a high percentage of mutant allele.
Fig 3. Correlation between percentage allele frequency and post-cycle 1 tumor response of at least 25%.
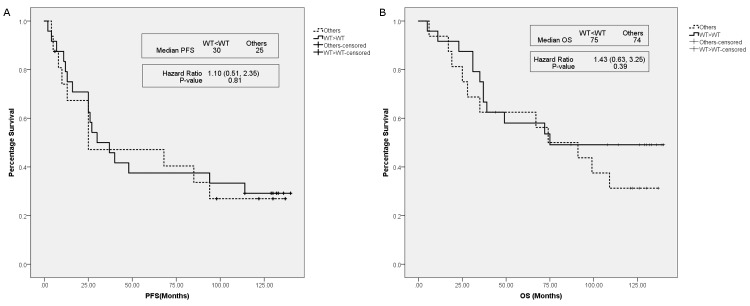
The PFS of WT→WT patients was not significantly different compared to the other 3 subgroups (hazard ratio {HR} 1.10 [95% CI 0.51–2.35]; P = 0.81) (Fig 4A). Similarly, no significant difference in OS was observed between WT→WT and the other 3 subgroups (HR 1.43 [95% CI 0.63–3.25]; P = 0.39) (Fig 4B). Multivariate analyses adjusting for T stage, presence or absence of metastases, ER and PgR status with respect to PFS and OS showed that the HR changed from 1.10 to 1.04 and 1.43 to 1.14 respectively, but the overall results remained unchanged.
Fig 4. Progression-free and overall survival between the WT→WT subgroup versus combination of other mutation pattern subgroups.
(A) Progression-free survival and (B) overall survival of the WT→WT subgroup and a combination of other mutation pattern subgroups (MT→MT, MT→WT, WT→MT). Bold line: WT→WT subgroup; broken line: combination of the other mutation subgroups MT→MT, MT→WT, WT→MT.
Discussion
To our knowledge, there have been limited studies that evaluated the influence of chemotherapy on mutational profiles in clinical breast cancer specimens. We examined the mutation profiles of 40 paired primary breast tumor biopsy samples pre- and post-chemotherapy 3 weeks apart after only one cycle of doxorubicin or docetaxel. The major findings were variation in mutation status between treatment-naive baseline samples and post-treatment samples in about a quarter of patients (11/40 or 28% WT→MT or MT→WT). Overall, the majority of baseline samples were WT at baseline (30/40; 75%), and 6/30 (20%) of these originally WT samples had new mutations detected in the corresponding post-treatment sample. Interestingly, all 6 samples were exposed to docetaxel rather than to doxorubicin. The majority of post-chemotherapy mutations that were uncovered after docetaxel exposure were less common mutations (ABL, EGFR, MET, PDGFRA), in contrast to the predominance of PIK3CA mutations observed in pre-treatment samples (8/10 baseline MT tumors harboured PIK3CA mutations). Half of the baseline MT tumors retained the same mutation after chemotherapy (MT→MT); all possessed PIK3CA mutations, and all but one were treated with doxorubicin. The remaining 5 MT tumors at baseline reverted to WT status after chemotherapy, with no clear pattern of prior drug exposure (doxorubicin or docetaxel); pre-treatment mutations in these tumors were also more diverse, in PIK3CA (n = 3), EGFR (n = 1), and KIT (n = 1).
Differing somatic mutations seen pre- and post-treatment as demonstrated in our study may be attributable to several key reasons. Firstly, data suggest that topological clonal heterogeneity within primary tumors exists, consisting of diverse mutations and a large number of subclonal and de novo mutations [9,10,11], and new clones with different mutational profiles may have developed spontaneously even in the few weeks that separated the pre- and post-chemotherapy biopsy. Additionally, certain rapidly proliferating MT or WT tumor cells may have a better response to chemotherapy, and this selective pressure from chemotherapy may lead to expansion of resistance clones [12]. Cells with different mutational profiles may have differing sensitivities to different chemotherapeutic agents; the observed loss of mutations after chemotherapy in our study may have been caused by increased sensitivity of certain mutation-containing clones to the treatment, hence primarily removing the mutant cells and consequently shifting the genetic evolutionary landscape in favour of the non-mutant subclones. These mutant alleles could still be present after chemotherapy but have fallen to such low frequencies that they were not detectable with our assay, and hence the tumors were considered to have changed from MT to WT. Similarly, certain wildtype clones may be very susceptible to chemotherapy, resulting in mutant clones becoming dominant in the post-treatment samples in some patients (MT→WT).
A noteworthy observation from our study is that all 6 originally WT tumor samples which acquired mutations post-treatment were exposed to docetaxel, instead of doxorubicin. One possible hypothesis for this observation is selective sensitivity of certain WT subclones to docetaxel thus enriching the post-treatment samples for non-WT cells, or pre-existence of docetaxel-resistant MT clones in low frequency in baseline samples which became more readily detectable after other subclones were eradicated with docetaxel.
The phenomenon of mutation variations post-treatment has been reported in other cancer types such as non-small cell lung cancer, where EGFR mutant to wild type changes were observed after chemotherapy and postulated to be related to intratumoral heterogeneity and differing chemosensitivity levels of mutant and wild-type cells[13]. It is unique that in our study, mutation changes were detected as early as 3 weeks after a single cycle of chemotherapy which could offer an early evaluation of the evolving mutational landscape of the tumor in response to drug treatment. The value of detecting potentially actionable mutations earlier on is in identifying new therapeutic targets which can provide insights on developing rational novel combinations or optimal sequencing of chemotherapy and targeted drugs to treat residual resistant disease. For example, the common PIK3CA hotspot mutations in our samples, namely H1047R, E545K and E542K are the targets of pan-PI3K inhibitors such as GDC-0941, BKM120, BYL719 and XL-147 which are being actively evaluated in combination with chemotherapy, endocrine therapy, and/or anti-HER2 agents in advanced breast cancers [14,15]. c-MET signalling inhibition has demonstrated activity in metastatic breast cancer using the drug cabozantinib, which targets MET mutations such as Y1248H, D1246N and K1262R [16,17,18]. EGFR mutations have been identified in triple-negative breast cancers in a small study highlighting the possible application of oral EGFR tyrosine kinase inhibition therapy in selected breast cancer patients [19]. Of interest is the EGFR S768I mutation identified in a baseline sample in our study that has demonstrated greater sensitivity to AEE788, an oral multi-receptor tyrosine kinase inhibitor, compared to erlotinib and gefitinib in an in-vitro study [20].
The analytical sensitivity of mutations for the Sequenom assay is typically around 10%[6,21]. Mutant allele frequencies observed did not show any consistent direction shift pre- and post-chemotherapy in the MT→MT group. Two out of the 5 paired samples in this MT→MT group exhibited a decrease in allele frequency post-chemotherapy. However, we believe it is risky to interpret a decreased allele frequency as representing a reduction in tumor clones with the mutation. Allele frequency can be affected by tumour content, hence a decreased allele frequency could simply reflect a lower tumor content in the sample. This issue is not specific to Sequenom, and is an issue for next generation sequencing and other genotyping assays performed on tissue samples with mixtures of tumor and normal tissue.
Interestingly, our analyses demonstrated a lower likelihood of tumor response if there was a high mutant allele frequency in a baseline mutant sample. Notably 8 out of our 10 baseline samples contained PIK3CA mutations. There has been some prospective data in the neoadjuvant setting reporting on the presence of PIK3CA mutations being associated with a poorer response and clinical outcome. This was observed in the neoadjuvant GeparQuattro, GeparQuinto, and GeparSixto studies[22], as well as the Neoadjuvant Lapatinib and/or Trastuzumab Treatment Optimization trial (NeoALTTO) study[23], where patients carrying a PIK3CA mutation had decreased pathologic complete responses compared with those who had wild-type PIK3CA. Although our sample size is small, our findings appear to be concordant with these reports.
In our study, we used core biopsy to obtain tumor specimens. Single core biopsy samples with limited tissue might not represent the complete genomic diversity of the tumor, and minute proportions of mutant clones may be overlooked due to sampling bias. In particular, all the patients in our study had large clinical T3 or T4 tumors, and a single core biopsy in such large tumors will inevitably result in sampling bias. Interestingly in our study, only one mutation was identified per biopsy which highlights the limitation of single biopsies which contain small amount of material which could lead to under-estimation of the tumor mutational landscape. However a core biopsy is still logistically most feasible in the clinic, although performing multiple baseline core biopsies or perhaps even an excisional biopsy, or repeating tumor biopsies for the primary and recurrent lesions during the course of treatment may be solutions to provide a better reflection of the entire tumor genomic landscape to guide therapy.
We recognise that our study has several important limitations. Firstly, our sample size is small, although it was reassuring that the frequencies and types of mutations detected in our samples at baseline were comparable to the Catalogue of Somatic Mutations in Cancer (COSMIC) database (PIK3CA, 8/40, 20%; EGFR, 1/40, 2.5%; KIT, 1/40, 2.5%) and other studies [24,25,26]. Secondly, the OncoCarta panel interrogates a finite number of mutations; for example, although TP53 is the second most frequently mutated gene after the PI3KCA proto-oncogene occurring in approximately 23% of breast cancer samples [27], it was not included in our assay panel. Furthermore, while Sequenom assays have high sensitivity with as little as 1ng of DNA, mutations are only detectable if present in at least 5–10% of the total DNA [6]. This assay is thus far less sensitive compared with current technology such as deep sequencing which albeit cost prohibitive and difficult to apply routinely, enhances mutation detection rate in low purity, highly polyclonal tumors.
Our findings highlight that tumor mutational profiles can change as quickly as after one cycle of chemotherapy in breast cancer, underscoring the problem of intra-tumoral heterogeneity and the evolving tumor as potential major barriers to using a single predictive biomarker test and at a single timepoint to guide treatment. Potential strategies to overcome the hurdle of tumor heterogeneity include tissue collection at multiple tumor sites, routine tissue collection at relapse, profiling of circulating tumor cells, or more recently measurement of circulating free tumor DNA as surrogates, which could allow repeated and more comprehensive profiling of heterogeneity and an improved understanding of potential resistance mechanisms [28,29]. Emerging technologies such as single-cell genome and exome sequencing or deep sequencing of tumors may further assist in characterizing intra-tumoral heterogeneity [28].
Conclusion
In conclusion, our study highlighted differences in mutation status in pre- and post-chemotherapy breast tumours. This phenomenon may be due to pre-existing intra-tumoral heterogeneity detected by sampling bias, cytotoxic therapy applying selective pressure and leading to an enhanced tumor evolutionary rate, or direct drug-induced genetic aberrations. Mutations detected pre- and especially early on post-chemotherapy exposure could guide us in the identification and development of additional druggable targets to enhance therapeutic response.
Supporting Information
(DOC)
(PDF)
(PDF)
Acknowledgments
We are grateful to all patients who participated.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Medical Research Council (NMRC), Singapore [grant numbers NMRC/CG/NCIS/2010 [NPRC10/NM16O]; NMRC/CSI/0015/2009].
References
- 1. Osborne C, Wilson P, Tripathy D. Oncogenes and tumor suppressor genes in breast cancer: potential diagnostic and therapeutic applications. Oncologist. 2004; 9: 361–377. [DOI] [PubMed] [Google Scholar]
- 2. Tan SH, Lee SC. Clinical implications of chemotherapy-induced tumor gene expression in human breast cancers. Expert Opin Drug Metab Toxicol. 2010; 6: 283–306. 10.1517/17425250903510611 [DOI] [PubMed] [Google Scholar]
- 3. Buchholz TA, Stivers DN, Stec J, Ayers M, Clark E, Bolt A, et al. Global gene expression changes during neoadjuvant chemotherapy for human breast cancer. Cancer J. 2002; 8: 461–468. [DOI] [PubMed] [Google Scholar]
- 4. Lee SC, Xu X, Lim YW, Iau P, Sukri N, Lim SE, et al. Chemotherapy-induced tumor gene expression changes in human breast cancers. Pharmacogenet Genomics. 2009; 19: 181–192. 10.1097/FPC.0b013e32831ebb5d [DOI] [PubMed] [Google Scholar]
- 5. Jiang YZ, Yu KD, Bao J, Peng WT, Shao ZM. Favorable prognostic impact in loss of TP53 and PIK3CA mutations after neoadjuvant chemotherapy in breast cancer. Cancer Res. 2014; 74: 3399–3407. 10.1158/0008-5472.CAN-14-0092 [DOI] [PubMed] [Google Scholar]
- 6. Fumagalli D, Gavin PG, Taniyama Y, Kim SI, Choi HJ, Paik S, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010; 10: 101 10.1186/1471-2407-10-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee SC, Xu X, Chng WJ, Watson M, Lim YW, Wong CI, et al. Post-treatment tumor gene expression signatures are more predictive of treatment outcomes than baseline signatures in breast cancer. Pharmacogenet Genomics. 2009; 19: 833–842. 10.1097/FPC.0b013e328330a39f [DOI] [PubMed] [Google Scholar]
- 8. Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981; 47: 207–214. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014; 512: 155–160. 10.1038/nature13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012; 366: 883–892. 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011; 472: 90–94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012; 12: 323–334. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- 13. Wang S, An T, Duan J, Zhang L, Wu M, Zhou Q, et al. Alterations in EGFR and related genes following neo-adjuvant chemotherapy in Chinese patients with non-small cell lung cancer. PLoS One. 2013; 8: e51021 10.1371/journal.pone.0051021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006; 94: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. https://clinicaltrials.gov/ (accessed 19th Sept 2015).
- 16. Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011; 10: 2298–2308. 10.1158/1535-7163.MCT-11-0264 [DOI] [PubMed] [Google Scholar]
- 17. Gordon MS, Vogelzang NJ, Schoffski P, Daud A, Spira AI, O'Keeffe BA, et al. Cabozantinib (XL184) has activity in both soft tissue and bone: Results of a phase II randomized discontinuation trial (RDT) in patients (pts) w/ advanced solid tumors. J Clin Oncol 29: 2011. (suppl; abstr 3010). [Google Scholar]
- 18. Winer EP, Tolaney S, Nechushtan H, Berger R, Kurzrock R, Ron I-G, et al. Activity of cabozantinib (XL184) in metastatic breast cancer (MBC): Results from a phase II randomized discontinuation trial (RDT). J Clin Oncol 30, 2012. (suppl; abstr 535). [Google Scholar]
- 19. Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, et al. Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res. 2011; 13: R35 10.1186/bcr2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009; 15: 460–467. 10.1158/1078-0432.CCR-08-1757 [DOI] [PubMed] [Google Scholar]
- 21.Ehrich M, Hogg G, Pearce M. Low abundant mutation profiling in tumor samples using the Sequenom OncoCarta Panel (Sequenom Application Note 12/11/2008).
- 22. Loibl S, von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014; 32: 3212–3220. 10.1200/JCO.2014.55.7876 [DOI] [PubMed] [Google Scholar]
- 23. Majewski IJ, Nuciforo P, Mittempergher L, Bosma AJ, Eidtmann H, Holmes E, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015; 33: 1334–1339. 10.1200/JCO.2014.55.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.COSMIC. Catalogue of somatic mutations in cancer. http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/ (accessed 21st Sept 2015)
- 25. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008; 68: 6084–6091. 10.1158/0008-5472.CAN-07-6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santarpia L, Qi Y, Stemke-Hale K, Wang B, Young EJ, Booser DJ, et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res Treat. 2012; 134: 333–343. 10.1007/s10549-012-2035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012; 33: 2007–2017. 10.1093/carcin/bgs232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ng CK, Pemberton HN, Reis-Filho JS. Breast cancer intratumor genetic heterogeneity: causes and implications. Expert Rev Anticancer Ther. 2012; 12: 1021–1032. 10.1586/era.12.85 [DOI] [PubMed] [Google Scholar]
- 29. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013; 368: 1199–1209. 10.1056/NEJMoa1213261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.