Abstract
Osteosarcoma (OS) is the most common primary tumor in humans and dogs affecting the skeleton, and spontaneously occurring OS in dogs serves as an extremely useful model. Unacceptable toxicities using current treatment protocols prevent further dose-intensification from being a viable option to improve patient survival and thus, novel treatment strategies must be developed. Histone deacetylase inhibitors (HDACi) have recently emerged as a promising class of therapeutics demonstrating an ability to enhance the anti-tumor activity of traditional chemotherapeutics. To date, gene expression analysis of OS cell lines treated with HDACi has not been reported, and evaluation of the resultant gene expression changes may provide insight into the mechanisms that lead to success of HDACi. Canine OS cells, treated with a clinically relevant concentration of the HDACi valproic acid (VPA), were used for expression analysis on the Affymetrix canine v2.0 genechip. Differentially expressed genes were grouped into pathways based upon functional annotation; pathway analysis was performed with MetaCore and Ingenuity Pathways Analysis software. Validation of microarray results was performed by a combination of qRT-PCR and functional/biochemical assays revealing oxidative phosphorylation, cytoskeleton remodeling, cell cycle, and ubiquitin-proteasome among those pathways most affected by HDACi. The mitomycin C-bioactivating enzyme NQ01 also demonstrated upregulation following VPA treatment, leading to synergistic reductions in cell viability. These results provide a better understanding of the mechanisms by which HDACi exert their effect in OS, and have the potential to identify biomarkers that may serve as novel targets and/or predictors of response to HDACi-containing combination therapies in OS.
Keywords: VALPROATE, OSTEOSARCOMA, PATHWAY ANALYSIS, EXPRESSION PROFILING
Osteosarcoma (OS) is a high-grade primary bone neoplasm of mesenchymal origin and represents the most common malignant tumor of the bone in both humans and dogs [Dernell et al., 2001; Kansara and Thomas, 2007]. There are remarkable similarities between human and canine OS, which include a bimodal peak incidence, a slight male predilection, similar anatomic site predilections (metaphyses of long bones), aggressive biological behavior, hematogenous metastasis to the lungs early in the course of disease, similar sensitivity to chemotherapeutics, and a nearly identical reported incidence of occult metastasis at the time of diagnosis [Dernell et al., 2001; Kansara and Thomas, 2007; Mueller et al., 2007; Cleton-Jansen et al., 2009]. In both cases, the addition of adjuvant chemotherapy results in an improved overall survival, although this does not hold true for patients presenting with metastatic disease at the time of diagnosis, and in humans the 5-year survival for these patients remains around 30% [Rosen et al., 1983; Link et al., 1986; Dernell et al., 2001]. Unacceptable toxicity prevents further dose intensification of current multimodal chemotherapy protocols from being a viable option to further improve survival, and thus there is a need to develop novel therapeutics for this disease. Generating meaningful data from clinical studies in humans with OS can be difficult because of the relatively low incidence; approximately 900 new cases are reported per year in the U.S. [Kansara and Thomas, 2007]. In contrast, the high incidence of OS in dogs (approximately 8–12,000 cases/year) [Dernell et al., 2001] presents a unique opportunity to not only study the biology of OS but also rigorously evaluate novel therapeutics.
Histone deacetylase inhibitors (HDACi) have emerged as a very promising novel class of therapeutics in cancer treatment. There are currently a number of HDACi in clinical evaluation for hematologic and solid tumors, and two (vorinostat and romidepsin) have received FDA approval as single agents in the treatment of relapsed or refractory T-cell lymphoma [Mann et al., 2007; Marsoni et al., 2008; Prince et al., 2009]. The exact mechanisms by which these drugs act in cancer therapy is not fully understood, but they appear to have pleiotropic anti-tumor effects including induction of differentiation, growth arrest, initiation of senescence, enhanced apoptosis, decreased angiogenesis, immunomodulatory activities, and an ability to synergize with traditional cytotoxic chemotherapies and radiation [Deroanne et al., 2002; Kim et al., 2003; Bolden et al., 2006; Minucci and Pelicci, 2006; Tomasi et al., 2006; Xu et al., 2007]. The clinical utility of agents that target HDAC enzymes is, in part, based upon the aberrant expression and/or function of HDACs in cancer initiation and progression, with overexpression of some HDACs correlating with poor prognosis in a number of tumor types including gastric, prostate, breast, pancreatic, lung, and hepatocellular cancers [Rikimaru et al., 2007; Miyake et al., 2008; Weichert et al., 2008a b c].
The specific role of HDAC enzymes in the pathogenesis of OS has not been elucidated; however, HDAC enzymes play crucial roles in the normal development of bone cells including osteoblasts and osteoclasts [Schroeder et al., 2004; Schroeder and Westendorf, 2005; Westendorf, 2007]. It has been shown that the treatment of normal osteoblasts by HDACi results in accelerated maturation and differentiation, in part due to the inhibition of the interaction between HDAC3 and the transcription factor Runx2 which controls expression of osteocalcin, a marker of osteoblast differentiation [Schroeder et al., 2004]. In addition, the HDACi-induced effect on osteoblast differentiation has been shown to act through the regulation of Wnt/β catenin pathway [Schroeder et al., 2007]. It has been reported that the majority of high-grade OS have a demonstrable inactivity of the Wnt pathway, determined by a lack of nuclear β-catenin staining in biopsies [Kansara and Thomas, 2007; Cai et al., 2009; Cleton-Jansen et al., 2009]. However, there are conflicting reports on the activity of the Wnt pathway in OS, and in some cases inhibition of the pathway in OS has been shown to reduce tumorigenesis and metastatic potential of some cell lines [Rubin et al., 2010]. In fact, one decoy receptor for the Wnt pathway, secreted frizzled-related protein 3 (SFRP3), has a potential tumor suppressor function in OS, showing an ability to reduce osteoblast proliferation and increase osteoblast differentiation. Furthermore, high-grade OS tumors appear to lack or have downregulated expression of SFRP3 [Chung et al., 2004; Mandal et al., 2007].
In addition to the effects on differentiation processes of osteoblasts, HDAC inhibition also results in alterations of chromatin structure, a property that has been utilized to sensitize cells to chemotherapeutic agents that target DNA. The chromatin decondensation resulting from HDAC inhibition has been shown to increase sensitivity to topoisomerase I and II inhibitors such as doxorubicin, epirubicin, and keranotecin in a variety of models of human and canine solid tumors, including OS [Marchion et al., 2004, 2005; Munster et al., 2007; Wittenburg et al., 2010].
Although microarray data exist on the effect of HDACi in osteoblasts, to our knowledge there are no reports of microarray data utilizing pathway analysis in HDACi-treated OS cells. Here, we attempt to further elucidate the molecular sequelae of HDAC inhibition in OS cells through pathway analysis of a canine OS cell line treated with the HDACi valproic acid (VPA), and use a combination of quantitative PCR and biochemical/functional assays in canine OS cells to validate the results of our microarray analysis.
MATERIALS AND METHODS
CELL LINES AND CONDITIONS
The D17 canine OS cell line was purchased from the American Type Culture Collection (Rockville, MD) The Abrams canine OS cell line was kindly provided by Dr. William Dernell (Colorado State University, Fort Collins, CO). Cells were cultured in minimum essential medium Eagle (Lonza, Walkersville, MD) supplemented with 10% fetal bovine serum (Atlas, Fort Collins, CO) and penicillin/ streptomycin (Invitrogen, Carlsbad, CA). For experimental procedures, cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
CHEMICALS AND ANTIBODIES
Valproic acid was purchased from Sigma Chemical, Co. (St. Louis, MO). Bovine serum albumin, 2,6-dichlorophenolindophenol (DCPIP), dicumarol, and NADH were purchased from Sigma–Aldrich (St. Louis, MO). Suc-Leu-Tyr-AMC fluorogenic substrate was purchased from AnaSpec (San Jose, CA). Bicinchoninic acid protein assay reagents and SuperSignal chemiluminescent substrate were purchased from Pierce Chemical Co. (Rockford, IL). The proteasome inhibitor ONX-912 was provided by Onyx Pharmaceuticals (South San Francisco, CA). Alamar Blue™ fluorogenic substrate was obtained from AbD Serotec (Oxford, UK).
GENE EXPRESSION MICROARRAY
Valproic acid treated (1 mM) and untreated D17 canine OS cells were used for analysis of differential gene expression using the Affymetrix™ Canine v2.0 gene chip, which was performed by the Microarray Core at the University of Colorado Health Sciences Center (Denver, CO). Briefly, cells in log-phase growth were treated in triplicate with 0 or 1 mM VPA for 48 h, after which cells were harvested and stored in RNAlater for transfer to the core. RNA was extracted using the Qiashredder and RNeasy Kit from Qiagen (Valencia, CA). RNA quantity and integrity were assessed using the Nanodrop 1000 (ThermoScientific, Waltham, MA) and Bioanalyzer (Agilent Technologies, Foster City, CA) respectively. Resulting RNA was then used for first- and second-strand cDNA synthesis followed by cDNA cleanup and overnight in vitro transcription. Following cRNA cleanup and fragmentation steps, samples were then hybridized to the Affymetrix chip and scanned.
MICROARRAY DATA ANALYSIS
Data normalization and selection of differentially expressed genes was performed as previously described by Ptitsyn et al. [2008]. Briefly, normalized data were used to select a preliminary set of differentially expressed genes using the University of Pittsburgh Gene Expression Data Analysis suite (GEDA, http://bioinformatics.upmc.edu/GE2/GEDA.html). To reduce the number of false-positive differential genes and provide a shortlist of genes deviating from the expected average value and enriched with differential genes, a standard J5 metric with threshold 4 and optional 4 iteration of jackknife procedure was applied. Then we prepared a proxy set of human genes based on homology of canine probe targets to human genes determined by BLAST and BLAT search tools. This curated set of nearest homologs was submitted to pathways analysis. The reason for using the proxy set is that gene interaction databases and analysis software support only human, mouse, and rat data input.
FUNCTIONAL ANNOTATION AND PATHWAY ANALYSIS
Analysis of biological pathways was performed using MetaCore software version 6.4 (GeneGo Inc., Division of Thomson Reuters), Ingenuity Pathways Analysis version 8.8 (IPA, Ingenuity Systems Inc.). Significance of a particular pathway represented in a given list of genes is estimated by Fisher’s exact test with adjustments to current database contents. The GeneGo and IPA databases are accessed online and the contents (including Canonic Maps, Molecular Functions, Gene Interactions, etc.) are frequently updated by the corps of curators reading research publications and extracting information related to all forms of interaction among genes and chemical compounds. Consequently, the results of pathway analyses performed at different times may differ in details. The maps of canonic signaling and metabolic pathways (groups of interacting genes and other factors such as metabolites and chemical compounds drawn by Ingenuity and Genego developers) are convoluted, but tested for significance independently. As a result, the list of significant pathways often enriched with redundant pathways overlapping by a majority of components. The interpretation of the meaning of statistically significant pathways relies on the knowledge of the biological function underlying the pathway maps and cross-comparison between two independent pathway databases (IPA and Metacore). We also applied DAVID web-based tools to provide preliminary functional annotation of all potentially differential genes [Huang da et al., 2007].
QUANTITATIVE REAL-TIME RT-PCR
For validation of microarray results, a subset of VPA-altered genes was chosen for qRT-PCR in canine OS cell lines treated with VPA. RNA was extracted using the Qiashredder and RNeasy Kit (Qiagen) according to manufacturer’s recommendations. Reverse transcription of RNA was performed using the Omniscript® RT Kit (Qiagen) with a no-RT control for each sample. Each PCR reaction contained iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA), 100 nM forward primer, 300 nM reverse primer, 50 nM reference dye (ROX), and 25 ng template cDNA. Primers for canine gene sequences used for microarray validation are listed in Table I. PCR reactions were carried out in duplicate on an Mx3000P™ thermal cycler (Stratagene, La Jolla, CA) programmed to run at 95°C for 10 min, 60°C for 1 min with fluorescence monitoring (42 cycles), then ramp back up to 95°C with continuous fluorescence monitoring for dissociation curves. Average threshold values (Ct) were then used to evaluate changes in gene expression using freely available Relative Expression Software Tool (REST) v2.0.13 (Qiagen).
TABLE I.
Primer Sequences of Canine Genes Selected From Pathway Analysis for Validation Using Quantitative Real-Time RT-PCR
| Gene | Forward | Reverse |
|---|---|---|
| Ezrin | AGC CAA TCA ACG TCC GAG TTA CCA | ACT GGA GGC CAA AGT ACC ACA CTT |
| Moesin | ATT GGC CAA GGA ACG TCA AGA AGC | AGA TTC GAG CTG TCA ACT CTG CCA |
| PSMD6 | AAG GAT GGT GCT CTG ACA GCC TTT | TTA GGC GGT TTC TCC TGT CCC AAT |
| HSP90 | TGT AAT TGC TGA CCC ACG AGG GAA | TTC TTC CAT GGG CTC CTC AAC AGT |
| NCAPH | ATG TGG AGC TTG CTG ACA AAG TGC | AGA CAG GCA AAG GCC AGA GGT ATT |
| MCM4 | TCC CAG CTG ATT CCA GAG ATG CAA | AAA CAG AAG GCT CAG CAA TGC GAC |
| AURKA | TTG GGT GGT CAG TAC ATG CTC CAT | AGG TCT CTT GGT ATG TGC TTG CCT |
| CCNA2 | AGC ACT CTA CAC AGT CAC AGG ACA | TCT GGT GGG TTG AGG AGA GAA ACA |
| THBS1 | CAA TGC CAA CCA AGC TGA CCA TGA | ACA AGT CTG CAG TTG TCC CTG TCA |
| HPRT | ACT TTG CTT TCC TTG GTC AGG CAG | GGC TTA TAT CCA ACA CTT CGT GGG |
LACTATE MEASUREMENT
Lactate generation by OS cells, measured by concentration in cell culture media, was evaluated as a surrogate for the overall activity of the oxidative phosphorylation pathway. Cells were plated in triplicate in 24-well plates and allowed to adhere overnight. VPA was added to the wells and incubated for 48 h at 37°C, after which media was aspirated and spun down to remove any remaining cells. Relative lactate concentrations were evaluated using a commercially available D-Lactate colorimetric assay (BioVision, Mountain View, CA) according to the manufacturer’s recommendations. Results were integrated from a standard curve generated at the same time experimental values were measured and normalized to cell number.
NQO1 ACTIVITY MEASUREMENT
The NQO1 activity in VPA treated and untreated cells was measured as previously described [Gustafson et al., 2003]. Briefly, canine OS cell lines were grown in the presence of 0 or 1 mM VPA, and, following treatment, were washed twice with PBS and pelleted by centrifugation. Pellets were resuspended in 100 μl of 25 mM Tris (pH 7.4) and disrupted by sonication using three 2-s bursts at 30% power; the resulting sonicates were centrifuged at 15,000g and the supernatant was collected and assayed for NQO1 activity and total protein content. For NQO1 activity measurement in lysates, a reaction mix containing 25 mM Tris (pH 7.4), 0.07% bovine serum albumin (w/v), 200 μM NADH, and 40 μM DCPIP was used and reactions were carried out in the presence and absence of 20 μM dicumarol. The NQO1 activity is described as the dicumarol inhibitable decrease in absorbance at 600 nm with DCPIP as a substrate and is expressed in nanomoles of DCPIP reduced per minute per milligram of protein. Total protein in cell lysates was determined by the bicinchoninic acid assay using BSA as a standard.
20S PROTEOSOME CHYMOTRYPSIN-LIKE ACTIVITY
Canine OS cells were plated in six-well plates and allowed to adhere overnight prior to treatment with 0 or 1 mM VPA for 48 h. After incubation, media was aspirated and cells were washed twice with PBS. Plates were then placed on ice and 90–100 μl of lysis buffer (20 mM Tris–HCl pH 8.0; 5 mM EDTA) was added to wells. Cells were scraped into the lysis buffer and placed into 1.5 ml Eppendorf tubes prior to freezing at −20°C. Lysates were thawed on ice and spun down at 3,000g for 15 min at 4°C and the supernatants were transferred to fresh ice-cold tubes. Twenty microliters of lysate was added, in duplicate, to wells of a black-walled 384-well plate followed by 20 μl of Assay Buffer (20 mM Tris pH 8.0; 0.5 mM EDTA) containing 120 μM Suc-Leu-Tyr-AMC substrate. Fluorescence was determined on a Synergy™ HT microplate reader (BioTek, Winooski, VT) using an excitation wavelength of 340 nm and an emission wavelength of 465 nm. The fluorescence reader was programmed to read relative fluorescence units (RFU) every 3.5 min for 12 cycles. Mean slopes of the linear portion of the curves were calculated as RFU/min. Protein content of the lysates was determined with the BCA assay using bovine serum albumin as a standard. Calpain activity was then determined as the RFU/min/μg of protein loaded. Results were then normalized to the untreated controls and expressed as a fold change in calpain activity.
ENDOTHELIN-1 MEASUREMENT
Canine cells were incubated in 0, 0.5, or 1.0 mM VPA for 48 h and the levels of endothelin in cell culture supernatants were evaluated using the ELISA-based Endothelin-1 Assay Kit (Immuno-Biological Laboratories Co., Ltd, Gunma, Japan) according to manufacturer’s recommendations. Briefly, cell supernatants were added to a precoated ELISA plate provided in the kit and allowed to incubate overnight at 4°C. Then labeled antibody solution was added to each well and incubated at 37°C for 30 min, followed by addition of a chromogenic substrate and measurement of absorbance at 450 nm. Endothelin levels were calculated by comparing to a standard curve prepared simultaneously with the measurement of test samples.
CELL GROWTH INHIBITION
For the endothelin measurement assays, parallel proliferation assays were set up to determine if changes in the levels of measured Et-1 were a result of decreased cell viability. Cells were plated in triplicate in 96-well plates and allowed to adhere overnight at 37°C. Cells were then incubated in 0, 0.25, 0.5, or 1.0 mM VPA for 48 h, after which the relative viable cell numbers were determined by fluorometric bioreductive assay (Alamar blue), normalized to untreated controls. For proliferation assays involving proteasome inhibitor therapy, cells were plated in drug-free media in triplicate into 96-well plates and allowed to adhere overnight at 37°C. Cells were then exposed to VPA and increasing concentrations of proteasome inhibitor and incubated for 48 h at 37°C. Following incubation, relative viable cell numbers were determined by Alamar blue assay, normalized to untreated controls. For combination treatments involving mitomycin C, cells were plated in triplicate in 96-well plates and incubated in 0, 0.5, or 1.0 mM VPA for 48 h, then media was removed and replaced with mitomycin C-containing media for an additional 24-h incubation. Relative viable cell number was determined by Alamar blue assay.
STATISTICAL ANALYSIS
For qRT-PCR experiments, results of three independent runs performed in duplicate were used for analysis of relative expression changes by entering average threshold values (Ct) into the REST v2.0.13 software program. Changes in expression for all bars represented graphically were considered statistically significant (P < 0.05). For calpain and NQ01 activity, results were generated from three independent experiments and represent mean ± SD. Fold change differences were compared by one-sample, two-tailed t-test and a P-value <0.05 was considered significant. Results of proliferation experiments are reported as mean ± SD of three replicates.
In order to determine whether the addition of VPA to proteasome inhibitor synergistically reduced viable cell number, the Bliss independence model was utilized. Briefly, the Bliss criterion is described by the following equation:
where E(x) is the fractional inhibition of concentration x of VPA (between 0 and 1), E(y) is the fractional inhibition of concentration y of chemotherapy, and E(x,y) is the combined effect. Theoretical growth inhibition curves were constructed using this equation, and standard deviations were estimated by error propagation of experimental SD. Differences between treatment groups (Bliss theoretical vs. experimental) were assessed using two-way ANOVA and a Bonferroni post-test. Using this model, if the experimental fractional inhibition across concentration was significantly higher than the theoretical value, the interaction was considered synergistic.
RESULTS
SELECTION OF GENES AND PATHWAY ANALYSIS
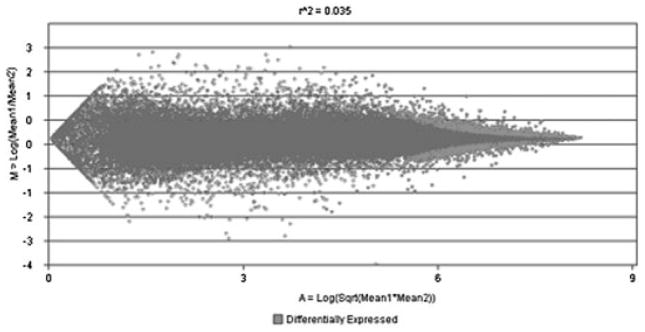
Analysis of our dataset from the microarray resulted in the selection of 1,570 genes that were differentially expressed (674 upregulated, 894 downregulated) between VPA-treated and control cells. As shown in Figure 1, the genes that were selected were a balanced group between those that were moderately and highly expressed, categories that are most appropriate for selection of potential biomarkers [Ptitsyn et al., 2008]. After selection of the differentially expressed genes, a proxy set of human genes was generated using DAVID web-based tools to provide functional annotation based on sequence homology to probe targets using BLAST and BLAT web-based tools (Supplementary Data 1), and this proxy set of human genes was then used for subsequent pathway analysis.
Fig. 1.
MA plot of genes selected from microarray analysis of VPA-treated canine OS cells; 1,570 genes were selected (light gray) based on those that were differentially expressed in response to VPA treatment and represent a balanced group of those that are moderately and highly expressed.
Analysis of gene expression to identify altered pathways revealed 43 that were statistically overrepresented (P < 0.05) in the list of genes; the top 15 are represented in Figure 2. The pathway demonstrating the highest gene enrichment in this dataset involves oxidative phosphorylation and, as shown in Supplementary Data 2, the overall result of VPA treatment is an upregulation of a number of genes involved in oxidative phosphorylation, resulting in an overall upregulation of respiratory complex I, respiratory complex II, cytochrome C oxidase, and ATP synthase. We also indirectly demonstrate the effect of VPA on oxidative phosphorylation by the reduction in the amount of lactate produced by the cells and secreted into the media (Fig. 3), implying a functional shift away from glycolysis. The regulation of oxidative phosphorylation may have important implications in the progression of metastasis as well as susceptibility to apoptosis in OS cells [Dey and Moraes, 2000; Harris et al., 2000; Ptitsyn et al., 2008]. Additional pathways with potential importance to OS initiation, progression, and response to therapy include development through the hedgehog pathway, cytoskeleton remodeling, antigen presentation by MHC class I, cell adhesion, cell cycle, and proteolysis (Supplementary Data 3 and 4). Interestingly, our results demonstrate similarities to a study that evaluated the effect of HDACi on cultured osteoblasts, as both studies have discovered that treatment with HDACi results in modulation of differentiation-associated genes involving the Wnt pathway, among others [Schroeder et al., 2007].
Fig. 2.
Top 15 biological pathways significantly overrepresented in the list of genes differentially expressed between VPA-treated and control OS cells.[Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/jcb]
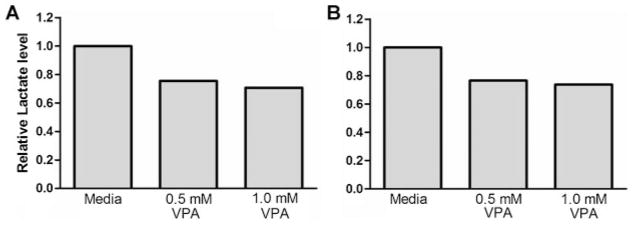
Fig. 3.
Measurement of lactate concentrations in cell culture media of VPA treated Abrams (A) and D17 (B) OS cells. Cells were incubated for 48 h in VPA and then cell culture media removed and evaluated for lactate content by colorimetric assay. Results demonstrate a reduction in the secreted concentrations of lactate by VPA treatment and are consistent with results obtained from the analysis of gene expression indicating an overall upregulation of the oxidative phosphorylation pathway.
VALIDATION BY QUANTITATIVE REAL-TIME RT-PCR
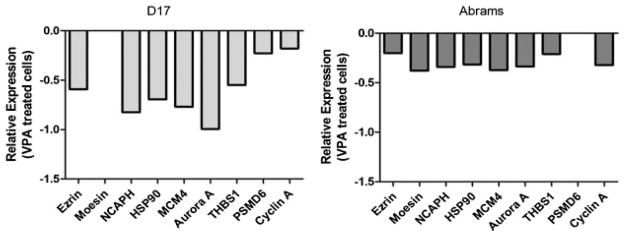
In order to validate the results of the microarray, a short list of genes chosen from the pathway maps as well as from a list of the most differentially expressed genes was generated for quantitative real-time PCR. These genes were chosen from the pathway maps because of their putative role in tumor initiation, progression, or response to therapy. These genes include the metastasis-related genes ezrin and moesin, the S-phase replication-associated protein MCM4, heat-shock protein HSP90, cell cycle-associated proteins cyclin A and aurora A, proteosome regulatory subunit PSMD6, and thrombospondin 1. The results of PCR analysis are shown in Figure 4 and serve to validate the gene expression changes and the selection criteria for these genes.
Fig. 4.
Validation of microarray by quantitative real-time RT-PCR of genes selected from significantly altered pathways as well as from a list of the most differentially expressed genes in response to VPA treatment in canine OS cells. PCR results are consistent with downregulation of genes seen in microarray pathway analysis. Moesin (D17) and PSMD6 (Abrams) were both downregulated but not considered statistically significant (P > 0.05).
PROTEASOME INHIBITION BY VALPROIC ACID
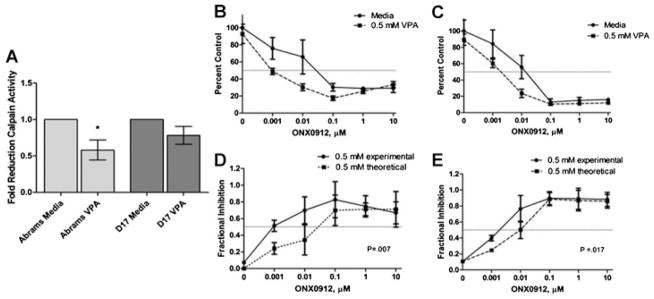
In addition to PCR analysis of the proteasomal regulatory subunit PSMD6, we also evaluated the inhibition of the proteasome pathway as a whole through evaluation of chymotrypsin-like activity in VPA-treated cells. As shown in Figure 5a, canine OS cells treated for 48 h with VPA exhibit an overall decrease in the chymotrypsin-like activity compared to controls, providing further validation of the pathway analysis.
Fig. 5.
(A) Evaluation of proteasome inhibition by VPA. Human and canine OS cells were treated with 0 or 1.0 mM VPA for 48 h and then proteasome inhibition was measured by evaluation of calpain activity in cell lysates, *P < 0.05. Proliferation evaluation of Abrams (B) and D17 (C) OS cells co-treated with 0 or 0.5 mM VPA and increasing concentrations of the proteasome inhibitor ONX0912 demonstrate sensitization to proteasome inhibition by VPA. Bliss analysis comparing fractional inhibition of theoretical additivity to experimental values reveals synergistic antiproliferative activity of the 0.5 mM VPA/ONX0912 combination in Abrams (D) and D17 (E) OS cells.
In addition, based upon the finding that VPA treatment appears to lead to proteasome inhibition, we evaluated the potential of a combination therapy utilizing VPA and the proteasome inhibitor ONX0912. The addition of VPA led to a synergistic reduction in viable cell numbers compared to treatment with ONX0912 alone (Fig. 5b).
ENHANCEMENT OF NQO1 ACTIVITY BY VALPROIC ACID
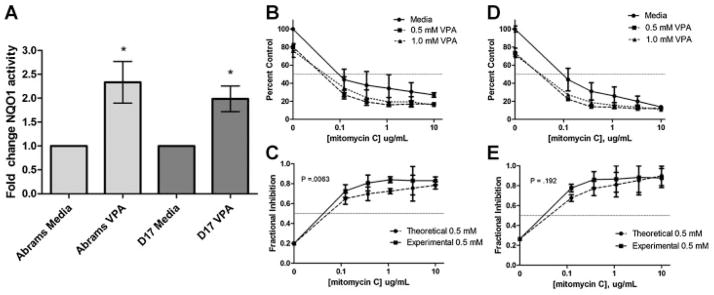
One of the pathways that was statistically overrepresented in our microarray analysis is the ubiquinone metabolism pathway, and NAD(P)H:quinone oxidoreductase 1 (NQO1) was also found to be amongst the most highly upregulated genes in response to VPA treatment. To validate this finding, we evaluated NQO1 activity in VPA-treated cells. As shown in Figure 6A, the specific activity of NQO1 was enhanced by treatment with VPA, which is consistent with upregulation of NQO1 gene transcription observed by microarray.
Fig. 6.
(A) NQO1 activity measured in lysates of VPA-treated (1 mM) and untreated OS cells. Both cell lines demonstrate increased NQO1 activity in response to VPA treatment, consistent with upregulation of NQO1 gene activity; *P < 0.05. Proliferation evaluation of Abrams (B) and D17 (C) OS cells co-treated with 0, 0.5, or 1.0 mM VPA and increasing concentrations of mitomycin C demonstrate sensitization by VPA. Bliss analysis comparing fractional inhibition of theoretical additivity to experimental values reveals synergistic anti-proliferative activity of the 0.5 mM VPA/mitomycin C combination in Abrams (D) and additive activity in D17 (E) OS cells.
In addition to increased NQO1 activity, we also assessed the ability of VPA to sensitize OS cells to the antiproliferative effects of the NQO1 substrate drug mitomycin C. As shown in Figure 6, canine OS cells treated for 48 h with VPA and mitomycin C show an enhanced anti-proliferative effect compared to mitomycin C alone.
DOWNREGULATION OF ENDOTHELIN-1 BY VPA
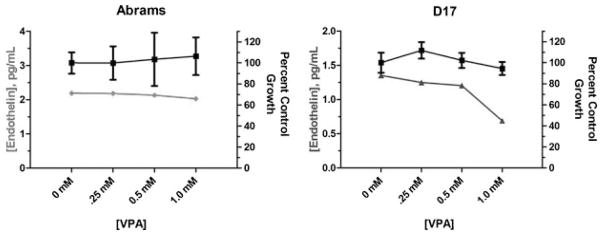
The pro-tumorigenic protein endothelin 1 (Et-1), which plays a role in promoting OS metastasis through production of matrix-degrading matrix metalloproteins, and is found at elevated levels in patients with metastatic disease [Grant et al., 2003; Felx et al., 2006], was found to be highly downregulated by VPA via gene expression microarray. We used a commercially available ELISA-based assay to evaluate the levels of Et-1 in supernatants of cells treated with a clinically achievable concentration of VPA for 48 h. As shown in Figure 7, the concentrations of Et-1 secreted by OS cells is reduced in the presence of 1 mM VPA, and this reduction is not the result of reduced cell viability as there is negligible anti-proliferative effect at that concentration of VPA (Fig. 7).
Fig. 7.
Measurement of endothelin-1 in supernatants of VPA-treated canine OS cells by ELISA. A marked reduction in endothelin-1 (gray lines) is observed in the D17 canine OS cell line upon 48-h exposure to 1 mM VPA. This reduction in endothelin is not the result of reduced cell viability as the percent viable cells remains relatively constant across the range of VPA doses (black lines).
DISCUSSION
We interrogated changes in global gene expression in a canine OS cell line treated for 48 h with the HDACi VPA, and then performed a panel of experiments designed to validate the individual genes and pathways identified by microarray data analysis. We chose an incubation in VPA of 48 h based upon previously published results by our group and others demonstrating that inhibition of HDAC is still evident at that time point and that 48-h exposure prior to the addition of a traditional cytotoxic chemotherapy is required for optimal synergistic activity [Marchion et al., 2004; Wittenburg et al., 2010]. This particular incubation condition was shown previously to result in only a modest decrease in colony forming ability, demonstrated by clonogenic assay, and a modest increase in caspase 3 activity and apoptosis of OS cells, neither of which were significantly different from controls [Wittenburg et al., 2010]. While many microarray experiments focus on the identification a few “key” differentially regulated genes, we have taken the additional step of placing all of the genes that were identified as differentially expressed in response to HDAC inhibition into functional pathways utilizing a novel data analysis pipeline [Ptitsyn et al., 2008]. Validation of our dataset was done, in part, through quantitative real-time RT-PCR of a few individual genes chosen from the pathways that were overrepresented, and we show that in two canine OS cell lines the downregulation of these selected genes is reproducible by RT-PCR.
While some of the individual genes may not be considered significantly altered when evaluated alone, functional annotation that groups them together may result in significant alteration of an entire pathway. In our study, we identified 1,570 genes that fit the criteria selected for determination of differentially expressed genes, and when placed into pathways we discover that the oxidative phosphorylation pathway is the most significantly overrepresented, with a large number of genes upregulated by VPA treatment. This finding has significant importance in cancer as it has been shown that loss of activity in this pathway, with concomitant upregulation of glucose utilization, results in increased aggressiveness and metastatic potential, referred to as the Warburg effect [Warburg, 1956; Amuthan et al., 2001; Gmeiner et al., 2008; Ptitsyn et al., 2008]. This may be partly explained by the finding that functional oxidative phosphorylation is required for apoptosis induced by the pro-apoptotic protein Bax in OS, providing evidence that evasion of apoptosis can be obtained by downregulation of oxidative phosphorylation [Dey and Moraes, 2000; Harris et al., 2000]. Our results suggest that one potential mechanism by which HDACi, and particularly VPA, exert an anti-tumor effect may be through re-sensitization of intrinsic apoptotic pathways via upregulation of oxidative phosphorylation.
An additional pathway that was overrepresented in our microarray analysis involves protein degradation via the ubiquitin-proteasome pathway. While evaluation of mRNA levels by quantitative real-time RT-PCR did show a reduction in the level of one regulatory subunit of the 20S proteasome (PSMD6), we also performed a functional assay that evaluated proteosomal activity in VPA-treated cells to determine the biological relevance. VPA-treated OS cells clearly demonstrate an inhibition of chymotrypsin-like activity, and this result is in agreement with another study describing a siRNA-mediated loss-of-function screen that identified the proteasome as playing an important role in HDACi-induced apoptosis [Fotheringham et al., 2009]. In fact, this same study suggests that one particular protein involved in shuttling of ubiquitinated cargo proteins may govern tumor sensitivity to drugs that target the proteasome, and found this particular protein overexpressed in cutaneous T-cell lymphoma, a cancer that appears particularly sensitive to HDACi. Proteasome inhibitors have been reported to be effective in the treatment of OS in vitro in a number of studies [Lauricella et al., 2003; Lim et al., 2004; Yan et al., 2007], possibly through regulation of cell cycle-associated proteins such as the cyclin-dependent kinases. This led us to test whether a combination of HDACi with VPA and proteasome inhibition with a novel compound, ONX0912, would provide an enhanced anti-tumor effect. We show that the combination of VPA and ONX0912 results in a synergistic reduction in viability of canine OS cells compared to cells treated with the proteasome inhibitor alone. It is important to note that, while these results suggest that enhanced inhibition of the proteasome results in improved reduction of cell viability, it is possible that this is not the only mechanism by which this effect occurs with the combination of these two drugs.
We also found ubiquinone metabolism to be one of the pathways altered by VPA treatment and NQO1 was additionally found to be one of the most significantly upregulated genes by the microarray. Upregulation of NQO1 could have important implications for the treatment of cancer as this particular enzyme plays a role in the bio-activation of anti-tumor quinines such as mitomycin C [Gustafson et al., 2003]. We proceeded to validate this finding through evaluation of NQO1-specific activity in canine OS cell lysates following 48-h treatment with VPA. We demonstrate that the ability of NQO1 to reduce DCPIP is enhanced following VPA treatment, consistent with increased levels of the enzyme. To further test the significance of this, we evaluated the ability of VPA to sensitize OS cells to the anti-proliferative effects of mitomycin C and show a sensitizing effect on Abrams and D17 canine OS cell lines. This is consistent with in vitro reports of synergy between VPA and mitomycin C in colon carcinoma cells [Friedmann et al., 2006]. However, as mitomycin C is also a DNA-binding drug [Verweij and Pinedo, 1990], the effect of VPA-induced chromatin decondensation and increased access to DNA by mitomycin C, as observed with other DNA-intercalating agents [Marchion et al., 2005; Wittenburg et al., 2010], cannot be ruled out as a contributing factor. Given that a number of identified pathways play important roles in DNA damage response, including protein turnover, cytoskeleton remodeling, and the electron transport chain, another possibility for the enhanced effect observed when VPA is combined with mitomycin C, doxorubicin, or other DNA damaging agents is an inhibition of the cellular ability to repair DNA damage. Indeed, the coordinated cellular response to DNA damage involves multiple levels of regulation involving protein–DNA interactions, protein–protein interactions, and multiple interconnected cellular pathways [Begley and Samson, 2004]. The discovery that DNA damage results in an extensive change in transcription factor promoter binding with some transcription factors binding significantly more genes in the presence of a DNA damaging agent [Workman et al., 2006] provides another potential explanation for increased sensitivity to DNA damage following HDACi therapy, as alterations in chromatin structure will change availability of promoter sites to transcription factors that bind specific regions in response to DNA damage. While the sensitization of OS cells to mitomycin C by VPA may result, in part, from the upregulation of NQO1, the effect of HDACi with VPA on the DNA damage response as a mechanism of sensitization to mitomycin C and other DNA-targeting anti-cancer agents certainly warrants future investigation.
An additional gene identified by microarray analysis as one significantly downregulated by VPA treatment is Et-1. Et-1 is reported to have a variety of pro-tumorigenic properties that include acting as a mitogen for numerous human cancer cell lines as well as endothelial cells and vascular smooth muscle cells, protecting cells from Fas ligand-mediated apoptosis, and promoting metastasis through upregulation of stroma-degrading matrix metalloproteinases in OS [Grant et al., 2003; Felx et al., 2006]. In addition, elevated plasma levels of Et-1 have been found in tumor-bearing patients with the highest levels found in those with metastatic disease [Grant et al., 2003]. This provides rationale for targeting Et-1 in cancer therapy. We found that incubation of OS cells with clinically achievable levels of VPA results in decreased Et-1 secretion into culture media, consistent with the downregulation observed on microarray. This may provide additional support to the use of combinations of VPA and proteasome inhibitors for the treatment of OS, as the induction of pro-metastatic genes such as the MMPs by Et-1 appears to be mediated through NF-κB, a well-documented target of proteasome inhibitors [Felx et al., 2006; Orlowski and Kuhn, 2008].
One potential drawback of this study is the lack of secondary evaluation of HDAC inhibition, which might be performed through use of additional inhibitors or RNA interference studies, providing more detailed and mechanistic information of the effect HDACi as a whole. However, our main goal was to utilize a novel method of analyzing the gene expression information resulting from a 48-h exposure to VPA specifically, and to validate this method of pathway analysis. Additionally, future studies should focus on examining the interrelationships between the pathways that were identified and the potential mechanisms by which HDACi controls these pathways. While our results focused on changes in gene expression, they do not provide mechanistic details necessary to determine if altered gene transcription is due simply to altered chromatin structure or is secondary to an “off-target” effect such as alterations in acetylation and activity of other non-histone proteins, resulting in secondary changes in gene expression.
The results of this study shed further light onto the potential mechanisms by which HDACi with VPA exerts its antitumor effect in OS. We have identified oxidative phosphorylation as the most significantly altered metabolic pathway in response to VPA. In addition, we show that the proteasome pathway is altered through decreased chymotrypsin-like activity, leading us to test a combination of VPA and a proteasome inhibitor in OS cells. Our results suggest that this particular combination may prove beneficial in the treatment of OS. We show that VPA is capable of upregulating the oxidoreductase NQO1, enhancing its enzymatic activity leading to increased sensitivity of OS cells to the NQO1 substrate drug mitomycin C. In addition, we show that treatment of OS cells with VPA results in decreased secretion of Et-1, which has been shown to correlate with poor outcome in cancer when elevated serum levels are detected in patients. Taken together, the results of our pathway analysis in OS cells treated with an HDACi reveal a number of pathways that have already been identified as potential anti-tumor targets and provide further support for the design of VPA-containing combination protocols in the treatment of OS.
Supplementary Material
Acknowledgments
Grant sponsor: Morris Animal Foundation (CSU Fund); Grant number: 5-354500; Grant sponsor: National Institutes of Health; Grant number: NCRR T32-RR-007072-06; Grant sponsor: American Cancer Society; Grant number: RSG-04-219-01.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley TJ, Samson LD. Network responses to DNA damaging agents. DNA Repair (Amst) 2004;3:1123–1132. doi: 10.1016/j.dnarep.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Cai Y, Mohseny AB, Karperien M, Hogendoorn PC, Zhou G, Cleton-Jansen AM. Inactive Wnt/Beta-catenin pathway in conventional high-grade osteosarcoma. J Pathol. 2010;220(1):24–33. doi: 10.1002/path.2628. [DOI] [PubMed] [Google Scholar]
- Chung YS, Baylink DJ, Srivastava AK, Amaar Y, Tapia B, Kasukawa Y, Mohan S. Effects of secreted frizzled-related protein 3 on osteoblasts in vitro. J Bone Miner Res. 2004;19:1395–1402. doi: 10.1359/JBMR.040412. [DOI] [PubMed] [Google Scholar]
- Cleton-Jansen AM, Anninga JK, Briaire-de Bruijn IH, Romeo S, Oosting J, Egeler RM, Gelderblom H, Taminiau AH, Hogendoorn PC. Profiling of high-grade central osteosarcoma and its putative progenitor cells identifies tumourigenic pathways. Br J Cancer. 2009;101:2064. doi: 10.1038/sj.bjc.6605482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernell WS, Straw RC, Withrow SJ. Tumors of the skeletal system. In: Withrow SJ, MacEwen EG, editors. Small animal clinical oncology. Philadelphia: W.B. Saunders Company; 2001. pp. 378–417. [Google Scholar]
- Deroanne CF, Bonjean K, Servotte S, Devy L, Colige A, Clausse N, Blacher S, Verdin E, Foidart JM, Nusgens BV, Castronovo V. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- Dey R, Moraes CT. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J Biol Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- Felx M, Guyot M-C, Isler M, Turcotte RE, Doyon J, Khatib A-M, Leclerc S, Moreau A, Moldovan F. Endothelin-1 (ET-1) promotes MMP-2 and MMP-9 induction involving the transcription factor NF-κB in human osteosarcoma. Clin Sci. 2006;110:645–654. doi: 10.1042/CS20050286. [DOI] [PubMed] [Google Scholar]
- Fotheringham S, Epping MT, Stimson L, Khan O, Wood V, Pezzella F, Bernards R, La Thangue NB. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell. 2009;15:57–66. doi: 10.1016/j.ccr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Friedmann I, Atmaca A, Chow KU, Jager E, Weidmann E. Synergistic effects of valproic acid and mitomycin C in adenocarcinoma cell lines and fresh tumor cells of patients with colon cancer. J Chemother. 2006;18:415–420. doi: 10.1179/joc.2006.18.4.415. [DOI] [PubMed] [Google Scholar]
- Gmeiner WH, Hellmann GM, Shen P. Tissue-dependent and -independent gene expression changes in metastatic colon cancer. Oncol Rep. 2008;19:245–251. [PubMed] [Google Scholar]
- Grant K, Loizidou M, Taylor I. Endothelin-1: A multifunctional molecule in cancer. Br J Cancer. 2003;88:163–166. doi: 10.1038/sj.bjc.6700750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson DL, Siegel D, Rastatter JC, Merz AL, Parpal JC, Kepa JK, Ross D, Long ME. Kinetics of NAD(P)H:quinone oxidoreductase I (NQO1) inhibition by mitomycin C in vitro and in vivo. J Pharmacol Exp Ther. 2003;305:1079–1086. doi: 10.1124/jpet.103.050070. [DOI] [PubMed] [Google Scholar]
- Harris MH, Vander Heiden MG, Kron SJ, Thompson CB. Role of oxidative phosphorylation in Bax toxicity. Mol Cell Biol. 2000;20:3590–3596. doi: 10.1128/mcb.20.10.3590-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- Lauricella M, D’Anneo A, Giuliano M, Calvaruso G, Emanuele S, Vento R, Tesoriere G. Induction of apoptosis in human osteosarcoma Saos-2 cells by the proteasome inhibitor MG132 and the protective effect of pRb. Cell Death Differ. 2003;10:930–932. doi: 10.1038/sj.cdd.4401251. [DOI] [PubMed] [Google Scholar]
- Lim JH, Chang YC, Park YB, Park JW, Kwon TK. Transcriptional repression of E2F gene by proteasome inhibitors in human osteosarcoma cells. Biochem Biophys Res Commun. 2004;318:868–872. doi: 10.1016/j.bbrc.2004.04.103. [DOI] [PubMed] [Google Scholar]
- Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick JA, Ayala AG, Shuster JJ, Abelson HT, Simone JV, Vietti TJ. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314:1600–1606. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- Mandal D, Srivastava A, Mahlum E, Desai D, Maran A, Yaszemski M, Jalal SM, Gitelis S, Bertoni F, Damron T, Irwin R, O’Connor M, Schwartz H, Bolander ME, Sarkar G. Severe suppression of Frzb/sFRP3 transcription in osteogenic sarcoma. Gene. 2007;386:131–138. doi: 10.1016/j.gene.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, He K, Sridhara R, Abraham S, Booth BP, Verbois L, Morse DE, Jee JM, Pope S, Harapanhalli RS, Dagher R, Farrell A, Justice R, Pazdur R. Vorinostat for treatment of cutaneous manifestations of advanced primary cutaneous T-cell lymphoma. Clin Cancer Res. 2007;13:2318–2322. doi: 10.1158/1078-0432.CCR-06-2672. [DOI] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Richon V, Sullivan DM, Munster PN. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–237. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- Marchion DC, Bicaku E, Daud AI, Sullivan DM, Munster PN. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005;65:3815–3822. doi: 10.1158/0008-5472.CAN-04-2478. [DOI] [PubMed] [Google Scholar]
- Marsoni S, Damia G, Camboni G. A work in progress: The clinical development of histone deacetylase inhibitors. Epigenetics. 2008;3:164–171. doi: 10.4161/epi.3.3.6253. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Miyake K, Yoshizumi T, Imura S, Sugimoto K, Batmunkh E, Kanemura H, Morine Y, Shimada M. Expression of hypoxia-inducible factor-1 alpha, histone deacetylase 1, and metastasis-associated protein 1 in pancreatic carcinoma: Correlation with poor prognosis with possible regulation. Pancreas. 2008;36:e1–e9. doi: 10.1097/MPA.0b013e31815f2c2a. [DOI] [PubMed] [Google Scholar]
- Mueller F, Fuchs B, Kaser-Hotz B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007;27:155–164. [PubMed] [Google Scholar]
- Munster P, Marchion D, Bicaku E, Schmitt M, Lee JH, DeConti R, Simon G, Fishman M, Minton S, Garrett C, Chiappori A, Lush R, Sullivan D, Daud A. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: A clinical and translational study. J Clin Oncol. 2007;25:1979–1985. doi: 10.1200/JCO.2006.08.6165. [DOI] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: Lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15:3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- Ptitsyn AA, Weil MM, Thamm DH. Systems biology approach to identification of biomarkers for metastatic progression in cancer. BMC Bioinformatics. 2008;9(Suppl 9):S8. doi: 10.1186/1471-2105-9-S9-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikimaru T, Taketomi A, Yamashita Y, Shirabe K, Hamatsu T, Shimada M, Maehara Y. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72:69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- Rosen G, Marcove RC, Huvos AG, Caparros BI, Lane JM, Nirenberg A, Cacavio A, Groshen S. Primary osteogenic sarcoma: Eight-year experience with adjuvant chemotherapy. J Cancer Res Clin Oncol. 1983;106(Suppl):55–67. doi: 10.1007/BF00625054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EM, Guo Y, Tu K, Xie J, Zi X, Hoang BH. Wnt inhibitory factor 1 decreases tumorigenesis and metastasis in osteosarcoma. Mol Cancer Ther. 2010;9:731–741. doi: 10.1158/1535-7163.MCT-09-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. J Bone Miner Res. 2005;20:2254–2263. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Nair AK, Staggs R, Lamblin AF, Westendorf JJ. Gene profile analysis of osteoblast genes differentially regulated by histone deacetylase inhibitors. BMC Genomics. 2007;8:362. doi: 10.1186/1471-2164-8-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi TB, Magner WJ, Nazmul A, Khan H. Epigenetic regulation of immune escape genes in cancer. Cancer Immunol Immunother. 2006;55:1159–1184. doi: 10.1007/s00262-006-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij J, Pinedo HM. Mitomycin C: Mechanism of action, usefulness and limitations. Anticancer Drugs. 1990;1:5–13. [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Rocken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008a;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008b;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: Specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008c;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ. Histone deacetylases in control of skeletogenesis. J Cell Biochem. 2007;102:332–340. doi: 10.1002/jcb.21486. [DOI] [PubMed] [Google Scholar]
- Wittenburg L, Bisson L, Rose B, Thamm DH. The histone deacetylase inhibitor valproic acid sensitizes human and canine osteosarcoma to doxorubicin. Cancer Chemother Pharmacol. 2011;67(1):83–92. doi: 10.1007/s00280-010-1287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, Mak HC, McCuine S, Tagne J-B, Agarwal M, Ozier O, Begley TJ, Samson LD, Ideker T. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Yan XB, Yang DS, Gao X, Feng J, Shi ZL, Ye Z. Caspase-8 dependent osteosarcoma cell apoptosis induced by proteasome inhibitor MG132. Cell Biol Int. 2007;31:1136–1143. doi: 10.1016/j.cellbi.2007.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.