Abstract
Although articular cartilage degeneration represents a major public health problem, the underlying molecular mechanisms are still poorly characterized. We have previously utilized genome-wide expression analysis to identify specific markers of porcine articular cartilage, one of them being Thrombospondin-4 (Thbs4). In the present study we analyzed Thbs4 expression in mice, thereby confirming its predominant expression in articular cartilage, but also identifying expression in other tissues, including bone. To study the role of Thbs4 in skeletal development and integrity we took advantage of a Thbs4-deficient mouse model that was analyzed by undecalcified bone histology. We found that Thbs4-deficient mice do not display phenotypic differences towards wildtype littermates in terms of skeletal growth or bone mass acquisition. Since Thbs4 has previously been found over-expressed in bones of Phex-deficient Hyp mice, we additionally generated Thbs4-deficient Hyp mice, but failed to detect phenotypic differences towards Hyp littermates. With respect to articular cartilage we found that Thbs4-deficient mice display transient thinning of articular cartilage, suggesting a protective role of Thbs4 for joint integrity. Gene expression analysis using porcine primary cells revealed that Thbs4 is not expressed by synovial fibroblasts and that it represents the only member of the Thbs gene family with specific expression in articular, but not in growth plate chondrocytes. In an attempt to identify specific molecular effects of Thbs4 we treated porcine articular chondrocytes with human THBS4 in the absence or presence of conditioned medium from porcine synovial fibroblasts. Here we did not observe a significant influence of THBS4 on proliferation, metabolic activity, apoptosis or gene expression, suggesting that it does not act as a signaling molecule. Taken together, our data demonstrate that Thbs4 is highly expressed in articular chondrocytes, where its presence in the extracellular matrix is required for articular cartilage integrity.
Introduction
Osteoarthritis is a highly prevalent disorder characterized by loss of articular cartilage, a unique avascular and hypocellular tissue covering the joints [1,2]. The current treatment options are limited to surgical procedures, such as microfracture, osteochondral implantation, and, in the end-stage, total joint replacement [3,4]. In addition, autologous transplantation of articular chondrocytes after ex vivo expansion has emerged as a promising alternative therapeutic approach over the last two decades [5,6]. Since it is commonly accepted however, that any of these treatments only leads to formation of a fibrocartilagenous tissue with inferior quality compared to native articular cartilage, it is of utmost clinical importance to understand the molecular mechanisms controlling the function of articular chondrocytes. This may not only help to optimize ongoing tissue-engineering approaches, but also to identify potential molecular targets for pharmacological stimulation of matrix production by articular chondrocytes.
One potential explanation for the paucity of knowledge regarding specific regulators of articular chondrogenesis is the shortness of articular cartilage in mice, which hinders the identification of unexpected osteoarthritis phenotypes in genetically modified mouse models, unlike it was the case for other skeletal disorders [7]. For the same reason, it is extremely difficult to isolate primary articular chondrocytes from mice, which explains why many experiments related to arthritis have been performed with rib or epiphyseal growth plate chondrocytes. Regardless of these limitations several studies have been performed to identify specific markers of articular cartilage in mice, and there is an increasing number of mouse models displaying an arthritis phenotype [8–14]. Given the potential importance of identifying molecular differences between chondrocytes from articular and non-articular cartilage, we have previously performed genome-wide expression analysis with native tissues and cultured cells of porcine origin [15]. Here it was possible to separate articular and growth plate cartilage from the bone matrix and to culture a sufficient number of chondrocytes from both sources ex vivo. The major disadvantage of this approach was related to the use of porcine Gene Chips, which did not provide the same genetic coverage compared to the murine system.
Despite this limitation however, we made at least three important observations [15]. First, we identified common markers of cartilage, but also genes with specific expression in either growth plate or articular cartilage. Second, we found that the molecular differences between the two types of chondrocytes persisted after 10 and 20 days of ex vivo culture. Third, we defined 19 markers of articular chondrocytes, thereby raising the question, which of these are relevant regulators of articular cartilage integrity. Here we analyzed the physiological role of Thbs4, one of these previously identified markers, by studying the skeletal phenotype of a mouse deficiency model. We found that Thbs4-deficient mice do not display defects of skeletal growth or bone mass acquisition, but that articular cartilage thickness is transiently reduced.
Materials and Methods
1. Mouse models
Thbs4-deficient mice on a C57Bl/6 genetic background were purchased from the Jackson Laboratories (#005845). Their genotyping was performed with the primers 5´-GGG TGG GAT TAG ATA AAT GCC TGC TCT-3´, 5´-GGA GAG AGA ATA GCA AGA TCA GCT C-3´, and 5´-AAC AAG CAA TGG AAG GCA GAC CCT G-3´, giving rise to a 412 bp and a 544 bp fragment for the wildtype and mutant allele, respectively. To evaluate the impact of Thbs4 in the context of X-linked hypophosphatemic rickets, Thbs4-deficient mice were crossed with Hyp mice (C57Bl/6 genetic background), which were also obtained from the Jackson Laboratories (#000528). To induce experimental arthritis, Thbs4-deficient mice were crossed with transgenic mice expressing human TNFα (Tg197 on a C57Bl/6 genetic background), which have been described elsewhere [16]. Joint swelling of the foot paws was assessed between 5 and 10 weeks of age using a clinical score graded from 0 (no swelling) to 3 (severe swelling of toes and ankle) as described previously [17]. Grip strength was analyzed on wire (diameter of 3 mm) using a score from 0 (normal grip strength) to -4 (no detectable grip strength) as described [17]. All mice were fed ad libitum and housed in a regular light/dark cycle under SPF-conditions. Animal experiments were approved by the animal facility of the University Medical Center Hamburg Eppendorf and by the “Amt für Gesundheit und Verbraucherschutz” (Org529).
2. Cell culture
Primary murine osteoclasts were generated by differentiating bone marrow cells for 10 days with 1,25-dihydroxyvitamin D3 (10 nM) added for the whole period, and M-Csf (20 ng/ml) and Rankl (40 ng/ml) added from day 3 until day 10 [18]. Primary murine osteoblasts were isolated by collagenase digestion of calvariae from newborn mice and differentiated for 20 days in the presence of ascorbic acid (10 mM) and ß-glycerophosphate (50 μg/ml) as described [18]. Porcine synovial tissue was obtained from the right and the left knee of 6 weeks old minipigs. Digestion of the prepared synovial membrane was performed with 1 mg/dl collagenase type 1a solution (Sigma-Aldrich, Germany) for 60–75 min at 37°C. Isolated synovial fibroblasts were seeded and cultured in Synoviocyte Basal Medium (Cell Applications, USA) supplemented with 10% heat-inactivated Synoviocyte Growth Supplement (Cell Application, USA) at normal cell culture conditions. Conditioned medium of these cells was collected for 24 hours in basal medium. To isolate chondrocytes bone-cartilage cylinders were harvested from the medial and lateral condyle of knee joints of 6 weeks old minipigs. The cylinders were separated in articular and growth plate cartilage under the dissecting microscope. Chondrocytes were released by collagenase type la solution (Sigma-Aldrich, Germany) as described [15]. The cells were cultured in DMEM/Hams’F12 (Biochrom, Germany) supplemented with 10% (v/v) FBS (Lonza, Germany) at 37°C under an atmosphere of 5% (v/v) O2 and 5% (v/v) CO2. These experiments were approved by the animal facility of the University Medical Center Hamburg Eppendorf and by the “Amt für Gesundheit und Verbraucherschutz” (Org356).
3. Expression analysis
RNA from murine tissues, cultured murine osteoclasts and osteoblasts, or porcine chondrocytes and synoviocytes was isolated using the RNeasyMini kit (Qiagen, Germany). DNase digestion was performed according to manufacturer’s instructions. Concentration and quality of RNA were measured using a NanoDrop ND-1000 system (NanoDrop Technology, USA). For RT-PCR expression analysis, 1μg of RNA was reversed transcribed using SuperScriptIII (Invitrogen, Germany) according to manufacturer’s instructions. Predesigned TaqMan gene expression assays (Applied Biosystems, Germany) were used to quantify expression of all murine genes, as well as for the porcine genes ACAN, FN1, COL2A1, COL10A1, ASPN, FRZB, and SDC4. For the other porcine gene the resulting cDNA was used for a PCR reaction with gene-specific primers (PRG4: 5´-CAT CTC TCT TTG ACG GTG AGG G-3´ and 5´-GCT CCA TAG TGC AGA CTT TCT TGA-3´; THBS1: 5´-CCA GCA GCC GTT TCT ATG TTG T -3’ and 5´-cct atg tga cga gga tca tgc-3´; THBS2: 5´-gac gag ttt ggg tct gtg ga-3´and 5´-cca gcg tag gtt tgg tca ta-3´; THBS3: 5´-cag gta cga ctg ctg tgg ac-3´and 5´-ggc act gtg tca ttg cat cg-3´; THBS4: 5’-ATC CAG GCG ATC GAA ATT CTG-3’ and 5’-AGG TGT CCT ATC GCT GGT TCC T-3’; THBS5/COMP: 5’-GGA TGC CTG TGA CAA CTG TC-3’ and 5’-AAG GCC CTG AAG TCG GTG AG-3’, MATN3: 5´-ACC CAC GCG CCC TAT TCT-3´and 5´-CGA GTG GGT CTG GAG ATG GA-3´; GAPDH: 5’-CTT CGT CAA GCT CAT TTC CTG G-3’ and 5’-AGT CAG GAG ATG CTC GGT GTG-3’) and SYBR Green Master Mix (Applied Biosystems, Germany). GAPDH expression was used as an internal control. Relative quantification was performed according to the ΔΔCT method, and results were expressed in linear form using the formula 2-ΔΔCT for both RT-PCR assays.
4. Skeletal analysis
After sacrifice the dissected skeletons were fixed in 3.7% PBS-buffered formaldehyde for 18 hours at 4°C, before they were stored in 80% ethanol. All skeletons were analyzed by contact radiography using a Faxitron Xray cabinet (Faxitron Xray Corp., USA) to measure the length of the lumbar spine and femora. For bone histology, the lumbar vertebral bodies L3 to L6 and one tibia of each mouse were dehydrated in ascending alcohol concentrations and then embedded in methylmetacrylate as described previously [18]. Sections of 5 μm thickness were cut in the sagittal plane on a Microtec rotation microtome (Techno-Med GmbH, Germany). For articular cartilage histology, knee joints were processed in the same way. All sections were stained by toluidine blue and von Kossa/van Gieson staining procedures as described [18]. Histomorphometry was performed according to the ASBMR guidelines [19] using the OsteoMeasure histomorphometry system (Osteometrics Inc., USA). Articular chondrocyte apoptosis was assessed on 5 μm thin decalcified paraffin sections by in situ TUNEL technology (Roche, #11684795910) according to the manufacturer´s instructions.
5. Cellular assays
To assess proliferation, metabolic activity and apoptosis, porcine articular chondrocytes were seeded into 96-well plates at a density of 1.000 cells per ml. On the next day cells were incubated for 24 hours with human THBS4 (R&D Systems, #2390-TH) at different concentrations in serum-free medium or in serum-free medium mixed with an equal amount of conditioned medium from porcine synovial fibroblasts. BrdU incorporation, MTT conversion and Caspase 3/7 activities were determined with commercially available systems (GE Healthcare Amersham, #RPM250, Sigma Aldrich, #M2158, and Promega, #G8090, respectively) according to the manufacturer´s instructions. To analyze cell adhesion 96-well-microtest plates were coated with 5 μg/ml THBS4 or 5 μg/ml THBS5/COMP (R&D Systems, #3134-CP) at 4°C overnight. Porcine articular chondrocytes were washed twice with HBBS/C (Hank´s Balanced Salt Solution + 1 mM calcium chloride), pre-treated with tosylphenylalanyl chloromethyl ketone-treated trypsin (0.1 mg/ml) in HBBS/C, before trypsinization was stopped after incubation for 5 min at 37°C. The cells were then washed three times with HBBS/C and resuspended in HBBS/C containing 1% heat inactivated BSA. Were indicated, a monoclonal antibody to integrin ß1 (BD Biosciences, #552828) was added to a final concentration of 16 μg/ml and incubated with the cells for 30 min at room temperature. Coated wells were washed four times with HBBS/C before addition of 3 x 104 cells per well. After overnight incubation at 37°C under an atmosphere of 5% (v/v) O2 and 5% (v/v) CO2 the number of attached cells was quantified. To assess the effects of THBS4 and/or conditioned medium from porcine synovial fibroblasts on gene expression, cells were treated for 6 hours, before RNA was isolated for qRT-PCR expression analysis.
6. Serum analysis of individuals with osteoarthritis
Patients with osteoarthritis were recruited from the Department of Orthopaedics at the University Medical Center Hamburg-Eppendorf. Patient selection was based on a careful clinical examination and history according to the pattern of osteoarthritis (OA). Only individuals with primary OA were included. OA secondary to any known cause as well as any other arthropathy were exclusion criteria. All patients had severe, symptomatic primary OA of at least one large joint of the lower or upper extremity (knee, hip or shoulder; index joint) with radiographic joint space narrowing to less than a residual 1/3, and the request for total joint arthroplasty as the primary reason for consultation. Patients with knee or hip OA without signs and symptoms of OA in one or more additional joints were considered to have a relatively confined local disease and were classified as mono-osteoarthritis (mOA). Patients who, in addition to severe OA of the index joint, displayed clinically obvious hand OA of multiple interphalangeal joints and further joints of the lower and or upper extremities were considered to be more severely affected on a systemic level and were classified as poly-osteoarthritis (pOA). As an approach to distinguish between individuals with a presumably rather small total volume of articular cartilage affected locally by OA (mOA) versus individuals with a comparatively larger total volume of articular cartilage affected at multiple sites (pOA), we used these clinical differentiation criteria to build two distinct subpopulations of OA from our routine inpatient and outpatient clinic. Serum concentration of THBS4 was quantified using the antibody-based detection kit (Qayee-Bio, China). After the blood draw, serum samples were kept at room temperature for a maximum of two hours before storage at -70°C until analysis without additional freeze-thaw cycles. All participants provided written informed consent. This study and consent procedure was approved by the local ethics committee (Aerztekammer Hamburg, PV4037).
7. Statistical analysis
All data presented in the manuscript were obtained from the analysis of littermates (n ≥ 5) and are presented as means ± standard deviations. Statistical analysis was performed using unpaired, two-tailed Student’s t test, and p-values below 0.05 were considered statistically significant.
Results
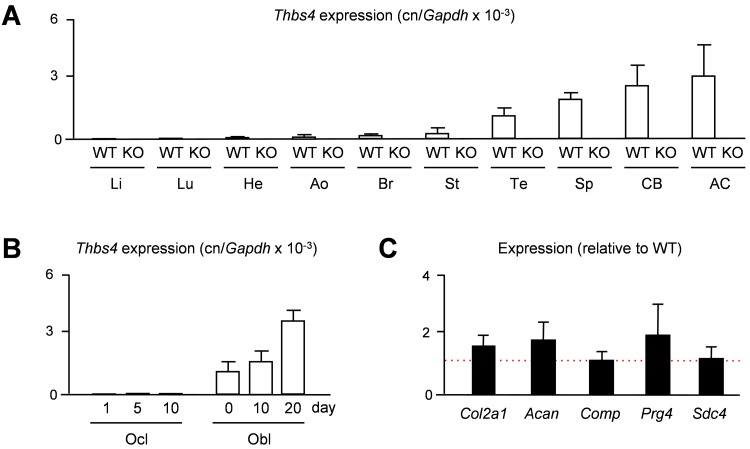
Given the previously observed predominant expression of THBS4 in porcine articular cartilage [15], we first addressed the question, whether the same is the case in mice. We therefore isolated RNA from different tissues of 15 weeks old wildtype and Thbs4-deficient mice to monitor Thbs4 expression by qRT-PCR. Here we found, as expected only in wildtype mice, that Thbs4 is highly expressed in articular cartilage, yet there was also strong expression in other tissues, such as tendon, spleen and cortical bone (Fig 1A). We additionally analyzed, which of the two bone remodeling cell types is the primary source of Thbs4 expression and performed qRT-PCR with RNA from primary osteoclasts and osteoblasts at different stages of differentiation. Here we found that Thbs4 was differentially expressed in osteoblast cultures, whereas Thbs4 transcripts were undetectable in osteoclast cultures (Fig 1B). Since articular cartilage displayed the highest expression of Thbs4 in wildtype mice, we additionally analyzed if Thbs4-deficiency would affect the expression of articular chondrocyte marker genes. Here we did not observe significant differences in the expression of Col2a1, Acan, Comp, Prg4 and Sdc4 between wildtype and Thbs4-deficient mice, suggesting that Thbs4 does not play a major role as a regulator of articular cartilage gene expression (Fig 1C). Nevertheless, given the high expression of Thbs4 in skeletal tissues, we went on to study the skeletal phenotype of Thbs4-deficient mice.
Fig 1. Murine Thbs4 is highly expressed in skeletal tissues.
(A) qRT-PCR monitioring Thbs4 expression in various tissues of 15 weeks old widltype (WT) and Thbs4-deficient mice (KO). Li, liver; Lu, lung; He, heart; Ao, aorta; Br, brain; St, stomach; Te, tendon; Sp, spleen; CB, cortical bone; AC, articular cartilage. Bars represent mean ± SD (n = 4). Thbs4 expression was undetectable in all KO tissues. (B) qRT-PCR monitoring Thbs4 expression in primary osteoclasts (Ocl) and osteoblasts (Obl) from wildtype mice at different stages of differentiation. Bars represent mean ± SD (n = 3). (C) qRT-PCR monitioring expression of the indicated genes in articular cartilage of 15 weeks Thbs4-deficient mice. The dotted red line indicates the expression in wildtype littermates. Bars represent mean ± SD (n = 4).
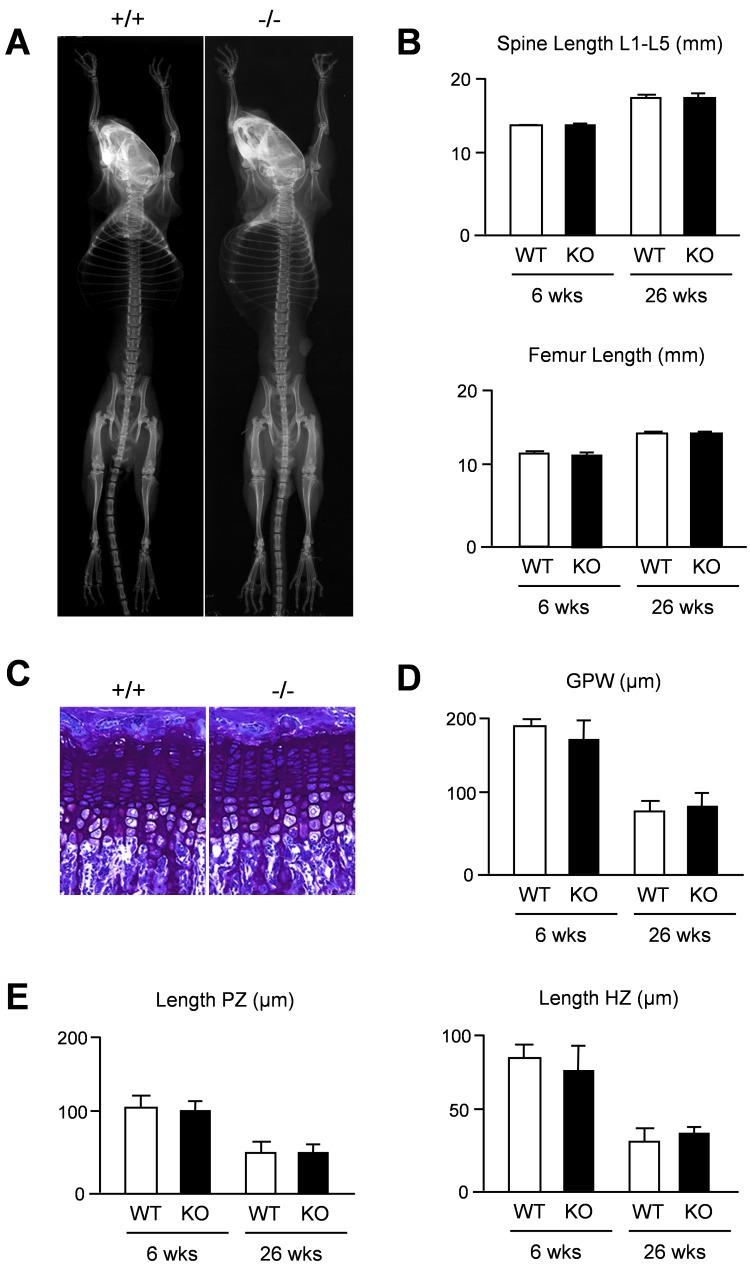
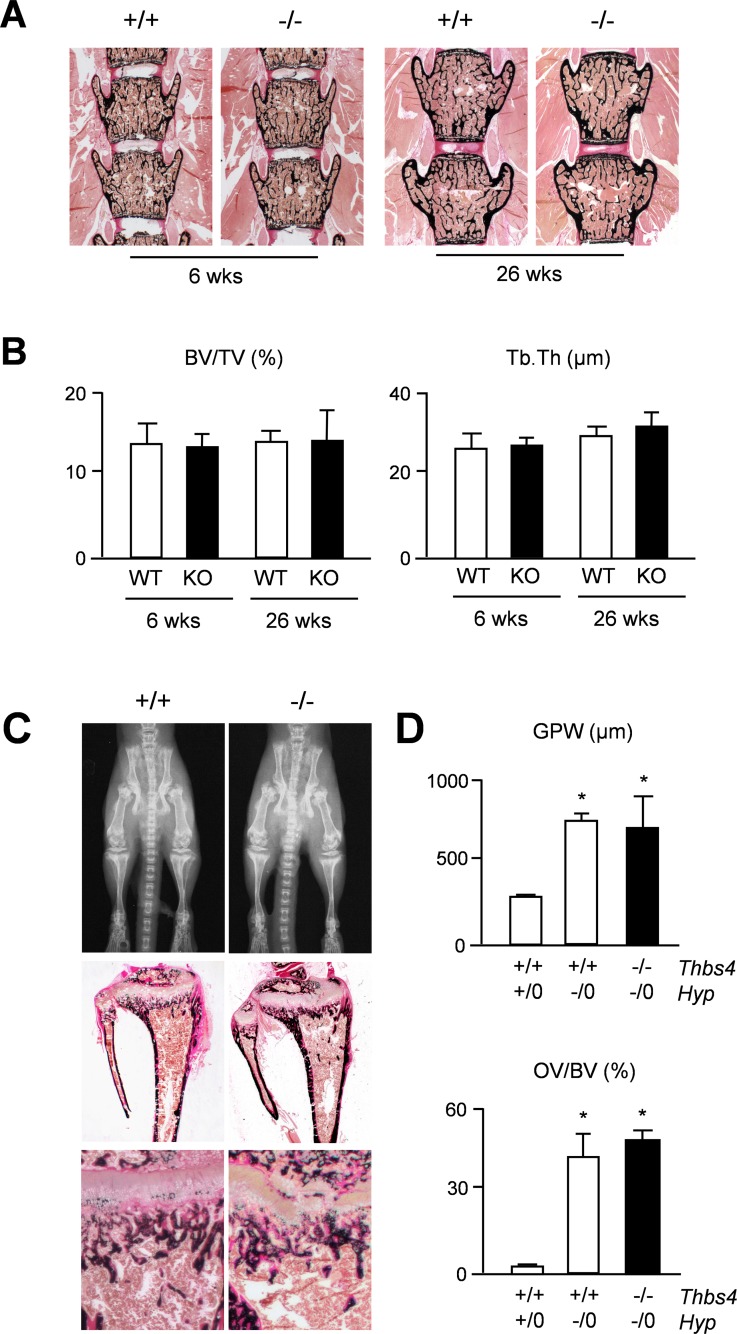
We first applied Xray analysis, but failed to detect major differences towards wildtype littermates (Fig 2A). Likewise, quantification of spine and femur length did not reveal a significant difference between wildtype and Thbs4-deficient mice at 6 and 26 weeks of age (Fig 2B). We additionally applied non-decalcified histology of tibia sections to analyze for potential differences in the growth plate (Fig 2C). Histomorphometric quantification of the growth plate thickness did not reveal statistically significant differences between wildtype and Thbs4-deficient mice at 6 and 26 weeks of age (Fig 2D). Likewise, the lengths of the proliferative and hypertophic zones within the tibia growth plate were not affected by Thbs4-deficiency (Fig 2E). We additionally analyzed spine sections in the same groups of mice with respect to a potential bone phenotype (Fig 3A). By quantifying trabecular bone parameters we again found no difference between wildtype and Thbs4-deficient littermates (Fig 3B). Collectively, these data suggested that Thbs4 does not function as a physiologically relevant regulator of skeletal growth, bone formation or remodeling.
Fig 2. Intact skeletal growth in Thbs4-deficient mice.
(A) Xray analysis demonstrates absence of gross skeletal abnormalities in 6 weeks old Thbs4-deficient mice. (B) Length of lumbar spine and femur in 6 and 26 weeks old wildtype (WT) and Thbs4-deficient (KO) mice. (C) Toluidine blue staining of the tibia growth plates from 6 weeks old wildtype (+/+) and Thbs4-deficient (-/-) mice. (D) Quantification of the tibial growth plate width (GPW) in wildtype (WT) and Thbs4-deficient (KO) mice at 6 and 26 weeks of age. (E) Quantification of the lengths of the proliferative zone (PZ) and hypertrophic zone (HZ) in the same sections. All bars represent mean ± SD (n = 6 per group).
Fig 3. No impact of Thbs4-deficiency on bone mass or matrix mineralization on a wildtype or Hyp genetic background.
(A) Von Kossa/van Gieson staining of spine sections from 6 and 26 weeks old wildtype (+/+) and Thbs4-deficient (-/-) mice. (B) Quantification of the trabecular bone volume per tissue volume (BV/TV) and trabecular thickness (Tb.Th.) in wildtype (WT) and Thbs4-deficient (KO) mice at both ages. Bars represent mean ± SD (n = 6 per group). (C) Xray analysis (top panels) and von Kossa/van Gieson staining of tibia (middle panels) or spine sections (bottom panels) from 6 weeks old Hyp mice with (+/+) or without (-/-) Thbs4. (D) Quantification of the growth plate width (GPW) in the tibia (top) or the osteoid volume per bone volume (OV/BV) from mice of the indicated genotypes. Bars represent mean ± SD (n = 5 per group). Asterisks indicate statistically significant differences towards WT controls (p<0.05).
Another question to be addressed was based on previously reported findings in Phex-deficient Hyp mice, a model of X-linked hypohosphatemic rickets [20,21]. These mice display defects of skeletal growth and bone matrix mineralization, which are only partially explained by hypophosphatemia [22,23]. A genome-wide expression analysis revealed that Thbs4 is markedly over-expressed in cortical bone of these mice, similar to Fgf23, whose increased expression in Hyp mice is known to cause their renal phosphate loss [24]. Since it was reasonable to speculate that a higher abundance of Thbs4 in the bone matrix could interfere with mineralization, we generated Thbs4-deficient Hyp mice and analyzed their skeletal phenotype. Here we found, as expected, that Hyp mice displayed a severe skeletal phenotype with pathological enrichment of non-mineralized osteoid, yet this pathology was not affected by Thbs4-deficiency (Fig 3C). Subsequent quantification of growth plate thickness and osteoid volume confirmed that the skeletal phenotype of Hyp mice was not significantly altered by additional Thbs4-deficiency (Fig 3D), thereby demonstrating that increased Thbs4 expression by Phex-deficient osteoblasts is not involved in the pathogenesis of X-linked hypophosphatemic rickets.
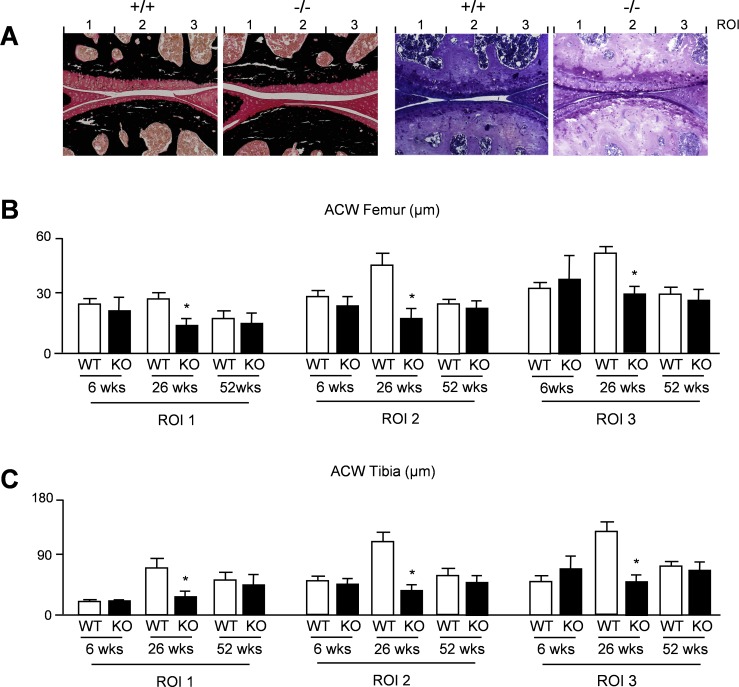
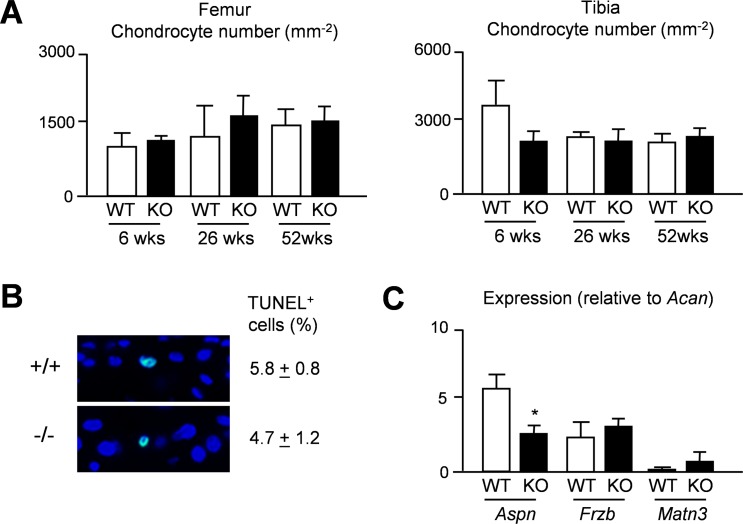
To assess the articular cartilage phenotype of Thbs4-deficient mice we analyzed sections from the knee joints of 6, 26 and 52 weeks old mice and determined the thickness of the articular cartilage layer in three different regions of interest (Fig 4A). Here we found that 26 weeks old Thbs4-deficient mice displayed significantly reduced articular cartilage thickness in all regions of interest, both in the femur (Fig 4B) and in the tibia (Fig 4C). This genotype-dependent difference was not observed at the age of 52 weeks, where the articular cartilage thickness was similar to 26 weeks old Thbs4-deficient mice in both genotypes. We also determined the number of chondrocytes per cartilage area in femur and tibia from the respective mice (Fig 5A). Here we did not observe statistically significant differences, and the same was the case for the percentage of apoptotic cells as assessed by TUNEL assay at the age of 26 weeks (Fig 5B). Finally, in an attempt to obtain a molecular explanation for the transient phenotype of Thbs4-deficient mice, we monitored expression of known osteoarthritis susceptibility (OAS) genes [25–28] in articular cartilage from 26 weeks old mice. Whereas Gdf5 expression was not detectable by qRT-PCR in samples from either genotype, we found no significant changes between wildtype and Thbs4-deficient mice in terms of Frzb or Matn3 expression (Fig 5C). Interestingly however, Aspn, encoding a small leucine-rich proteoglycan potentially inhibiting TGFß-dependent matrix synthesis [28,29], was expressed at lower levels in articular cartilage of 26 weeks old Thbs4-deficient mice.
Fig 4. Transient thinning of articular cartilage in Thbs4-deficient mice.
(A) Von Kossa/van Gieson (left panels) or toluidine blue staining (right panels) of articular cartilage from the knee joints of 26 weeks old wildtype (+/+) and Thbs4-deficient (-/-) mice. The three regions of interest for quantification of articular cartilage width are indicated. (B) Quantification of the articular cartilage width (ACW) in femora from wildtype (WT) and Thbs4-deficient (KO) mice at 6, 26 and 52 weeks of age. (C) Quantification of the articular cartilage width (ACW) in femur and tibia sections from the same mice. All bars represent mean ± SD (n = 6 per group). Asterisks indicate statistically significant differences between WT and KO (p<0.05).
Fig 5. Chondrogenesis is unaffected in articular cartilage in Thbs4-deficient mice.
(A) Quantification of the chondrocyte number per cartilage area in femur and tibia sections from 6, 26 and 52 weeks wildtype and Thbs4-deficient mice. Bars represent mean ± SD (n = 6 per group). (B) Representative images showing TUNEL-positive cells in articular cartilage from the femora of 26 weeks old wildtype and Thbs4-deficient mice. The percentage of TUNEL-positive cells is givenon the right. Values represent mean ± SD (n = 6 per group). (C) qRT-PCR monitioring expression of the indicated genes in articular cartilage of 26 weeks wildtype Thbs4-deficient mice. Bars represent mean ± SD (n = 4). Asterisks indicate statistically significant differences between WT and KO (p<0.05).
To address the question, if Thbs4-deficiency would affect the severity of joint destruction in a mouse model of rheumatoid arthritis, we additionally crossed Thbs4-deficient mice with mice carrying a transgene causing over-expression of human TNFα [16,17]. Here we found that the presence of the transgene caused progressive joint swelling of the foot paws together with a decline in grip strength until the age of 12 weeks, yet Thbs4-deficiency did not significantly affect these two clinical scores (S1A Fig). When we histologically analyzed the knee joints at 12 weeks of age however, we found enhanced destruction of subchondral bone specifically in Thbs4-deficient TNFα-transgenic mice (S1B Fig). Taken together, these findings revealed that Thsb4 has a protective role in articular cartilage, although its deficiency does not affect the affect proliferation or apoptosis of articular chondrocytes.
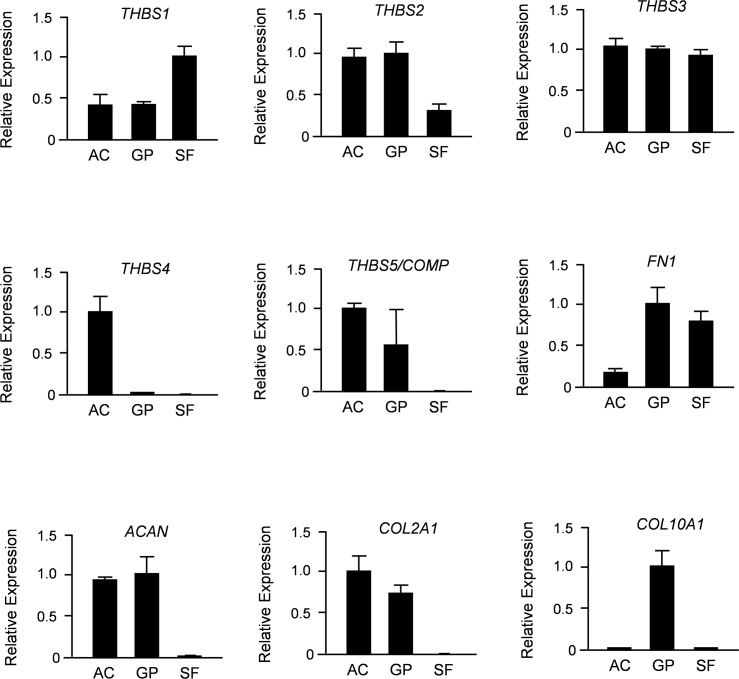
Since Thbs4 is only one member of a protein family, we next compared expression of all five thrombospondin-encoding genes in primary chondrocytes from articular and growth plate cartilage, as well as in synovial fibroblasts. To avoid any cross-contamination of these cell populations we again utilized minipigs, where the two types of cartilage, as well as synovial fibroblasts can be undoubtedly separated. Using qRT-PCR expression analysis we found that THBS1, THBS2, THBS3 and COMP/THBS5 were all expressed in both types of chondrocytes (Fig 6). With the exception of COMP/THBS5, we also detected their expression in synovial fibroblasts. In sharp contrast, THBS4 expression was only detected in articular chondrocytes and not in any of the other cell types.
Fig 6. Porcine THBS4 is specifically expressed in articular chondrocytes.
Shown are the results of qRT-PCR expression analyses for all members of the THBS family, as well as markers for synovial fibroblasts (FN1), chondrocytes (ACAN, COL2A1) and hypertrophic chondrocytes (COL10A1). Primary cells (AC, articular chondrocytes; GP, growth plate chondrocytes; SF, synovial fibroblasts) were derived from 6 weeks old minipigs. Shown is the relative expression (after normalization to GAPDH) towards the cell type displaying the highest expression level. All bars represent mean ± SD (n = 3).
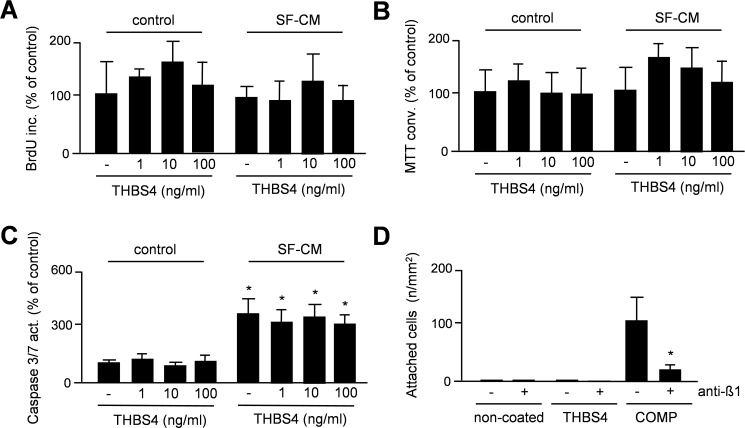
To analyze a potential impact of THBS4 on the behavior of articular chondrocytes we also used porcine cells. More specifically, we assessed cellular proliferation (Fig 7A), metabolic activity (Fig 7B) and apoptosis (Fig 7C) over 24 hours in primary articular chondrocytes in the presence of increasing concentrations of human THBS4. As a control we performed the same assays with conditioned medium (50% final concentration) from cultured porcine synovial fibroblasts (SF-CM), and again co-administered increasing concentrations of human THBS4. Here we found that THBS4 did not cause a significant influence on any of the parameters. Interestingly however, while SF-CM did not affect proliferation or metabolic activity of the articular chondrocytes, it significantly increased the Caspase-3/7 activity, suggesting a pro-apoptotic influence, which was however unaffected by THBS4. Finally, since COMP/THBS5 has been shown to mediate chondrocyte attachment in an integrin-dependent manner [30], we analyzed if THBS4 would serve a similar function. To address this possibility we coated non-tissue culture plates with THBS4 or COMP, before adding porcine articular chondrocytes in the presence or absence of an antibody against ß1-integrin. After 24 hours we counted the adherent cells and found that they attached to COMP-coated plates in a ß1-integrin-dependent manner (Fig 7D). In contrast, we failed to detect adherent articular chondrocytes on THBS4-coated plates.
Fig 7. THBS4 does not affect the molecular behavior of porcine articular chondrocytes.
(A) BrdU incorporation monitoring cellular proliferation of porcine articular chondrocytes in the presence of increasing concentrations of human THBS4 and/or conditioned medium from porcine synovial fibroblasts (SF-CM), as indicated. Bars represent mean ± SD (n = 5). (B) MTT conversion monitoring metabolic activity of porcine articular chondrocytes under the same conditions. Bars represent mean ± SD (n = 5). (C) Caspase-3/7 activity monitoring apoptosis of porcine articular chondrocytes under the same conditions. Bars represent mean ± SD (n = 5). (D) Cell attachment to non-coated plates or plates coated with THBS4 or THBS5/COMP in the absence or presence of a ß1-integrin antibody. Bars represent mean ± SD (n = 6). The asterisk indicates a statistically significant of the antibody (p<0.05).
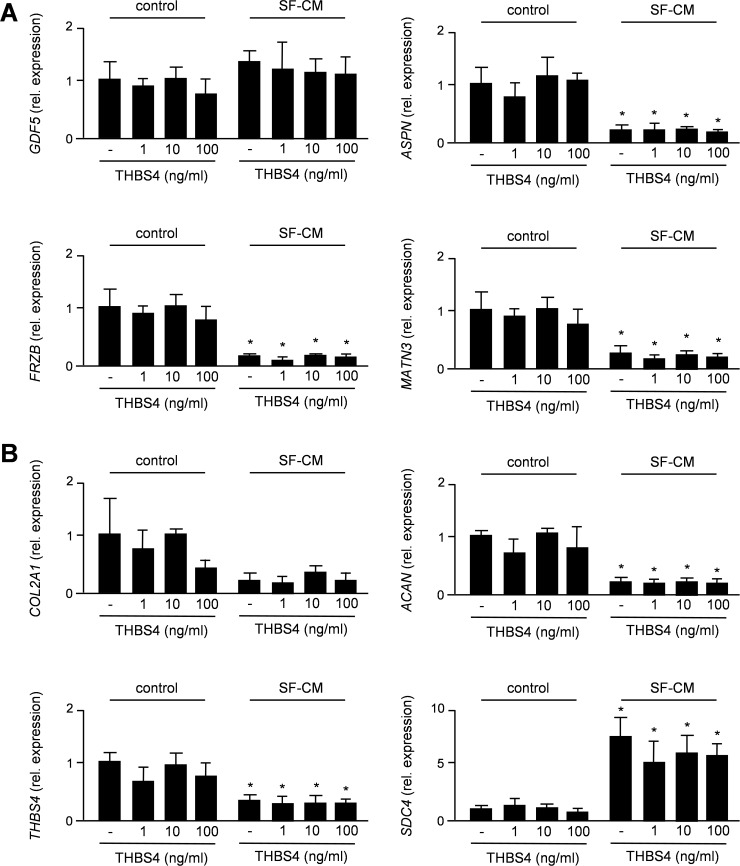
As we observed reduced expression of Aspn in articular cartilage of 26 weeks old Thbs4-deficient, we additionally treated articular chondrocytes for 6 hours with THBS4 and/or SF-CM, before isolating RNA for qRT-PCR expression analysis. When monitoring expression of the OAS genes we observed no significant influences of THBS4, either alone or in the presence of SF-CM (Fig 8A). Importantly however, expression levels of ASPN, FRZB and MATN3 (but not of GDF5) were remarkably reduced by SF-CM, thus suggesting a direct transcriptional influence by yet unidentified SF-derived molecules. Based on these findings we additionally monitored expression of COL2A1, ACAN, THBS4 and SDC4, the latter gene encoding a negative regulator of articular chondrocytes [9]. Again, we failed to detect a significant influence of THBS4, either alone or in the presence of SF-CM (Fig 8B). In contrast, whereas SF-CM caused a transcriptional repression of ACAN and THBS4, SDC4 expression was more than 5-fold induced by SF-CM, thus underscoring the suspected influence of SF-derived molecules on gene expression in articular chondrocytes. With respect to THBS4 however, our combined results suggest that it does not act as a signaling molecule directly regulating articular proliferation, differentiation or gene expression.
Fig 8. THBS4 does not affect gene expression in porcine articular chondrocytes.
(A) qRT-PCR monitoring OAS gene expression in porcine articular chondrocytes treated with human THBS4 and/or SF-CM for 6 hours. (B) qRT-PCR monitoring expression of COL2A1, ACAN, THBS4 and SDC4 expression in the same samples. Bars represent mean ± SD (n = 4). Asterisks indicate statistically significant differences towards untreated cells (p<0.05).
Discussion
The thrombospondins represent a family of secreted matricellular proteins potentially regulating various processes of tissue remodeling [31]. The five Thbs family members can be divided into two subgroups based on their domain structure and mode of multimerization. Thbs4, together with Thbs3 and Thbs5, belongs to the second subgroup considered to form a pentameric stucture [32]. While mutations of Thbs5, better known as cartilage-oligomeric matrix protein (Comp), cause two different forms of skeletal dysplasia, the role of Thbs3 and Thbs4 in the skeleton are still poorly defined [33,34]. More specifically, while a transiently accelerated endochondral ossification has been reported for mice lacking Thbs3, the skeletal phenotype of Thbs4-deficient mice has not been analyzed previously. Interestingly however, two recent studies have identified a specific function of Thbs4 in myocardial remodeling [35,36], thereby underscoring the relevance of previous findings showing that Thbs4 expression is specifically induced in hypertrophic or failing hearts [37,38]. More recently, Thbs4-deficient mice were found to display an altered composition of extracellular matrices in tendons and skeletal muscles, which also affected the physiological functions of both tissues [39].
Since we have previously identified Thbs4 as a marker of articular cartilage [15], our main interest was an in-depth skeletal phenotyping of Thbs4-deficient mice, thereby also addressing the question, whether its increased expression in bones from Hyp mice is relevant for the pathogenesis of X-linked hypophosphatemic rickets [24]. Through the use of undecalcified histology with subsequent histomorphometry we found that Thbs4-deficiency has no impact on skeletal growth, bone mass acquisition or skeletal remodeling, and we were able to rule out a contribution of Thbs4 to the skeletal phenotype of Phex-deficient Hyp mice. We did however observe a significant reduction of articular cartilage thickness in 26 weeks old Thbs4-deficient mice when compared to wildtype littermates, although there was no difference found in 6 or 52 weeks old animals. More specifically, it appeared that the age-related gain of articular cartilage thickness is abolished in Thbs4-deficient mice, whereas the ageing-associated articular cartilage degeneration was not accelerated [40–42]. These data indicate that Thbs4 has a protective role for articular cartilage integrity and suggest that its absence is partially compensated by other molecules, possibly Thbs family members, thereby preventing complete loss of joint surfaces in Thbs4-deficient mice. We additionally crossed the Thbs4-deficiency into a TNF-transgenic background, which is commonly used to study the impact of specific molecules on the severity of rheumatoid arthritis [16,43,44]. Here we did not observe a significant impact of the Thbs4-deficiency on two clinical scores of rheumatoid arthritis, i.e. paw swelling and grip strength, yet loss of subchondral bone was apparently enhanced in Thbs4-deficient TNF-transgenic mice, thereby supporting the concept that Thbs4 has a protective role in articular cartilage.
With respect to the underlying molecular mechanisms we performed experiments with porcine articular chondrocytes, thereby avoiding the principal problem to obtain primary murine articular chondrocytes at sufficient quantity and without contaminating additional cell types. Here we administered recombinant human THBS4 to study its potential effects on different cellular parameters. Using the porcine system additionally allowed us to introduce a control, i.e. conditioned medium from synovial fibroblasts (SF-CM), since these cells appear to secrete factors modulating activities of articular chondrocytes [45]. We found, unexpectedly, that short-term treatment with SF-CM significantly increased Caspase-3/7 activity and SDC4 expression in articular chondrocyte cultures, while it reduced the expression of genes associated with osteoarthritis and/or encoding components of the cartilage extracellular matrix, including THBS4. Albeit interesting and worth being further investigated, the most important finding related to the present study however was that THBS4 administration did not affect any of the tested parameters, and it did not protect against the negative influence of SF-CM. Therefore, although we observed a specific reduction of Aspn expression in 26 weeks old Thbs4-deficient mice, it is unlikely that this alteration is directly caused by Thbs4-deficiency, since our combined analyses essentially rule out that THBS4 acts as a signaling molecule directly regulating transcription in articular chondroctes. We additionally performed cell adhesion assays, thereby confirming that COMP/THBS5 mediates chondrocyte attachment in an integrin-dependent manner [30], unlike THBS4. Albeit these findings are principally consistent with the lack of differences regarding cellular density in articular cartilage between wildtype and Thbs4-deficient mice, they failed to provide a molecular explanation for the observed differences. Therefore, we can only speculate about the causes of the transient reduction of articular cartilage thickness in Thbs4-deficient mice. However, since Thbs4 has been shown to interact with various matrix molecules [46], this phenotype might be related to subtle differences in extracellular matrix integrity, similar to the tendons, where Thbs4-deficiency affects collagen fibrillogenesis [39].
The absence of evidence supporting a function of THBS4 as a signaling molecule essentially rules out the possibility that the development of drugs activating THBS4 is a possible approach for the treatment of osteoarthritis. However, since THBS4, in contrast to the other THBS family members or additional matrix proteins, such as type-II-collagen or aggrecan, is specifically expressed by articular and not by growth plate chondrocytes, monitoring THBS4 expression could still be useful for the quality control of tissue-engineered articular cartilage [15]. In the same line of thought, it is reasonable to hypothesize that THBS4 or THBS4 fragments could serve as biomarkers to monitor joint destruction. In fact, while it is obvious that the introduction of disease-specific biomarkers, such as PSA for prostatic hyperplasia, has revolutionized disease management in the last decades, a screening or treatment monitoring for osteoarthritis is still not possible, since specific markers of articular cartilage remain to be identified [47]. We therefore measured THBS4 serum concentrations in individuals with mono-osteoarthritis (S2A Fig) or poly-osteoarthritis (S2B Fig) using a commercially available ELISA against intact THBS4. Although we found that the latter group displayed higher circulating levels of intact THBS4, the difference towards individuals with mono-osteoarthritis or controls was not significant (S2C Fig). This implies that THBS4 is most likely not a valid biomarker of human osteoarthritis, yet it might be useful to analyze a larger number of individuals, also including cases with other causes of articular cartilage loss (such as rheumatoid arthritis), and to analyze for the presence of THBS4 cleavage products that are potentially generated in specific pathological settings.
Supporting Information
(A) Quantification of foot paw swelling (left) and grip strength (right) over time in TNF-transgenic mice with (WT) or without (KO) a functional Thbs4 allele. Values represent mean ± SD (n = 4 per group). (B) Von Kossa/van Gieson staining of knee joints from 12 weeks old TNF-transgenic mice with (+/+) or without (-/-) a functional Thbs4 allele. The quantification of the subchondral bone volume is given on the right. Bars represent mean ± SD (n = 4 per group). Asterisks indicate statistically significant differences between WT and KO (p<0.05).
(TIF)
(A) Age and gender distribution of individuals with mono-osteoarthritis (n = 20). (B) Age and gender distribution of individuals with poly-osteoarthritis (n = 21). (C) THBS4 concentrations in the sera from patients with mono-osteoarthritis (mOA) or poly-osteoarthritis (pOA). The dotted red line indicates the mean serum concentration measured in 6 control individuals without osteoarthritis.
(TIF)
Acknowledgments
We thank Dr. George Kolias (Fleming Institute, Vari, Greece) for kindly providing Tg197 TNFtg mice. This work was supported by grants from the Deutsche Forschungsgemeinschaft (AM103/21-1, SPP1468-IMMUNOBONE), the European Community’s Seventh Framework Programme under grant agreement n°602300 (SYBIL), and the IMI-funded project BTCure.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by the following: Deutsche Forschungsgemeinschaft (AM103/21-1, SPP1468-IMMUNOBONE) [http://www.dfg.de/]; Seventh Framework Programme under grant agreement n°602300 (SYBIL) [http://www.sybil-fp7.eu/]; and IMI-funded project BTCure [http://www.imi.europa.eu/].
References
- 1. Triche R, Mandelbaum BR. (2013) Overview of cartilage biology and new trends in cartilage stimulation. Foot Ankle Cli. 18: 1–12. [DOI] [PubMed] [Google Scholar]
- 2. Conaghan PG, Kloppenburg M, Schett G, Bijlsma JW. (2014) Osteoarthritis research priorities: a report from a EULAR ad hoc expert committee. Ann Rheum Dis 73: 1442–1445. 10.1136/annrheumdis-2013-204660 [DOI] [PubMed] [Google Scholar]
- 3. van der Kraan PM. (2012) Osteoarthritis year 2012 in review: biology. Osteoarthritis Cartilage 20: 1447–1450. 10.1016/j.joca.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 4. Magnussen RA, Dunn WR, Carey JL, Spindler KP. (2008) Treatment of focal articular cartilage defects in the knee: a systematic review. Clin Orthop Relat Res 466: 952–962. 10.1007/s11999-007-0097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tuan RS, Chen AF, Klatt BA. (2013) Cartilage regeneration. J Am Acad Orthop Surg 21: 303–311. 10.5435/JAAOS-21-05-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paetzold H, Goepfert C, Huber G, Hoenig E, Pörtner R, Schilling AF, et al. (2012) The development of the collagen fibre network in tissue-engineered cartilage constructs in vivo. Engineered cartilage reorganises fibre network. Eur Cell Mater 23: 209–221. [DOI] [PubMed] [Google Scholar]
- 7. Elefteriou F, Yang X. (2011) Genetic mouse models for bone studies—strengths and limitations. Bone 49: 1242–1254. 10.1016/j.bone.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, et al. (2005) The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest 115: 622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Echtermeyer F, Bertrand J, Dreier R, Meinecke I, Neugebauer K, Fuerst M, et al. (2009) Syndecan-4 regulates ADAMTS-5 activation and cartilage breakdown in osteoarthritis. Nat Med 15: 1072–1076. 10.1038/nm.1998 [DOI] [PubMed] [Google Scholar]
- 10. Settle SH Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. (2003) Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol 254, 116–130. [DOI] [PubMed] [Google Scholar]
- 11. Raducanu A, Hunziker EB, Drosse I, Aszódi A. (2009) Beta1 integrin deficiency results in multiple abnormalities of the knee joint. J Biol Chem 284: 23780–23792. 10.1074/jbc.M109.039347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang H, Beier F. (2014) Mouse models of osteoarthritis: modelling risk factors and assessing outcomes. Nat Rev Rheumatol 10: 413–421. 10.1038/nrrheum.2014.46 [DOI] [PubMed] [Google Scholar]
- 13. Vo N, Niedernhofer LJ, Nasto LA, Jacobs L, Robbins PD, Kang J, et al. (2013) An overview of underlying causes and animal models for the study of age-related degenerative disorders of the spine and synovial joints. J Orthop Res 31: 831–837. 10.1002/jor.22204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schelbergen RF, de Munter W, van den Bosch MH, Lafeber FP, Sloetjes A, Vogl T, et al. (2014) Alarmins S100A8/S100A9 aggravate osteophyte formation in experimental osteoarthritis and predict osteophyte progression in early human symptomatic osteoarthritis. Ann Rheum Dis [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 15. Hissnauer TN, Baranowsky A, Pestka JM, Streichert T, Wiegandt K, Goepfert C, et al. (2010) Identification of molecular markers for articular cartilage. Osteoarthritis Cartilage 18: 1630–1638. 10.1016/j.joca.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 16. Keffer J, Probert L, Cazlaris H, Georgopoulos S, Kaslaris E, Kioussis D, et al. (1991) Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. EMBO J 10: 4025–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stock M, Böhm C, Scholtysek C, Englbrecht M, Fürnrohr BG, Klinger P, et al. (2013) Wnt inhibitory factor 1 deficiency uncouples cartilage and bone destruction in tumor necrosis factor α-mediated experimental arthritis. Arthritis Rheum 65: 2310–2322. 10.1002/art.38054 [DOI] [PubMed] [Google Scholar]
- 18. Albers J, Keller J, Baranowsky A, Beil FT, Catala-Lehnen P, Schulze J, et al. (2013) Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin. J Cell Biol 200: 537–549. 10.1083/jcb.201207142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, et al. (1987) Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2: 595–610. [DOI] [PubMed] [Google Scholar]
- 20. Strom TM, Francis F, Lorenz B, Böddrich A, Econs MJ, Lehrach H, et al. (1997) Pex gene deletions in Gy and Hyp mice provide mouse models for X-linked hypophosphatemia. Hum Mol Genet 6: 165–171. [DOI] [PubMed] [Google Scholar]
- 21. Beck L, Soumounou Y, Martel J, Krishnamurthy G, Gauthier C, Goodyer CG, et al. (1997) Pex/PEX tissue distribution and evidence for a deletion in the 3' region of the Pex gene in X-linked hypophosphatemic mice. J Clin Invest 99: 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ecarot B, Glorieux FH, Desbarats M, Travers R, Labelle L. (1992) Defective bone formation by Hyp mouse bone cells transplanted into normal mice: evidence in favor of an intrinsic osteoblast defect. J Bone Miner Res 7: 215–220. [DOI] [PubMed] [Google Scholar]
- 23. Seitz S, Rendenbach C, Barvencik F, Streichert T, Jeschke A, Schulze J, et al. (2013) Retinol deprivation partially rescues the skeletal mineralization defects of Phex-deficient Hyp mice. Bone 53: 231–238. 10.1016/j.bone.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 24. Liu S, Tang W, Fang J, Ren J, Li H, Xiao Z, et al. (2009) Novel regulators of Fgf23 expression and mineralization in Hyp bone. Mol Endocrinol 23: 1505–1518. 10.1210/me.2009-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, et al. (2007) A functional polymorphism in the 5' UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 39: 529–533. [DOI] [PubMed] [Google Scholar]
- 26. Stefánsson SE, Jónsson H, Ingvarsson T, Manolescu I, Jónsson HH, Olafsdóttir G, et al. (2003) Genomewide scan for hand osteoarthritis: a novel mutation in matrilin-3. Am J Hum Genet 72: 1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loughlin J, Dowling B, Chapman K, Marcelline L, Mustafa Z, Southam L, et al. (2004) Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci U S A 101: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kizawa H, Kou I, Iida A, Sudo A, Miyamoto Y, Fukuda A, et al. (2005) An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet 37: 138–144. [DOI] [PubMed] [Google Scholar]
- 29. Xu L, Li Z, Liu SY, Xu SY, Ni GX. (2015) Asporin and osteoarthritis. Osteoarthritis Cartilage 23: 933–939. 10.1016/j.joca.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 30. Chen FH, Thomas AO, Hecht JT, Goldring MB, Lawler J. (2005) Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J Biol Chem 280: 32655–32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stenina-Adognravi O. (2013) Thrombospondins: old players, new games. Curr Opin Lipidol 24: 401–409. 10.1097/MOL.0b013e3283642912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stenina OI, Topol EJ, Plow EF. (2007) Thrombospondins, their polymorphisms, and cardiovascular disease. Arterioscler Thromb Vasc Biol 27: 1886–1894. [DOI] [PubMed] [Google Scholar]
- 33. Posey KL, Yang Y, Veerisetty AC, Sharan SK, Hecht JT. (2008) Model systems for studying skeletal dysplasias caused by TSP-5/COMP mutations. Cell Mol Life Sci 65: 687–699. 10.1007/s00018-007-7485-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hankenson KD, Hormuzdi SG, Meganck JA, Bornstein P. (2005) Mice with a disruption of the thrombospondin 3 gene differ in geometric and biomechanical properties of bone and have accelerated development of the femoral head. Mol Cell Biol 25: 5599–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frolova EG, Sopko N, Blech L, Popovic ZB, Li J, Vasanji A, et al. (2012) Thrombospondin-4 regulates fibrosis and remodeling of the myocardium in response to pressure overload. FASEB J 26: 2363–2373. 10.1096/fj.11-190728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, et al. (2012) A thrombospondin-dependent pathway for a protective ER stress response. Cell 149: 1257–1268. 10.1016/j.cell.2012.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gabrielsen A, Lawler PR, Yongzhong W, Steinbrüchel D, Blagoja D, Paulsson-Berne G, et al. (2007) Gene expression signals involved in ischemic injury, extracellular matrix composition and fibrosis defined by global mRNA profiling of the human left ventricular myocardium. J Mol Cell Cardiol 42: 870–883. [DOI] [PubMed] [Google Scholar]
- 38. Mustonen E, Aro J, Puhakka J, Ilves M, Soini Y, Leskinen H, et al. (2008) Thrombospondin-4 expression is rapidly upregulated by cardiac overload. Biochem Biophys Res Commun 373: 186–191. 10.1016/j.bbrc.2008.05.164 [DOI] [PubMed] [Google Scholar]
- 39. Frolova EG, Drazba J, Krukovets I, Kostenko V, Blech L, Harry C, et al. (2014) Control of organization and function of muscle and tendon by thrombospondin-4. Matrix Biol 37: 35–48. 10.1016/j.matbio.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okamura N, Hasegawa M, Nakoshi Y, Iino T, Sudo A, Imanaka-Yoshida K, et al. (2010) Deficiency of tenascin-C delays articular cartilage repair in mice. Osteoarthritis Cartilage 18: 839–848. 10.1016/j.joca.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 41. Martin JA, Buckwalter JA. (2001) Roles of articular cartilage aging and chondrocyte senescence in the pathogenesis of osteoarthritis. Iowa Orthop J 21: 1–7. [PMC free article] [PubMed] [Google Scholar]
- 42. Coles JM, Zhang L, Blum JJ, Warman ML, Jay GD, Guilak F, et al. (2010) Loss of cartilage structure, stiffness, and frictional properties in mice lacking PRG4. Arthritis Rheum 62: 1666–1674. 10.1002/art.27436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zwerina J, Redlich K, Schett G, Smolen JS. (2005) Pathogenesis of rheumatoid arthritis: targeting cytokines. Ann N Y Acad Sci 1051: 716–729. [DOI] [PubMed] [Google Scholar]
- 44. Schett G, Gravallese E. (2012) Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol 8: 656–664. 10.1038/nrrheum.2012.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steinhagen J, Bruns J, Niggemeyer O, Fuerst M, Rüther W, Schünke M, et al. (2010) Perfusion culture system: Synovial fibroblasts modulate articular chondrocyte matrix synthesis in vitro. Tissue Cell 42: 151–157. 10.1016/j.tice.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 46. Narouz-Ott L, Maurer P, Nitsche DP, Smyth N, Paulsson M. (2000) Thrombospondin-4 binds specifically to both collagenous and non-collagenous extracellular matrix proteins via its C-terminal domains. J Biol Chem 275: 37110–37117. [DOI] [PubMed] [Google Scholar]
- 47. Lotz M, Martel-Pelletier J, Christiansen C, Brandi ML, Bruyère O, Chapurlat R, et al. (2013) Value of biomarkers in osteoarthritis: current status and perspectives. Ann Rheum Dis 72: 1756–1763. 10.1136/annrheumdis-2013-203726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Quantification of foot paw swelling (left) and grip strength (right) over time in TNF-transgenic mice with (WT) or without (KO) a functional Thbs4 allele. Values represent mean ± SD (n = 4 per group). (B) Von Kossa/van Gieson staining of knee joints from 12 weeks old TNF-transgenic mice with (+/+) or without (-/-) a functional Thbs4 allele. The quantification of the subchondral bone volume is given on the right. Bars represent mean ± SD (n = 4 per group). Asterisks indicate statistically significant differences between WT and KO (p<0.05).
(TIF)
(A) Age and gender distribution of individuals with mono-osteoarthritis (n = 20). (B) Age and gender distribution of individuals with poly-osteoarthritis (n = 21). (C) THBS4 concentrations in the sera from patients with mono-osteoarthritis (mOA) or poly-osteoarthritis (pOA). The dotted red line indicates the mean serum concentration measured in 6 control individuals without osteoarthritis.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.