Abstract
Introduction
MicroRNAs (miRNAs) regulate messenger RNAs (mRNAs) and as such have been implicated in a variety of diseases, including cancer. MiRNAs regulate mRNAs through binding of the miRNA 5’ seed sequence (~7–8 nucleotides) to the mRNA 3’ UTRs; polymorphisms in these regions have the potential to alter miRNA-mRNA target associations. SNPs in miRNA genes as well as miRNA-target genes have been proposed to influence cancer risk through altered miRNA expression levels.
Methods
MiRNA-SNPs and miRNA-target gene-SNPs were identified through the literature. We used SNPs from Genome-Wide Association Study (GWAS) data that were matched to individuals with miRNA expression data generated from an Agilent platform for colon tumor and non-tumor paired tissues. These samples were used to evaluate 327 miRNA-SNP pairs for associations between SNPs and miRNA expression levels as well as for SNP associations with colon cancer.
Results
Twenty-two miRNAs expressed in non-tumor tissue were significantly different by genotype and 21 SNPs were associated with altered tumor/non-tumor differential miRNA expression across genotypes. Two miRNAs were associated with SNP genotype for both non-tumor and tumor/non-tumor differential expression. Of the 41 miRNAs significantly associated with SNPs all but seven were significantly differentially expressed in colon tumor tissue. Two of the 41 SNPs significantly associated with miRNA expression levels were associated with colon cancer risk: rs8176318 (BRCA1), ORAA 1.31 95% CI 1.01, 1.78, and rs8905 (PRKAR1A), ORGG 2.31 95% CI 1.11, 4.77.
Conclusion
Of the 327 SNPs identified in the literature as being important because of their potential regulation of miRNA expression levels, 12.5% had statistically significantly associations with miRNA expression. However, only two of these SNPs were significantly associated with colon cancer.
Introduction
MicroRNAs (miRNAs) are small, non-coding regulatory RNA molecules [1–5] that have the ability to alter gene expression and thus biological pathway function. MiRNAs have been associated with several diseases, including colorectal cancer risk and survival [6–8]. The importance of miRNAs in the carcinogenic process is under intense study. It has been proposed that SNPs located in miRNA gene loci and shown to be associated with colorectal cancer may be operating through their influence on miRNA expression levels [9]. Others have proposed that SNPs within miRNA target genes (i.e. mRNAs) can influence miRNA binding of these genes through the alteration of miRNA binding sites within the mRNA, causing altered expression of mRNA [10]. Mature miRNA levels have been proposed to be correlated with the presence of their target mRNAs in vitro within model organisms [11], and as such, SNPs within target genes could alter miRNA expression due to altered mRNA levels. Thus, it also has been proposed that SNPs within miRNA target genes (mRNAs) that are associated with disease risk could be operating through their influence on miRNAs. The majority of miRNA regulation occurs through the binding of the miRNA seed region (~7–8 nucleotides at the 5’ UTR end of the miRNA) to the 3’ UTR of the mRNA [12]; as such, SNPs within the 3’ UTR of miRNA-target genes may create or destroy miRNA binding sites [10], and are of particular interest to this investigation.
We are in a unique position to assess whether SNPs in miRNA gene loci and SNPs in miRNA target genes influence miRNA expression in colon tissue, and in turn investigate whether these same variants influence risk of colon cancer. To test the hypothesis that SNPs alter miRNA expression levels, we use non-tumor tissue and evaluate differences in expression across genotype. However, since a SNP could alter miRNA expression levels equally in both tumor and non-tumor tissue, such an association would not necessarily contribute to cancer risk. To determine if SNPs are associated with the carcinogenic process through miRNA regulation, we also evaluate if miRNA expression is different across genotypes between tumor and non-tumor tissues. Additionally, we assess if miRNAs that are associated with SNPs are differentially expressed in colon tumors. Finally, we evaluate the association between SNPs associated with miRNA expression and risk of colon cancer.
Methods
Study population
The study population consisted of individuals previously enrolled in a study of Diet, Lifestyle, and Colon cancer at the University of Utah and the Kaiser Permanente Medical Research Program (KPMRP) [13] for whom GWAS data as well as miRNA from both tumor and non-tumor tissue was available, and at the Twin Cities metropolitan area in Minnesota for GWAS data only (Table 1). Study subjects included incident cases of colon cancer between the ages of 30 and 79 who were non-Hispanic white, Hispanic, or African American, and were able to provide a signed informed consent prior to participation in the study. The study was approved by the University of Utah Institutional Review Board for Human Subjects.
Table 1. Descriptive table of included subjects.
| GWAS | miRNA | GWAS and miRNA | ||||||
|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Cases | Cases | |||||
| N | % | N | % | N | % | N | % | |
| Sex | ||||||||
| Male | 643 | 54.8 | 618 | 55.4 | 606 | 52.7 | 193 | 56.1 |
| Female | De | 45.2 | 497 | 44.6 | 544 | 47.3 | 151 | 43.9 |
| Center | ||||||||
| Kaiser | 290 | 24.7 | 356 | 31.9 | 585 | 50.9 | 191 | 55.5 |
| Minn. | 609 | 51.9 | 527 | 47.3 | 0 | 0.0 | 0 | 0.0 |
| Utah | 274 | 23.4 | 232 | 20.8 | 565 | 49.1 | 153 | 44.5 |
| Stage | ||||||||
| I | 0 | 0.0 | 346 | 34.7 | 296 | 27.9 | 90 | 26.4 |
| II | 0 | 0.0 | 291 | 29.2 | 286 | 27.0 | 103 | 30.2 |
| III | 0 | 0.0 | 270 | 27.1 | 283 | 26.7 | 115 | 33.7 |
| IV | 0 | 0.0 | 91 | 9.1 | 195 | 18.4 | 33 | 9.7 |
| Site | ||||||||
| Proximal | 0 | 0.0 | 505 | 49.1 | 557 | 49.1 | 176 | 51.2 |
| Distal | 0 | 0.0 | 523 | 50.9 | 578 | 50.9 | 168 | 48.8 |
| Age | ||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| 65.3 | 9.9 | 65.0 | 9.9 | 65.4 | 9.5 | 65.8 | 9.0 | |
miRNA processing
RNA (miRNA) was extracted from formalin-fixed paraffin embedded tissues. We assessed slides and tumor blocks that were prepared over the duration of the study prior to the time of miRNA isolation to determine their suitability. The study pathologist reviewed slides to delineate tumor and non-tumor tissue. Cells were dissected from 1–4 sequential sections on aniline blue stained slides using an H&E slide for reference. Total RNA containing miRNA was extracted, isolated, and purified using the RecoverAll Total Nucleic Acid isolation kit (Ambion); RNA yields were determined using a NanoDrop spectrophotometer. 100 ng total RNA was labeled with Cy3 and hybridized to Agilent Human miRNA Microarray V19.0 and were scanned on an Agilent SureScan microarray scanner model G2600D. The Agilent Human microarray was generated using known miRNA sequence information compiled in the Sanger miRBase database v19.0. The microarray contains probes for 2006 unique human miRNAs, with one to four unique probes for each of the known miRNAs. The miRNA array contains 60,000 unique human sequences and averages 30 replicates per probe sequence. Data were extracted from the scanned image using Agilent Feature Extract software v.11.5.1.1. Data were required to pass stringent QC parameters established by Agilent that included tests for excessive background fluorescence, excessive variation among probe sequence replicates on the array, and measures of the total gene signal on the array to assess low signal. If samples failed to meet quality standards for any of these parameters, the sample was re-labeled, hybridized to arrays, and scanned. If a sample failed QC assessment a second time the sample was deemed to be of poor quality and the individual was excluded from down-stream analysis. To minimize differences that could be attributed to the array, amount of RNA, location on array, or other factors that could erroneously influence expression, total gene signal is normalized by multiplying each sample by a scaling factor stratified by tumor site. Within each tumor site, the scaling factor [14] (http://genespring-support.com/files/gs_12_6/GeneSpring-manual.pdf) is the median of the 75th percentiles of all the samples divided by the individual 75th percentile of each sample.
We refer to miRNAs using standard nomenclature used in the miRBase database [15]. In this notation, the first three letters signifies the organism, and these are followed by a unique number. The number is followed by a dash and number (i.e., −1) if more than one loci codes for the miRNA. A lettered suffix denotes closely related miRNAs. If two miRNAs are coded by the same precursor product then the minor product is assigned the suffix (*). If predominant/minor product status is not known then the suffix −5p and −3p are used to denote 5′ and 3′ arm respectively. One example would be, let-7 may be reported in the literature previously, however since then let-7 has been further delineated to several closely related mature sequences and genomic loci, including let-7a-3p, let-7a-5p, and let-7b-3p.
We previously tested the reliability of the Agilent Microarray over time, repeating eight tumor and five matched normal samples (for a total of 13 samples) that had be scanned over the course of the study, and found repeatability correlation coefficient of 0.98 [16]; previous comparison of Agilent miRNA expression for select miRNAs to that generated by qPCR showed 100% agreement in terms of both direction of change and fold change [17].
GWAS
Blood was drawn for participants who provided informed consent; these participants included those from the aforementioned study at the University of Utah, KPMRP, and in the Twin Cities metropolitan area in Minnesota [13]. GWAS data were obtained using Illumina HumanHap 550K, 610K as part of the GECCO study and has been described previously [18]. Imputation to HapMap2 Release 24 was performed using MACH which was imputed to HapMap Release 22 using BEAGLE. GWAS samples obtained from Utah and Minnesota are being uploaded in NCBI’s dbGaP (http://www.ncbi.nlm.nih.gov/gap) by the Fred Hutchinson’s Cancer Research Center.
Selection of miRNA-related SNPs
A literature search was conducted to identify all miRNA-related SNPs shown to be associated with susceptibility to any type of cancer and more specifically to colorectal cancer [9,19–51]. We excluded SNPs from consideration that failed Illumina quality measures or standard quality control procedures [52]. Specifically, SNPs were excluded if any of the following criteria were satisfied: i) Illumina GenTrain score < 0.6 or cluster separation < 0.4; ii) discordant genotype calls in any pair of duplicate study samples; iii) Mendelian error in either one of the HapMap QC trios or a small number of families identified in the BEACON data; iv) significant departure from Hardy-Weinberg Equilibrium (P<104).
Statistical Analysis
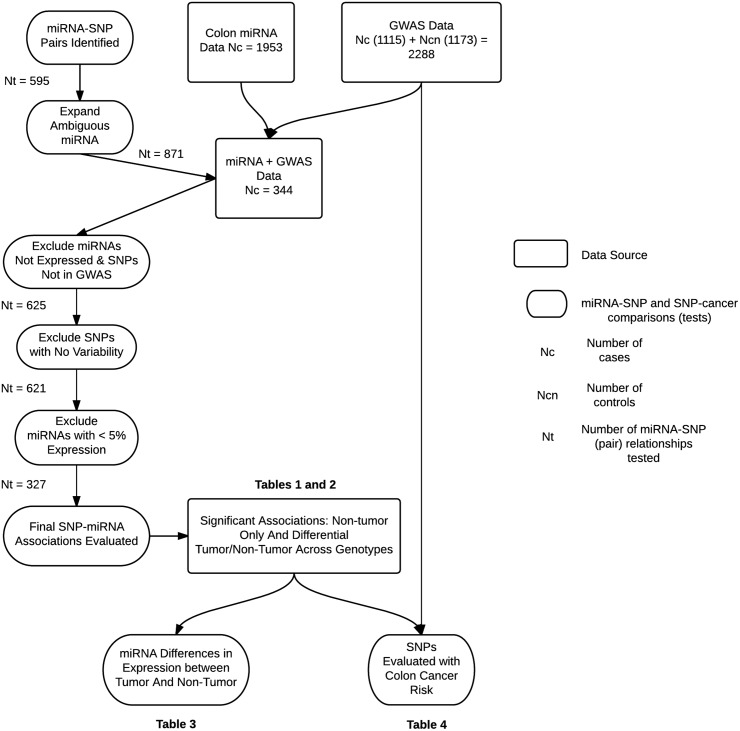
The study data flow is shown in Fig 1 and illustrates the sequence leading to the final analysis of miRNAs and their related SNP. Our sample consisted of 344 subjects who had both miRNA data and GWAS data and 1,115 cases and 1,173 controls with SNP data for colon risk assessment. We started with a total 559 miRNA-SNP pairs. These pairs were expanded to encompass specific miRNA terminology (i.e. 3p, 5p, etc) to yield 835 miRNA-SNP pairs for evaluation. From this number, we excluded miRNAs that were not expressed in at least one colon sample leaving 622 pairs. We further excluded any SNPs for which we did not have GWAS information, leaving 552 pairs. We also excluded SNPs that did not have variation (i.e. few with minor allele in our sample), leaving 548 pairs. Our final analysis included 327 pairs after we further excluded any miRNAs, and their associated SNP, that did not have expression in non-tumor colon tissue in at least 5% of the population.
Fig 1. Illustration of the study flow.
We utilized various bioinformatics tools. DbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) was utilized to identify SNP coordinates: all SNP chromosomal coordinates are from the assembly GRCh37 (GRCh37.p13). Ensembl’s Variant Effect Predictor (VEP) tool was utilized through the Ensembl GRCh37 site (http://grch37.ensembl.org/info/docs/tools/vep/index.html) to yield variant location and effect. VEP was utilized using all default settings. All SNPs were found in VEP. MiRNASNP v2.0, based on miRBase build 19 and dbSNP version 137 (http://bioinfo.life.hust.edu.cn/miRNASNP2/index.php) [53] was utilized to identify predicted lost and gained regulating miRNAs as a result of 3’ UTR changes in mRNA genes due to SNPs associated with colon cancer risk. These miRNAs were then also evaluated with their associated SNPs for significant alterations in miRNA expression.
We evaluated miRNA expression in both non-tumor colonic tissue and expression differences between tumor and non-tumor tissue to obtain a better understanding of how the SNPs relate to miRNA expression. Looking at non-tumor tissue allowed us to evaluate the effect of the SNP on miRNA expression. A SNP associated with miRNA expression across genotypes in non-tumor tissue could influence the baseline expression level of the miRNA but not influence cancer risk if it equally altered miRNA expression in tumors. Thus, changes in expression between tumor and non-tumor tissue were evaluated because of their potential importance for the carcinogenic process.
Statistical analysis was performed in three stages. First, we evaluated the miRNA expression for linear trend across genotypes adjusting for age, study center and sex using a linear regression model. P-values were generated from the bootstrap method [54] by creating a distribution of 10,000 β coefficients of the genotypes and evaluating H0: β = 0 vs. H1: β≠0 using R 3.1.2 (cran.r-project.org). All statistical analysis was performed using miRNA expression data that was both normalized and log2 transformed. The mean expressions reported in Tables 2, 3 and 4 are normalized but not log2 transformed in order to present the data in a more intuitive manner. All subsequent analysis was performed in SAS 9.4 4 (SAS Institute, Cary, NC). We report crude p-values, since each paired analysis was specifically hypothesized, as well as an adjusted p-value, taking into consideration all 327 comparisons made using the false discovery rate (FDR) [55]. Secondly, we evaluated miRNAs and SNPs from previously identified pairs to determine their association with colon cancer. In this analysis, we evaluated differential miRNA expression between tumor and non-tumor colonic tissue to determine if miRNA expression was differentially expressed in colon tumors. We used a paired t-test for this analysis and calculated P-values using 10,000 bootstrap samples. Finally, we evaluated colon cancer risk associated with SNPs that significantly influenced miRNA expression using logistic regression analysis. We report Odds Ratios (OR) and 95% Confidence Intervals (CI) that have been adjusted age, sex, and study center.
Table 2. Associations between miRNA expression and SNPs in miRNA genes and miRNA-target genes by genotype in non-tumor tissue (adjusted for age, sex and center).
| AA | AB | BB | p-values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene(s) | SNP | miRNA | N | Mean | % 0 Exp | N | Mean | % 0 Exp | N | Mean | % 0 Exp | Raw | FDR | Variant Type 3 |
| miRNA | ||||||||||||||
| CD22, MIR5196 | rs10406069, 0 = GG 1 = GA 2 = AA | hsa-miR-5196-5p | 218 | 64.14 | 0.00 | 105 | 60.93 | 0.00 | 21 | 61.38 | 0.00 | 0.029 | 0.52 | IN, DS, MS, 3' UTR, of CD22. NC, NCE of MIR5196. |
| MCM7, MIR106B, MIR25, MIR93 | rs1527423, 0 = AA 1 = AG 2 = GG | hsa-miR-25-3p | 86 | 14.51 | 9.30 | 182 | 12.40 | 10.44 | 76 | 10.41 | 21.05 | 0.004 | 0.49 | DS of COPS6. IN, NC, DS, NMD of MCM7. UP of MIR106B, MIR25, MIR93. |
| GNN, HSP90B1, MIR3652 | rs17797090, 0 = GG 1 = GA 2 = AA | hsa-miR-3652 | 288 | 159.68 | 0.00 | 56 | 145.86 | 0.00 | 0 | NA | NA | 0.032 | 0.52 | RR. TFB, 5' UTR, NMD, NC, NCE, IN of HSP90B1. NCE, NC of MIR3652. UP of RP11-642P15. |
| MIR146A | rs2910164, 0 = GG 1 = GC 2 = CC | hsa-miR-146a-5p | 197 | 8.24 | 32.49 | 129 | 8.29 | 31.01 | 18 | 6.61 | 5.56 | 0.038 | 0.52 | NC, NCE. |
| MIR1307, USMG5 | rs7911488, 0 = AA 1 = AG 2 = GG | hsa-miR-1307-3p | 154 | 13.03 | 0.00 | 153 | 13.03 | 0.65 | 37 | 11.27 | 0.00 | 0.019 | 0.52 | NC, NCE of MIR1307. UP of PDCD11. IN, 5' UTR of USMG5. |
| RPL17, RPL17-C18orf32, SNORD58A, SNORD58 | rs1943676, 0 = AA 1 = AG 2 = GG | hsa-miR-1539 | 147 | 4.24 | 36.73 | 167 | 4.10 | 53.89 | 30 | 4.02 | 56.67 | 0.004 | 0.49 | RR. UP of C18orf32. IN, NC, DS of MIR1539. SP, IN, NC, NCE, UP of RPL17. UP, DS of SNORD58A, SNORD58B, SNORD58C. DS of SRP72P1. |
| miRNA-Target | ||||||||||||||
| NRSN1 | rs1053047, 0 = GG 1 = GA 2 = AA | hsa-miR-143-5p | 92 | 3.96 | 94.57 | 175 | 3.39 | 92.57 | 77 | 5.36 | 84.42 | 0.029 | 0.52 | 3' UTR, DS, IN. |
| KIAA0423 (FAM179B)* | rs1053667, 0 = TT 1 = TC 2 = CC | hsa-miR-19b-3p | 305 | 11.19 | 14.10 | 39 | 13.44 | 28.21 | 0 | NA | NA | 0.028 | 0.52 | DS, 3' UTR, NMD. |
| SLC10A7 | rs1057560, 0 = GG 1 = GA 2 = AA | hsa-miR-25-3p | 97 | 11.54 | 14.43 | 171 | 12.39 | 13.45 | 76 | 14.09 | 7.89 | 0.039 | 0.52 | RR. 3' UTR, DS. |
| DNM2 | rs12232826, 0 = GG 1 = GT 2 = TT | hsa-miR-638 | 314 | 3980.05 | 0.00 | 30 | 4412.26 | 0.00 | 0 | NA | NA | 0.013 | 0.52 | IN. |
| CSK | rs1378940, 0 = AA 1 = AC 2 = CC | hsa-miR-4513 | 159 | 27.91 | 0.00 | 139 | 30.08 | 0.00 | 46 | 30.68 | 0.00 | 0.027 | 0.52 | RR. UP, IN, NC. |
| hsa-miR-1207-5p | 212 | 2043.96 | 0.00 | 117 | 1911.72 | 0.00 | 15 | 1699.90 | 0.00 | 0.016 | 0.52 | |||
| ICOS | rs1559931, 0 = GG 1 = GA 2 = AA | hsa-miR-196b-5p | 212 | 9.00 | 24.53 | 117 | 8.24 | 16.24 | 15 | 6.35 | 0.00 | 0.037 | 0.52 | RR. 3' UTR. |
| hsa-miR-2117 | 212 | 4.53 | 50.47 | 117 | 4.61 | 46.15 | 15 | 4.74 | 20.00 | 0.033 | 0.52 | |||
| LAD1 | rs16848494, 0 = CC 1 = CT 2 = TT | hsa-miR-143-5p | 328 | 4.28 | 90.85 | 16 | 0.00 | 100.00 | 0 | NA | NA | 0.027 | 0.52 | DS, 3' UTR, IN of LAD1. UP of TNNT2. |
| AFF1 | rs17703261, 0 = AA 1 = AT 2 = TT | hsa-miR-648 | 211 | 18.23 | 0.00 | 121 | 17.48 | 0.00 | 12 | 14.98 | 0.00 | 0.022 | 0.52 | 3' UTR, DS. |
| hsa-miR-1207-5p | 212 | 2043.96 | 0.00 | 117 | 1911.72 | 0.00 | 15 | 1699.90 | 0.00 | 0.020 | 0.52 | |||
| ICOS | rs4404254, 0 = TT 1 = TC 2 = CC | hsa-miR-196b-5p | 212 | 9.00 | 24.53 | 117 | 8.24 | 16.24 | 15 | 6.35 | 0.00 | 0.034 | 0.52 | 3' UTR. |
| hsa-miR-2117 | 212 | 4.53 | 50.47 | 117 | 4.61 | 46.15 | 15 | 4.74 | 20.00 | 0.035 | 0.52 | |||
| NKAIN4, FLJ16779 | rs720607, 0 = GG 1 = GA 2 = AA | hsa-miR-3196 | 112 | 1415.24 | 0.00 | 163 | 1326.52 | 0.00 | 69 | 1269.88 | 0.00 | 0.027 | 0.52 | RR. IN. |
| CD80 | rs7628626, 0 = CC 1 = CA 2 = AA | hsa-miR-2117 | 233 | 4.67 | 42.06 | 98 | 4.40 | 62.24 | 13 | 3.69 | 38.46 | 0.010 | 0.52 | RR. DS, 3' UTR, of CD80. DS of TIMMDC1. |
| FGF2 | rs7683093, 0 = CC 1 = CG 2 = GG | hsa-miR-92b-3p | 249 | 1.34 | 93.17 | 84 | 2.01 | 82.14 | 11 | 0.73 | 81.82 | 0.008 | 0.52 | RR. DS, 3' UTR of FGF2. IN, NMD, DS, US of NUDT6. |
| BRCA1 | rs8176318, 0 = CC 1 = CA 2 = AA | hsa-miR-525-5p | 165 | 3.20 | 46.67 | 130 | 3.16 | 51.54 | 49 | 3.85 | 69.39 | 0.035 | 0.52 | 3' UTR, DS. |
| PARP1 | rs8679, 0 = AA 1 = AG 2 = GG | hsa-miR-630 | 212 | 364.38 | 0.00 | 106 | 392.44 | 0.00 | 26 | 541.85 | 0.00 | 0.013 | 0.52 | 3' UTR, DS. |
| SELS (VIMP)* | rs9874, 0 = TT 1 = TC 2 = CC | hsa-miR-181a-5p | 268 | 25.24 | 0.00 | 69 | 29.71 | 0.00 | 7 | 25.02 | 0.00 | 0.027 | 0.52 | 3' UTR, DS. |
| MCM7, AP4M1 | rs999885, 0 = AA 1 = AG 2 = GG | hsa-miR-25-3p | 88 | 14.45 | 9.09 | 180 | 12.36 | 10.00 | 76 | 10.51 | 22.37 | 0.003 | 0.49 | IN, NMD, UP, NC, DS of AP4M1. UP of MCM7. DS of TAF6. |
1Related SNPs are those in linkage disequilibrium to SNPs within the dataset.
2EX: Exon; DS: Downstream; IN: intron; MS: Missense; NC: Non-coding; NCE: Non-coding exon; NMD: Nonsense-mediated decay; RR: Regulatory region; SN: Synonymous; SPR: Splice Region; TFB: Transcription Factor Binding site; US: Upstream.
3Variant Analysis was performed with Ensembl's VEP (GRCh37.p13). Multiple type of variants are listed for a given gene when these variant occur in different transcripts.
4This SNP has now merged into a new SNP (GRCh38).
5For miRNA Gene Regions this column has the miRNA with the SNP; for miRNA-target genes this column has the associated miRNA for the mRNA with the SNP.
6Association seen in literature.
*Names in parentheses are alternatives.
Table 3. Associations of miRNA expression by SNPs in miRNA genes and miRNA-target genes between tumor and non-tumor tissues (adjusted for age, sex, and center).
| AA | AB | BB | p-values | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene(s) | SNP | miRNA | N | Mean | T % 0 Exp | N % 0 Exp | N | Mean | T % 0 Exp | N % 0 Exp | N | Mean | T % 0 Exp | N % 0 Exp | Raw | FDR | Variant Type 3 |
| miRNA | |||||||||||||||||
| MIR605, PRKG1 | rs2043556, 0 = TT 1 = TC 2 = CC | hsa-miR-605 | 259 | -1.07 | 79.92 | 51.35 | 110 | -0.36 | 75.45 | 61.82 | 14 | 0.53 | 64.29 | 57.14 | 0.007 | 0.36 | RR. NC, NCE of MIR605. IN of PRKG1. US of RP11-539E19.2. |
| MIR548AP | rs2344843, 0 = AA 1 = AG 2 = GG | hsa-miR-548ap-5p | 163 | 0.39 | 96.93 | 88.96 | 167 | -0.42 | 98.80 | 85.03 | 53 | -0.66 | 100.00 | 84.91 | 0.029 | 0.54 | DS of MIR548AP. |
| MIR146A | rs2910164, 0 = GG 1 = GC 2 = CC | hsa-miR-146a-5p | 218 | 3.62 | 26.61 | 27.52 | 146 | -0.21 | 43.15 | 26.71 | 19 | 2.96 | 31.58 | 5.26 | 0.002 | 0.30 | NC, NCE. |
| MIR143 | rs353292, 0 = GG 1 = GA 2 = AA | hsa-miR-143-5p | 105 | 4.46 | 88.57 | 96.19 | 204 | -0.28 | 91.18 | 87.25 | 74 | -0.22 | 94.59 | 87.84 | 0.019 | 0.54 | US of MIR143. DS, NC, NCE of |
| hsa-miR-145-3p | 105 | 2.95 | 95.24 | 98.10 | 204 | -0.44 | 96.57 | 93.14 | 74 | -2.22 | 100.00 | 91.89 | 0.008 | 0.36 | MIR143HG. UP of MIR145. | ||
| MIR182 | rs76481776 (related SNP) 1 | hsa-miR-182-5p | 311 | 2.16 | 88.10 | 100.00 | 72 | 3.22 | 97.22 | 100.00 | 0 | NA | NA | NA | 0.024 | 0.54 | INTERGENIC VARIANT. |
| rs2693737, 0 = AA 1 = AG 2 = GG | |||||||||||||||||
| MIR143 | rs353293, 0 = CC 1 = CT 2 = TT | hsa-miR-143-5p | 106 | 4.46 | 88.68 | 96.23 | 203 | -0.28 | 91.13 | 87.19 | 74 | -0.22 | 94.59 | 87.84 | 0.014 | 0.47 | US of MIR143. DS, NC, NCE, IN of |
| hsa-miR-145-3p | 106 | 2.95 | 95.28 | 98.11 | 203 | -0.44 | 96.55 | 93.10 | 74 | -2.22 | 100.00 | 91.89 | 0.008 | 0.36 | MIR143HG. UP of MIR145. | ||
| miRNA-Target | |||||||||||||||||
| CAMK1D | rs10508445, 0 = AA 1 = AG 2 = GG | hsa-miR-4481 | 112 | -9.41 | 0.00 | 0.00 | 193 | -4.02 | 0.00 | 0.00 | 78 | 0.84 | 0.00 | 0.00 | 0.022 | 0.54 | IN, NC. |
| SLC10A7 | rs1057560, 0 = GG 1 = GA 2 = AA | hsa-miR-25-3p | 104 | 13.75 | 3.85 | 13.46 | 189 | 14.00 | 4.23 | 11.11 | 90 | 12.33 | 10.00 | 6.67 | 0.007 | 0.36 | RR. 3' UTR, DS. |
| GRINL1A | rs1062707, 0 = TT 1 = TC 2 = CC | hsa-miR-424-3p | 226 | 13.46 | 0.00 | 0.44 | 131 | 9.62 | 0.00 | 0.00 | 26 | 7.90 | 0.00 | 0.00 | 0.025 | 0.54 | IN, NC, DS, MND, 3' UTR, SN of GCOM1 7 . 3' UTR, NMD, SN, IN, |
| hsa-miR-424-5p | 226 | 4.37 | 66.37 | 89.82 | 131 | 2.65 | 68.70 | 87.02 | 26 | -0.05 | 76.92 | 88.46 | 0.022 | 0.54 | NC, NCE of POLR2M7. | ||
| KSR2 | rs11068503, 0 = TT 1 = TC 2 = CC | hsa-miR-149-3p | 126 | 1.33 | 0.00 | 0.00 | 178 | -1.79 | 0.00 | 0.00 | 79 | -4.84 | 0.00 | 0.00 | 0.007 | 0.36 | 3' UTR, DS. |
| EGFL7, LOC101928-612 | rs1332793, 0 = TT 1 = TC 2 = CC | hsa-miR-126-3p | 151 | 0.87 | 6.62 | 9.27 | 183 | -1.63 | 10.93 | 5.46 | 49 | 0.07 | 10.20 | 4.08 | 0.011 | 0.39 | US, IN of EGFL7. US of RP11-251M1.1. |
| VTCN1 | rs13505, 0 = AA 1 = AC 2 = CC | hsa-miR-196a-5p | 207 | 1.66 | 42.03 | 25.12 | 147 | 2.52 | 40.82 | 27.89 | 29 | 3.51 | 17.24 | 27.59 | 0.036 | 0.64 | RR. 3' UTR, DS. |
| CD86 | rs17281995, 0 = GG 1 = GC 2 = CC | hsa-miR-212-3p | 274 | 0.35 | 3.65 | 4.74 | 100 | -0.53 | 9.00 | 8.00 | 9 | -2.73 | 22.22 | 0.00 | 0.047 | 0.73 | 3’ UTR, DS. |
| IL6ST | rs2228043, 0 = GG 1 = GC 2 = CC | hsa-miR-221-3p | 298 | 8.21 | 15.10 | 35.57 | 85 | 11.62 | 12.94 | 45.88 | 0 | NA | NA | NA | 0.027 | 0.54 | 3' UTR, US, NC, NCE, MS, NMD, IN. |
| WWP2 | rs2270841, 0 = CC 1 = CT 2 = TT | hsa-miR-140-3p | 199 | -3.36 | 24.12 | 12.56 | 154 | -1.51 | 18.83 | 11.69 | 30 | -0.79 | 16.67 | 13.33 | 0.025 | 0.54 | DS of MIR140. SN, US, NC, NCE, DS of WWP2. |
| TAF1C | rs2288024, 0 = TT 1 = TC 2 = CC | hsa-miR-103a-3p | 358 | 14.46 | 3.07 | 0.84 | 25 | 22.63 | 0.00 | 0.00 | 0 | NA | NA | NA | 0.009 | 0.39 | 3' UTR, DS, IN, NC, NCE of DNAAF1. DS, 3' UTR, NC, NCE of TAF1C. |
| TNFRSF4 | rs2298209, 0 = GG 1 = GC 2 = CC | hsa-miR-200b-3p | 374 | 14.66 | 2.14 | 0.27 | 9 | 57.69 | 0.00 | 0.00 | 0 | NA | NA | NA | 0.043 | 0.70 | DS, NC, NCE, 3’ UTR of TNFRSF4. UP of TNFRSF18. |
| IL22RA2 | rs276466, 0 = AA 1 = AG 2 = GG | hsa-miR-30c-2-3p | 212 | 0.47 | 9.43 | 13.21 | 149 | -0.31 | 11.41 | 8.72 | 22 | -1.07 | 22.73 | 4.55 | 0.001 | 0.22 | 3' UTR. |
| ZNF396 | rs2909339, 0 = GG 1 = GA 2 = AA | hsa-miR-145-3p | 267 | -0.67 | 97.75 | 93.26 | 105 | 1.84 | 94.29 | 97.14 | 11 | -0.22 | 100.00 | 90.91 | 0.038 | 0.64 | DS, 3' UTR. |
| PRKAR1A | rs8905, 0 = TT 1 = TG 2 = GG | hsa-miR-214-3p | 285 | 5.60 | 15.79 | 13.33 | 87 | 6.97 | 6.90 | 17.24 | 11 | 9.22 | 9.09 | 27.27 | 0.006 | 0.36 | DS of FAM20. 3' UTR, DS, IN, NMD of PRKR1A. |
| ZNF257 | rs9304994, 0 = AA 1 = AG 2 = GG | hsa-miR-557 | 135 | 8.03 | 0.00 | 0.00 | 183 | -0.62 | 0.00 | 0.00 | 65 | -2.80 | 0.00 | 0.00 | 0.029 | 0.54 | 3' UTR, NMD, DS. |
1Related SNPs are those in linkage disequilibrium to SNPs within the dataset.
2EX: Exon; DS: Downstream; IN: intron; MS: Missense; NC: Non-coding; NCE: Non-coding exon; NMD: Nonsense-mediated decay; RR: Regulatory region; SN: Synonymous; SPR: Splice Region; TFB: Transcription Factor Binding site; US: Upstream.
3Variant Analysis was performed with Ensembl's VEP (GRCh37.p13). Multiple type of variants are listed for a given gene when these variant occur in different transcripts. Some additional genes not analyzed are listed by VEP.
4This SNP has now merged into a new SNP (GRCh38).
5For miRNA Gene Regions this column has the miRNA with the SNP; for miRNA-target genes this column has the associated miRNA for the mRNA with the SNP.
6Association seen in literature.
7Related genes/proteins of gene analyzed.
Table 4. MiRNAs significantly expressed between tumor and non-tumor colonic tissue that were identified with non-tumor and tumor/non-tumor SNP associations.
| Tumor | Normal | |||||
|---|---|---|---|---|---|---|
| miRNA | Associated Gene(s) | Mean | % 0 Expr. | Mean | % 0 Expr. | p-value |
| Non-tumor | ||||||
| hsa-miR-1207-5p | ICOS | 1832.06 | 0 | 2002.56 | 0 | <0.01 |
| hsa-miR-1307-3p | MIR1307, USMG5 | 11.04 | 0.4 | 12.87 | 0.1 | <0.01 |
| hsa-miR-1539 | RPL17, RPL17-C18orf32, SNORD58A, SNORD58 | 3.21 | 48.1 | 3.09 | 41.2 | 0.43 |
| hsa-miR-181a-5p | SELS | 35.4 | 0.3 | 25.35 | 0.2 | <0.01 |
| hsa-miR-196b-5p | ICOS | 15.72 | 23.2 | 6.04 | 22.3 | <0.01 |
| hsa-miR-19b-3p | KIAA0423 (FAM179B) | 23.81 | 8.8 | 8.19 | 17.4 | <0.01 |
| hsa-miR-2117 | ICOS, CD80 | 1.47 | 75.2 | 3.68 | 45.5 | <0.01 |
| hsa-miR-3196 | NKAIN4, FLJ16779 | 1207.4 | 0 | 1373.03 | 0 | <0.01 |
| hsa-miR-3652 | GNN, HSP90B1, MIR3652 | 149.06 | 0 | 158.9 | 0 | <0.01 |
| hsa-miR-4513 | CSK | 28.51 | 0 | 29.25 | 0 | <0.01 |
| hsa-miR-5196-5p | CD22, MIR5196 | 73.87 | 0 | 64.64 | 0 | <0.01 |
| hsa-miR-525-5p | BRCA1 | 1.87 | 56.1 | 2.38 | 46.6 | <0.01 |
| hsa-miR-630 | PARP1 | 342.94 | 0 | 399.24 | 0 | <0.01 |
| hsa-miR-638 | DNM2 | 3584.65 | 0 | 4091.42 | 0 | <0.01 |
| hsa-miR-648 | AFF1 | 16.47 | 0 | 18.3 | 0 | <0.01 |
| hsa-miR-92b-3p | FGF2 | 0.99 | 89.1 | 0.6 | 89.1 | <0.01 |
| Tumor/Non-Tumor Differential | ||||||
| hsa-miR-103a-3p | TAF1C | 60.46 | 1.9 | 41.75 | 1 | <0.01 |
| hsa-miR-126-3p | EGFL7, LOC101928612 | 14.4 | 9.9 | 14.36 | 8.3 | 0.32 |
| hsa-miR-140-3p | WWP2 | 5.29 | 21.6 | 7.35 | 15.3 | <0.01 |
| hsa-miR-145-3p | MIR143, ZNF396 | 1.05 | 98.1 | 1.67 | 94.2 | 0.04 |
| hsa-miR-149-3p | KSR2 | 33.69 | 0 | 35.89 | 0 | <0.01 |
| hsa-miR-182-5p | MIR182 | 2.15 | 91.5 | 0.01 | 99.9 | <0.01 |
| hsa-miR-200b-3p | TNFRSF4 | 143.75 | 1.6 | 120.84 | 0.7 | 0.44 |
| hsa-miR-212-3p | CD86 | 9.67 | 5 | 9.55 | 5 | 0.52 |
| hsa-miR-214-3p | PRKAR1A | 11.35 | 12.4 | 5.13 | 17 | <0.01 |
| hsa-miR-221-3p | IL6ST | 11.57 | 17.6 | 2.62 | 40.3 | <0.01 |
| hsa-miR-30c-2-3p | IL22RA2 | 4.84 | 11.3 | 4.92 | 10.7 | 0.27 |
| hsa-miR-424-3p | GRINL1A | 36.22 | 0.2 | 24.48 | 0.1 | <0.01 |
| hsa-miR-424-5p | GRINL1A | 3.97 | 69.2 | 0.53 | 89.5 | <0.01 |
| hsa-miR-4481 | CAMK1D | 78.1 | 0 | 84.13 | 0 | <0.01 |
| hsa-miR-548ap-5p | MIR548AP | 0.83 | 97.8 | 0.96 | 86.5 | <0.01 |
| hsa-miR-557 | ZNF257 | 74.59 | 0 | 74.44 | 0 | 0.34 |
| hsa-miR-605 | MIR605, PRKG1 | 1.54 | 79.2 | 2.03 | 59.1 | <0.01 |
| hsa-miR-196a-5p | VTCN1 | 6.27 | 39 | 3.66 | 31.9 | <0.01 |
| Both Tumor/Non-tumor and Non-tumor | ||||||
| hsa-miR-143-5p | MIR143, NRSN1, LAD1 | 2.46 | 92.7 | 2.35 | 90.5 | 0.81 |
| hsa-miR-146a-5p | MIR146A | 8.45 | 33.1 | 5.69 | 28.3 | 0.01 |
| hsa-miR-25-3p | MCM7, MIR106B, MIR25, MIR93, MCM7, AP4M1, SLC10A7 | 24.17 | 5.4 | 9.73 | 12.4 | <0.01 |
Results
Prior to adjustment for multiple comparisons, six SNPs within miRNA genes and 16 SNPs within miRNA-target genes were associated with differentially expressed miRNAs in non-tumor colonic tissue by genotype (Table 2). Twelve of the 16 miRNA-target gene SNPs were within the 3’ UTR. After adjustment for multiple comparisons, none of the identified associations remained statistically significant.
Six SNPs within miRNA genes and 15 SNPs within miRNA-targets genes were identified as being associated with differential miRNA levels between tumor and non-tumor tissue (Table 3); 12 of the 15 SNPs within miRNA-target genes occurred in the 3’ UTR. As was seen with the non-tumor associations, these miRNA-SNP associations were not significant after adjustment for multiple comparisons.
Further evaluation of miRNAs that were significantly influenced by SNPs prior to adjustment for multiple comparison in either non-tumor tissue or when comparing differences between tumor/non-tumor (see Tables 1 and 2 respectively) showed that all but seven miRNAs were significantly differentially expressed between colon tumor and non-tumor tissue (Table 4). Of those miRNAs associated with SNPs in non-tumor tissue (Table 2), nine miRNA were statistically significantly downregulated in tumor tissue and six were upregulated. Evaluation of miRNAs associated with SNPs in Table 3 (differentially expressed across genotype between tumor/non-tumor tissue types) showed that six were statistically significantly upregulated and six were downregulated in tumor tissue relative to non-tumor tissue. Two of the three miRNAs that were seen in both Tables 1 and 2, hsa-miR-146a-5p and hsa-miR-25-3p, were both statistically significantly upregulated in tumor tissue relative to non-tumor tissue (Table 4).
Evaluation of associations between colon cancer and SNPs that were significantly associated with miRNA expression levels showed two SNPs that were also significantly associated with increased risk of colon cancer (Table 5). Within the 3’ UTR of BRCA1, the homozygote variant genotype of rs8176318 was associated with increased risk of colon cancer (ORAA 1.34 95% CI 1.01, 1.78. The associated miRNA, hsa-miR-525-5p, was not expressed in a large portion of the population with the homozygous-rare genotype; therefore, differences in expression across genotypes could be attributable to the reduction in the percent expressing. A second SNP, rs8905, within the 3’ UTR of PRKAR1A was associated with over a twofold increased risk of colon cancer (ORGG 2.31 95% CI 1.11, 4.77). A third SNP, rs276466, was associated with increased risk of colon cancer with the heterozygote genotype (ORAG 1.21 95% CI 1.02, 1.44) only and not the homozygote rare genotype and therefore may be a spurious association.
Table 5. Associations between SNPs associated with miRNAs in non-tumor and tumor/non-tumor differential expression and risk of colon cancer.
| AA | AB | BB | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Associated Gene(s) | Controls | Cases | Controls | Cases | OR | (95% CI) | Controls | Cases | OR | (95% CI) |
| Non-tumor | |||||||||||
| rs10406069, 0 = GG 1 = GA 2 = AA | CD22, MIR5196 | 767 | 737 | 350 | 330 | 1 | (0.80, 1.16) | 56 | 48 | 0.9 | (0.57, 1.28) |
| rs1053047, 0 = GG 1 = GA 2 = AA | NRSN1 | 321 | 323 | 591 | 544 | 0.9 | (0.75, 1.11) | 261 | 248 | 0.9 | (0.74, 1.19) |
| rs1053667, 0 = TT 1 = TC 2 = CC | KIAA0423 | 1058 | 998 | 115 | 117 | 1.1 | (0.83, 1.43) | ||||
| rs12232826, 0 = GG 1 = GT 2 = TT | DNM2 | 1087 | 1026 | 86 | 89 | 1.1 | (0.81, 1.50) | ||||
| rs1378940, 0 = AA 1 = AC 2 = CC | CSK | 502 | 501 | 520 | 476 | 0.9 | (0.77, 1.09) | 151 | 138 | 0.9 | (0.70, 1.18) |
| rs1527423, 0 = AA 1 = AG 2 = GG | MCM7, MIR106B, MIR25, MIR93 | 332 | 299 | 574 | 557 | 1.1 | (0.89, 1.31) | 267 | 259 | 1.1 | (0.86, 1.37) |
| rs1559931, 0 = GG 1 = GA 2 = AA | ICOS | 659 | 652 | 441 | 395 | 0.9 | (0.76, 1.07) | 73 | 68 | 0.9 | (0.65, 1.31) |
| rs16848494, 0 = CC 1 = CT 2 = TT | LAD1 | 1125 | 1063 | 48 | 52 | 1.1 | (0.74, 1.65) | ||||
| rs17703261, 0 = AA 1 = AT 2 = TT | AFF1 | 776 | 711 | 353 | 369 | 1.1 | (0.94, 1.35) | 44 | 35 | 0.8 | (0.53, 1.33) |
| rs17797090, 0 = GG 1 = GA 2 = AA | GNN, HSP90B1, MIR3652 | 981 | 939 | 192 | 176 | 0.9 | (0.75, 1.18) | ||||
| rs1943676, 0 = AA 1 = AG 2 = GG | RPL17, RPL17-C18orf32, SNORD58A, SNORD58 | 506 | 499 | 526 | 498 | 1 | (0.80, 1.14) | 141 | 118 | 0.9 | (0.66, 1.14) |
| rs4404254, 0 = TT 1 = TC 2 = CC | ICOS | 658 | 652 | 442 | 395 | 0.9 | (0.75, 1.07) | 73 | 68 | 0.9 | (0.65, 1.31) |
| rs720607, 0 = GG 1 = GA 2 = AA | NKAIN4, FLJ16779 | 362 | 364 | 574 | 547 | 1 | (0.79, 1.15) | 237 | 204 | 0.9 | (0.67, 1.08) |
| rs7628626, 0 = CC 1 = CA 2 = AA | CD80 | 805 | 726 | 336 | 351 | 1.2 | (0.97, 1.40) | 32 | 38 | 1.3 | (0.80, 2.10) |
| rs7683093, 0 = CC 1 = CG 2 = GG | FGF2 | 856 | 823 | 291 | 265 | 1 | (0.78, 1.15) | 26 | 27 | 1.1 | (0.61, 1.84) |
| rs7911488, 0 = AA 1 = AG 2 = GG | MIR1307, USMG5 | 533 | 504 | 524 | 491 | 1 | (0.83, 1.17) | 116 | 120 | 1.1 | (0.83, 1.46) |
| rs8176318, 0 = CC 1 = CA 2 = AA | BRCA1 | 560 | 484 | 504 | 504 | 1.2 | (0.96, 1.36) | 109 | 127 | 1.3 | (1.01, 1.78) |
| rs8679, 0 = AA 1 = AG 2 = GG | PARP1 | 704 | 684 | 414 | 373 | 0.9 | (0.78, 1.11) | 55 | 58 | 1.1 | (0.73, 1.58) |
| rs9874, 0 = TT 1 = TC 2 = CC | SELS | 885 | 837 | 265 | 255 | 1 | (0.83, 1.24) | 23 | 23 | 1.1 | (0.60, 1.95) |
| rs999885, 0 = AA 1 = AG 2 = GG | MCM7, AP4M1 | 327 | 298 | 577 | 555 | 1.1 | (0.87, 1.28) | 269 | 262 | 1.1 | (0.86, 1.36) |
| Tumor/Non-tumor | |||||||||||
| rs10508445, 0 = AA 1 = AG 2 = GG | CAMK1D | 307 | 303 | 566 | 555 | 1 | (0.82, 1.22) | 300 | 257 | 0.9 | (0.70, 1.11) |
| rs1062707, 0 = TT 1 = TC 2 = CC | GRINL1A | 680 | 671 | 414 | 379 | 0.9 | (0.78, 1.10) | 79 | 65 | 0.8 | (0.58, 1.16) |
| rs11068503, 0 = TT 1 = TC 2 = CC | KSR2 | 366 | 339 | 555 | 544 | 1.1 | (0.88, 1.29) | 252 | 232 | 1 | (0.80, 1.27) |
| rs1332793, 0 = TT 1 = TC 2 = CC | EGFL7, LOC101928612 | 462 | 433 | 542 | 522 | 1 | (0.86, 1.23) | 169 | 160 | 1 | (0.77, 1.28) |
| rs13505, 0 = AA 1 = AC 2 = CC | VTCN1 | 623 | 609 | 450 | 427 | 1 | (0.81, 1.15) | 100 | 79 | 0.8 | (0.59, 1.12) |
| rs17281995, 0 = GG 1 = GC 2 = CC | CD86 | 848 | 809 | 304 | 277 | 1 | (0.80, 1.17) | 21 | 29 | 1.4 | (0.79, 2.48) |
| rs2043556, 0 = TT 1 = TC 2 = CC | MIR605, PRKG1 | 737 | 707 | 374 | 366 | 1 | (0.84, 1.21) | 62 | 42 | 0.7 | (0.48, 1.08) |
| rs2228043, 0 = GG 1 = GC 2 = CC | IL6ST | 898 | 841 | 275 | 274 | 1.1 | (0.88, 1.29) | ||||
| rs2270841, 0 = CC 1 = CT 2 = TT | WWP2 | 637 | 577 | 454 | 445 | 1.1 | (0.91, 1.28) | 82 | 93 | 1.3 | (0.92, 1.75) |
| rs2288024, 0 = TT 1 = TC 2 = CC | TAF1C | 1080 | 1046 | 93 | 69 | 0.8 | (0.55, 1.05) | ||||
| rs2298209, 0 = GG 1 = GC 2 = CC | TNFRSF4 | 1132 | 1088 | 41 | 27 | 0.7 | (0.41, 1.10) | ||||
| rs2344843, 0 = AA 1 = AG 2 = GG | MIR548AP | 472 | 467 | 529 | 499 | 1 | (0.81, 1.16) | 172 | 149 | 0.9 | (0.67, 1.12) |
| rs2693737, 0 = AA 1 = AG 2 = GG | MIR182 | 968 | 913 | 205 | 202 | 1 | (0.84, 1.29) | ||||
| rs276466, 0 = AA 1 = AG 2 = GG | IL22RA2 | 720 | 642 | 385 | 419 | 1.2 | (1.02, 1.44) | 68 | 54 | 0.9 | (0.61, 1.29) |
| rs2909339, 0 = GG 1 = GA 2 = AA | ZNF396 | 871 | 792 | 278 | 295 | 1.2 | (0.97, 1.42) | 24 | 28 | 1.3 | (0.73, 2.23) |
| rs353292, 0 = GG 1 = GA 2 = AA | MIR143 | 341 | 309 | 609 | 573 | 1.1 | (0.86, 1.27) | 223 | 233 | 1.2 | (0.92, 1.49) |
| rs353293, 0 = CC 1 = CT 2 = TT | MIR143 | 340 | 310 | 610 | 572 | 1 | (0.85, 1.25) | 223 | 233 | 1.2 | (0.91, 1.48) |
| rs8905, 0 = TT 1 = TG 2 = GG | PRKAR1A | 928 | 836 | 234 | 256 | 1.2 | (0.98, 1.46) | 11 | 23 | 2.3 | (1.11, 4.77) |
| rs9304994, 0 = AA 1 = AG 2 = GG | ZNF257 | 380 | 386 | 599 | 534 | 0.9 | (0.73, 1.05) | 194 | 195 | 1 | (0.78, 1.27) |
| Both Tumor/Non-tumor and Non-tumor | |||||||||||
| rs1057560, 0 = GG 1 = GA 2 = AA | SLC10A7 | 316 | 308 | 590 | 555 | 1 | (0.79, 1.17) | 267 | 252 | 1 | (0.77, 1.23) |
| rs2910164, 0 = GG 1 = GC 2 = CC | MIR146A | 665 | 652 | 439 | 405 | 0.9 | (0.78, 1.11) | 69 | 58 | 0.9 | (0.59, 1.23) |
1Referent group is AA (homozygote common genotype); AB (heterozygote genotype); BB (homozygote rare genotype).
2Odds Ratios (OR) and 95% Confidence Intervals (CI) adjusted for age, sex and center.
Bolded items are significantly associated with risk of colon cancer.
Discussion
It has been suggested that SNPs associated with risk of colon cancer could be functioning through their impact on miRNA regulation [9]. In this study, we investigated SNPs within miRNA gene and miRNA-target gene regions previously reported in the literature as associated with cancer, to evaluate if these SNPs influence miRNA expression and alter colon cancer risk. Of the 327 SNP/miRNA pairs evaluated, 22 SNPs were associated with significant (P<0.05) differences in miRNA expression in non-tumor tissue by genotype, and 21 SNPs were associated with significant differential miRNA expression between tumor and non-tumor tissue by genotype. Evaluation of these SNPs with colon cancer showed only two SNPs were significantly associated with colon cancer risk. This suggests that the majority of the hypothesized associations are not supported when evaluated directly with miRNA data.
Two SNPs in miRNA-target genes were identified as associated with an increased risk of colon cancer; these SNPs also were associated with statistically significant mean differential miRNA expression across genotypes. Of these, rs8176318, located in the 3’ UTR of BRCA1 and predicted to be a binding site for hsa-miR-525-5p when the G allele (or C in our data) is present [21], was associated with a linear decrease in hsa-miR-525-5p expression in non-tumor tissue when comparing homozygote common (CC) to homozygote rare genotypes (AA). MiRNA hsa-miR-525-5p also was downregulated in tumor as compared to non-tumor tissue (p = 0.0044) and rs8176318 was significantly associated with an increase in risk of colon cancer (ORAA 1.34 95% CI 1.01, 1.78). This variant has been reported as being associated with decreased BRCA1 expression, increased breast cancer risk, and greater likelihood of having stage IV breast cancer [56]. Expression levels of this miRNA are somewhat low, and may therefore less precise, however, together these findings could support the claim that the SNP rs8176318 contributes to the incidence and progression of some breast and colon cancers through altered miRNA regulation within these tissues.
We also identified another SNP, rs8905, in the 3’ UTR of PRKAR1A, that was significantly associated with decreased expression in tumors relative to non-tumor tissue of the miRNA that targets PRKAR1A, hsa-miR-214-3p, in subjects of the homozygote common (TT) genotype as compared to the heterozygote and homozygote rare (TG, GG respectively) genotypes. Additionally, a much higher percentage of non-tumor tissue did not express hsa-miR-214-3p in the heterozygote genotype as compared to the TT (common) genotype, and even less expression was observed for the homozygote rare genotype. Hsa-miR-214-3p was statistically significantly upregulated in tumor versus non-tumor tissues, and the SNP rs8905 was seen to significantly increase the risk of colon cancer (ORGG 2.31 95% CI 1.11, 4.77). In this instance, it is likely that the alteration of the miRNA expression is caused by the SNP and this directly contributes to colon tumor risk.
Our data did support some of the literature in terms of SNP and miRNA associations. In a recent study, two SNPs within the promoter region of hsa-miR-143/145, rs353292 and rs353293, were significantly associated with an increased risk of colorectal cancer (CRC) in a predominately Chinese population [9]. As these SNPs are in the promoter region of miRNAs, Li et al. proposed that the mechanism by which these SNPs increased CRC risk was through the expression of hsa-miR143/145 [9]. In our investigation, both rs353292 and rs353293 were associated with differential expression of hsa-miR-145-3p and hsa-miR-143-5p between genotypes between tumor and non-tumor tissues (Table 3). In both instances, miRNA expression was greater among individuals with two copies of the common allele (i.e. homozygous common genotype) in non-tumor tissue. Having a copy of the variant allele resulted in a decrease in miRNA expression in tumor tissue compared to non-tumor tissue. Of these two, hsa-miR-145-3p was significantly differentially expressed between tumor and non-tumor tissue, however neither of these SNPs were significantly associated with colon cancer.
Dikaiakos et al. [44] reported higher incidences of CRC among individuals with the CC genotype and with the C allele of rs2910164, which is in the precursor miRNA region of hsa-miR-146a, and suggested that this increased risk is attributed to alterations in the miRNA expression and alterations to its structure [44]. We observed that miR-146a is differentially expressed across genotypes of rs2910164 for non-tumor tissue as well as between tumor and non-tumor tissues. Additionally, expression of hsa-miR-146a-5p was statistically significantly upregulated in tumor tissue compared to non-tumor tissue, suggesting that this SNP may impact miRNA expression. However, unlike Dikaiakos, we did not observe that this SNP was associated with colon cancer.
In a review by Ryan et al. [20], AFF1 rs17703261 and KIAA0423 (or FAM179B) rs1053667, were significantly associated with non-specified cancer risk. In that work, AFF1 rs17703261 was inversely associated with cancer risk (OR for the A/T was 0.34 95% CI 0.20, 0.58) and KIAA0423 rs1053667 was directly associated with cancer risk (OR for the C/T was 3.29 95% CI 1.72, 6.32) [20]. The miRNAs that target AFF1 are hsa-miRs-19a, -19b, -585, and -648. In our study, we identified a significant decrease in expression of hsa-miR-648 in the homozygote rare (TT) genotype in non-tumor tissue. Additionally, we found that expression of this miRNA is significantly downregulated in tumor tissue compared to non-tumor tissues. However, we did not observe an association between AFF1 rs17703261 with the risk of developing colon cancer. The miRNAs that were identified as targeting KIAA0423 in Ryan et al. were hsa-miRs-19a and -19b. We identified significantly lower expression of hsa-miR-19b-3p in the heterozygote genotype as compared to the homozygote common genotype in non-tumor colonic tissues. We also found expression of this miRNA to be upregulated in tumor versus non-tumor tissues. However, rs1053667 was not associated with risk of colon cancer.
Wu et al. [43] investigated several SNPs within the 3’ UTRs of genes in the B7/CD28 signaling pathway, involved in T cell stimulation, including rs7628626 (CD80), rs1305 (VTCN1), and rs44044254 and rs1559931 (ICOS). In their study, they found an increased risk of CRC with the homozygote rare genotype of rs1305 and a decreased risk of CRC with the codominant and dominant model for rs4404254 and rs1559931. We saw slightly reduced, non-significant risk for all of these SNPs with colon cancer.
It has been suggested that miRNA expression levels are associated with expression of their corresponding target genes, and that the presence of the target genes prevents miRNA degradation by nucleases [11]. To investigate this relationship, we used miRNASNP v2.0 to evaluate SNPs within 3’ UTRs of mRNAs for changes in predicted miRNA regulation. Many of the SNPs discussed previously had documented predicted loss or gain of miRNA targeting based on the changes the SNP makes in the 3’ UTR of the target mRNA, and thus the binding region of the miRNA. An example of this change is the SNP in AFF1, rs17703261 which is predicted to cause a loss of targeting by hsa-miR-648. This miRNA is slightly downregulated in tumor as compared to non-tumor tissues in our data. The homozygous rare genotype has significantly less expression of hsa-miR-648 than the other genotypes. Other examples are SLC10A7 rs105760 and KIAA0423 rs1053667, which are predicted to cause loss of targeting by hsa-miR-25-3p and hsa-miR-19b-3p, respectively. Rs1053667 and rs105760 were associated with reduced expression in tumor as compared to non-tumor tissues of the predicted lost miRNAs with the variant allele. A further example, rs9874 (SELS) is predicted to cause a gained targeting by hsa-miR-181a-5p, and we see an increase in mean expression between tumor and non-tumor tissue with the variant allele. Additionally, BRCA1 rs8176318, is predicted to have a gain of function for hsa-miR-20a-3p. We evaluated this miRNA/SNP pair and did not see an association between the predicted gained miRNA and this SNP. However, as mentioned previously, BRCA1 rs8176318 was associated with the G allele specific (C in our tables) miRNA hsa-miR-525-5p and we did see a reduction in expression of hsa-miR-525-5p in non-tumor tissues with the variant allele as well as a lowered level of mean miRNA expression in tumor tissue as compared to non-tumor tissue. These results would support the theory that a loss of the presence of the target (as a result of the SNP) causes a reduction in the level of miRNA expression.
There are several considerations when evaluating our results. First, our study is comprised of colon cancer cases while many studies included both colon and rectal cancer cases. Differences in associations with cancer could differ based on the differences in cases definition. Our findings elucidate the complexity of miRNA regulation, especially the role of SNPs in the carcinogenic process that involves miRNA. MiRNAs are involved in the regulation of multiple genes and, even though SNPs were associated with miRNAs, the influence of miRNAs on other genes may determine the association with cancer. For example, hsa-miRNA-25-3p, along with its regulation of SLC10A7, is associated with MCM7, MIR106B, MIR25, MIR93 and AP4M1 and several SNPs, rs105756, rs999885, and rs1527423. This can make the interpretation of results difficult. While there are study limitations, a major strength of this study is the ability to evaluate hypothesized SNP/miRNA associations and to evaluate associated SNPs and miRNAs with colon cancer.
Out of the 327 unique SNP-miRNA pairs evaluated only 41 were associated with miRNA expression levels. Of these, only two SNPs were associated with colon cancer risk. While it has been hypothesized that SNPs in miRNA regions can influence cancer risk through miRNA regulation, our data suggest that such associations are relatively rare when evaluating colon cancer.
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We would like to acknowledge Ulrike Peters for oversight and funding related to the GWAS data, Erika Wolff and Michael Hoffman for miRNA processing, and Sandie Edwards for her efforts in overall study monitoring and tumor tissue collection
Data Availability
Based on signed consent forms of study participants, that data can only be released for purposes stated in the consent form. Data request should be addressed to Dr. Slattery who will review and complete as appropriate. GWAS SNP data are being uploaded to dbGaP by the Fred Hutchinson Cancer Research Institute for both Utah and Minnesota subjects (data repository number not yet available).
Funding Statement
Funding for this study was received by the National Cancer Institute (http://www.cancer.gov). The grant numbers are: CA163683 and CA48998. Funding was received by MLS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 2. Murray BS C S, Woods M, Ryan TE, Liu W (2010) An in silico analysis of microRNAS: Mining the miRNAome. Molecular bioSystems 6: 1853–1862. 10.1039/c003961f [DOI] [PubMed] [Google Scholar]
- 3. Arora S R R, Chhabra A, Jaiswal A, Rani V (2013) miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Molecular Genetics and Genomics 288: 77–87. 10.1007/s00438-013-0734-z [DOI] [PubMed] [Google Scholar]
- 4. Gartel AL K E (2008) miRNAs: Little known mediators of oncogenesis. Seminars in Cancer Biology 18: 103–110. 10.1016/j.semcancer.2008.01.008 [DOI] [PubMed] [Google Scholar]
- 5. Nam S L M, Choi K, Balch C, Kim S, Nephew KP (2009) MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Research 37: W356–W362. 10.1093/nar/gkp294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436. 10.1001/jama.299.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, et al. (2009) Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res 15: 5878–5887. 10.1158/1078-0432.CCR-09-0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, et al. (2012) Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 18: 3054–3070. 10.1158/1078-0432.CCR-11-1078 [DOI] [PubMed] [Google Scholar]
- 9. Li L, Pan X, Li Z, Bai P, Jin H, et al. (2013) Association between polymorphisms in the promoter region of miR-143/145 and risk of colorectal cancer. Hum Immunol 74: 993–997. 10.1016/j.humimm.2013.04.019 [DOI] [PubMed] [Google Scholar]
- 10. Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104: 3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kai ZS, Pasquinelli AE (2010) MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 17: 5–10. 10.1038/nsmb.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 69: 3613–3634. 10.1007/s00018-012-0990-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slattery ML, Potter J, Caan B, Edwards S, Coates A, et al. (1997) Energy balance and colon cancer—beyond physical activity. Cancer Res 57: 75–80. [PubMed] [Google Scholar]
- 14. Agilent Technologies I (2013) Agilent GeneSpring User Manual. Santa Clara, CA: Aglient Technologies Inc. [Google Scholar]
- 15. Griffiths-Jones S (2006) miRBase: the microRNA sequence database. Methods Mol Biol 342: 129–138. [DOI] [PubMed] [Google Scholar]
- 16.Slattery ML, Herrick JS, Samowitz W, Pellatt DF, Stevans JR, et al. (2015) MicroRNA profiles in colorectal carcinomas, adenomas, and normal colonic mucosa: variations in miRNA expression and disease progression. Unpublished, under review. [DOI] [PMC free article] [PubMed]
- 17.Pellatt DF, Stevans JR, Wolff RK, Mullany LE, Herrick JS, et al. (2015) Expression profiles of miRNA subsets distinguish human colorectal carcinoma and normal colonic mucosa. Unpublished, under review. [DOI] [PMC free article] [PubMed]
- 18. Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, et al. (2012) Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nature genetics 44: 770–776. 10.1038/ng.2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, et al. (2008) Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 29: 579–584. 10.1093/carcin/bgm304 [DOI] [PubMed] [Google Scholar]
- 20. Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 10: 389–402. 10.1038/nrc2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pelletier C, Weidhaas JB (2010) MicroRNA binding site polymorphisms as biomarkers of cancer risk. Expert Rev Mol Diagn 10: 817–829. 10.1586/erm.10.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, et al. (2010) Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res 70: 2789–2798. 10.1158/0008-5472.CAN-09-3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang D, Meyer L, Chang DW, Lin J, Pu X, et al. (2010) Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 70: 9765–9776. 10.1158/0008-5472.CAN-10-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou X, Chen X, Hu L, Han S, Qiang F, et al. (2010) Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecol Oncol 117: 287–290. 10.1016/j.ygyno.2010.01.020 [DOI] [PubMed] [Google Scholar]
- 25. Kim JS, Choi YY, Jin G, Kang HG, Choi JE, et al. (2010) Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study. Mol Carcinog 49: 913–921. 10.1002/mc.20672 [DOI] [PubMed] [Google Scholar]
- 26. Zhang X, Yang H, Lee JJ, Kim E, Lippman SM, et al. (2010) MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis 31: 2118–2123. 10.1093/carcin/bgq177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen AX, Yu KD, Fan L, Li JY, Yang C, et al. (2011) Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet 7: e1002259 10.1371/journal.pgen.1002259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Z, Wei S, Ma H, Zhao M, Myers JN, et al. (2011) A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis 32: 1668–1674. 10.1093/carcin/bgr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, et al. (2011) LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res 71: 3896–3903. 10.1158/0008-5472.CAN-10-4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reshmi G, Surya R, Jissa VT, Babu PS, Preethi NR, et al. (2011) C-T variant in a miRNA target site of BCL2 is associated with increased risk of human papilloma virus related cervical cancer—an in silico approach. Genomics 98: 189–193. 10.1016/j.ygeno.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 31. Xiong F, Wu C, Chang J, Yu D, Xu B, et al. (2011) Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res 71: 5175–5181. 10.1158/0008-5472.CAN-10-4407 [DOI] [PubMed] [Google Scholar]
- 32. Sung H, Lee KM, Choi JY, Han S, Lee JY, et al. (2011) Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: a case-control study in Korea. Breast Cancer Res Treat 130: 939–951. 10.1007/s10549-011-1656-2 [DOI] [PubMed] [Google Scholar]
- 33. Xu Y, Liu L, Liu J, Zhang Y, Zhu J, et al. (2011) A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer 128: 412–417. 10.1002/ijc.25342 [DOI] [PubMed] [Google Scholar]
- 34. Zhang L, Liu Y, Song F, Zheng H, Hu L, et al. (2011) Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci U S A 108: 13653–13658. 10.1073/pnas.1103360108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng H, Song F, Zhang L, Yang D, Ji P, et al. (2011) Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer. Int J Oncol 38: 1153–1161. 10.3892/ijo.2011.940 [DOI] [PubMed] [Google Scholar]
- 36. Bae JS, Kim JH, Pasaje CF, Cheong HS, Lee TH, et al. (2012) Association study of genetic variations in microRNAs with the risk of hepatitis B-related liver diseases. Dig Liver Dis. [DOI] [PubMed] [Google Scholar]
- 37. Liu Y, Zhang Y, Wen J, Liu L, Zhai X, et al. (2012) A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One 7: e32230 10.1371/journal.pone.0032230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teo MT, Landi D, Taylor CF, Elliott F, Vaslin L, et al. (2012) The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis 33: 581–586. 10.1093/carcin/bgr300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang L, Li Y, Cheng M, Huang D, Zheng J, et al. (2012) A functional polymorphism at microRNA-629-binding site in the 3'-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis 33: 338–347. 10.1093/carcin/bgr272 [DOI] [PubMed] [Google Scholar]
- 40. Moore AE, Young LE, Dixon DA (2012) A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene 31: 1592–1598. 10.1038/onc.2011.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu Q, Zhou CX, Chen NS, Zheng SD, Shen LM, et al. (2012) A polymorphism within ErbB4 is associated with risk for hepatocellular carcinoma in Chinese population. World J Gastroenterol 18: 383–387. 10.3748/wjg.v18.i4.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S, Chen H, Zhao X, Cao J, Tong J, et al. (2012) REV3L 3'UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene. [DOI] [PubMed] [Google Scholar]
- 43. Wu D, Tang R, Qi Q, Zhou X, Zhou H, et al. (2015) Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cell Immunol 293: 41–48. 10.1016/j.cellimm.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 44. Dikaiakos P, Gazouli M, Rizos S, Zografos G, Theodoropoulos GE (2015) Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biomark 15: 163–168. [DOI] [PubMed] [Google Scholar]
- 45. Pardini B, Rosa F, Naccarati A, Vymetalkova V, Ye Y, et al. (2015) Polymorphisms in microRNA genes as predictors of clinical outcomes in colorectal cancer patients. Carcinogenesis 36: 82–86. 10.1093/carcin/bgu224 [DOI] [PubMed] [Google Scholar]
- 46. Xie WQ, Tan SY, Wang XF (2014) Effect of a common genetic variant microRNA-146a rs2910164 on colorectal cancer: a meta-analysis. J Dig Dis 15: 647–653. 10.1111/1751-2980.12201 [DOI] [PubMed] [Google Scholar]
- 47. Li L, Sheng Y, Lv L, Gao J (2013) The association between two microRNA variants (miR-499, miR-149) and gastrointestinal cancer risk: a meta-analysis. PLoS One 8: e81967 10.1371/journal.pone.0081967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma XP, Zhang T, Peng B, Yu L, Jiang de K (2013) Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS One 8: e79584 10.1371/journal.pone.0079584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ding L, Jiang Z, Chen Q, Qin R, Fang Y, et al. (2015) A functional variant at miR-520a binding site in PIK3CA alters susceptibility to colorectal cancer in a Chinese Han population. Biomed Res Int 2015: 373252 10.1155/2015/373252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mao Y, Li Y, Jing F, Cai S, Zhang Z, et al. (2014) Association of a genetic variant in microRNA-146a with risk of colorectal cancer: a population-based case-control study. Tumour Biol 35: 6961–6967. 10.1007/s13277-014-1916-y [DOI] [PubMed] [Google Scholar]
- 51. Buas MF, Onstad L, Levine DM, Risch HA, Chow WH, et al. (2015) MiRNA-Related SNPs and Risk of Esophageal Adenocarcinoma and Barrett's Esophagus: Post Genome-Wide Association Analysis in the BEACON Consortium. PLoS One 10: e0128617 10.1371/journal.pone.0128617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 34: 591–602. 10.1002/gepi.20516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gong J, Liu C, Liu W, Wu Y, Ma Z, et al. (2015) An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database (Oxford) 2015: bav029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Davison AC, Hinkley DV (1997) Bootstrap Methods and their Application. Cambridge, United Kingdom: Cambridge University Press. [Google Scholar]
- 55. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society 57: 289–300. [Google Scholar]
- 56. Dorairaj JJ, Salzman DW, Wall D, Rounds T, Preskill C, et al. (2014) A germline mutation in the BRCA1 3'UTR predicts Stage IV breast cancer. BMC Cancer 14: 421 10.1186/1471-2407-14-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Based on signed consent forms of study participants, that data can only be released for purposes stated in the consent form. Data request should be addressed to Dr. Slattery who will review and complete as appropriate. GWAS SNP data are being uploaded to dbGaP by the Fred Hutchinson Cancer Research Institute for both Utah and Minnesota subjects (data repository number not yet available).