Abstract
Purpose
Cancer stem cells (CSCs) are thought to be responsible for tumor progression, metastasis, and recurrence. HER2 overexpression is associated with increased CSCs, which may explain the aggressive phenotype and increased likelihood of recurrence for HER2+ breast cancers. Telomerase is reactivated in tumor cells, including CSCs, but has limited activity in normal tissues, providing potential for telomerase inhibition in anti-cancer therapy. The purpose of this study was to investigate the effects of a telomerase antagonistic oligonucleotide, imetelstat (GRN163L), on CSC and non-CSC populations of HER2+ breast cancer cell lines.
Methods
The effects of imetelstat on CSC populations of HER2+ breast cancer cells were measured by ALDH activity and CD44/24 expression by flow cytometry as well as mammosphere assays for functionality. Combination studies in vitro and in vivo were utilized to test for synergism between imetelstat and trastuzumab.
Results
Imetelstat inhibited telomerase activity in both subpopulations. Moreover, imetelstat alone and in combination with trastuzumab reduced the CSC fraction and inhibited CSC functional ability, as shown by decreased mammosphere counts and invasive potential. Tumor growth rate was slower in combination treated mice compared to either drug alone. Additionally, there was a trend toward decreased CSC marker expression in imetelstat treated xenograft cells compared to vehicle control. Furthermore, the observed decrease in CSC marker expression occurred prior to and after telomere shortening, suggesting imetelstat acts on the CSC subpopulation in telomere length independent and dependent mechanisms.
Conclusions
Our study suggests addition of imetelstat to trastuzumab may enhance the effects of HER2 inhibition therapy, especially in the CSC population.
Keywords: Telomerase, breast cancer, HER2+, cancer stem cells, trastuzumab
INTRODUCTION
The cancer stem cell (CSC) hypothesis postulates that many cancers, including breast cancer, are hierarchically organized and a subpopulation of cells within the tumor possess the basic properties of stem cells, the ability to self-renew and differentiate [1,2]. CSCs also have other stem cell-like properties including the active expression of telomerase, anti-apoptotic pathway activation, increased activity of membrane transporters, and an increased ability to migrate [3]. Evidence suggests that CSCs may be responsible for tumor progression, metastasis, chemotherapy and radiotherapy resistance, and subsequent tumor recurrence [4-8]. Breast CSCs can be identified using the cell surface marker expression CD44+/CD24− or elevated enzymatic activity of aldehyde dehydrogenase (ALDH) [1,9].
Multiple studies suggest Human Epidermal Growth Factor Receptor 2 (HER2) plays an important role in regulating the CSC population in HER2+ breast cancer. HER2 overexpression and ALDH expression are significantly correlated in human breast cancer patient samples [9]. The CSC subpopulation in HER2 overexpressing breast cancer cell lines expresses the highest levels of HER2 protein without HER2 gene amplification changes [10]. Additionally, HER2 overexpression expands the normal breast epithelial stem/early progenitor cell population, as well as the CSC population in malignant breast cells, resulting in increased tumorigenicity and invasiveness with HER2 amplification [11]. HER2 blockade via trastuzumab or HER2/EGFR blockade via lapatinib decreases the CSC population [10,11]. Neoadjuvant trastuzumab significantly increases pathologic complete response rate compared to chemotherapy alone, suggesting a reduction in the CSC population [12,13]. In contrast to chemotherapy, lapatinib reduced the CSC population in the neoadjuvant setting, although this decrease was not statistically significant [7].
A hallmark of the cancer cell is its limitless replicative potential, achieved almost exclusively by telomere maintenance via telomerase reactivation [14]. Telomerase, the reverse transcriptase enzyme, is absent or lowly expressed in most normal somatic cells, but is highly expressed in cancer and enables malignant cells to maintain their telomere length just above the critically short threshold and thereby avoid senescence and apoptosis [15-18]. The differential expression between normal cells and cancer cells and the shorter telomere length of cancer cells makes telomerase inhibition a striking target for potential cancer therapeutics [19]. This potential led to the development of imetelstat (GRN163L), a lipidated 13-mer oligonucleotide N3′→P5′ thio phosphoramidate [20,21]. Imetelstat binds with high affinity to the template region of the RNA component of human telomerase resulting in competitive inhibition of telomerase enzymatic activity [22,23]. Our group and others have shown imetelstat alone, and in combination with chemotherapeutic agents or irradiation, can inhibit telomerase in a variety of tumor cells and compromise cancer cell viability and growth, both in vitro and in vivo [24-32].
Telomerase is expressed in both bulk cancer cells and CSCs, suggesting CSCs could be sensitive to telomerase inhibition therapy [6,33]. Imetelstat has been shown to target the CSC population in a number of tumor types [34-37]. While these studies investigated changes in marker expression, spheroid formation, and tumor growth in vivo after imetelstat pretreatment, the effect of telomerase inhibition on invasion and metastases was not addressed nor the effect of imetelstat in combination with standard therapies on the CSC population. Telomerase inhibitors are most effective when used in combination, likely due to the long lag time to achieve telomere shortening [38]. Our laboratory has shown imetelstat can augment the effects of trastuzumab and restore sensitivity in trastuzumab-resistant breast cancer cell lines [27].
In this study, we investigated the effect of imetelstat and trastuzumab treatment in HER2+ breast cancer cell lines. CSCs have active telomerase that can be inhibited by imetelstat treatment. Imetelstat alone can decrease the percentage of CSCs, as well as inhibit mammosphere formation. Additionally, we found imetelstat and trastuzumab combination treatment decreases the CSC population, mammosphere formation, invasive potential, and tumor growth in vivo.
MATERIALS AND METHODS
Reagents
The telomerase template antagonist, Imetelstat (GRN163L, 5'-Palm-TAGGGTTAGACAA-NH2-3'), and its complimentary control oligonucleotide with the same chemistry (Sense, 5'-Palm-ATCCAATCTGTT-NH2-3') were provided by Geron Corp and were prepared as previously described [23]. Trastuzumab was provided by the Indiana University Simon Cancer Center (IUSCC) Infusion Pharmacy.
Cell Culture
HCC1569 and HCC1954 breast cancer cell lines were purchased from ATCC (CRL-2330 and CRL-2338) and cultured in RPMI media (Corning cellgro) containing 10% fetal bovine serum (FBS, Fisher Scientific). SKBR3 and trastuzumab-resistant SKBR3-R pool 1 cells were a gift from Dr. Francisco Esteva (MD Anderson Cancer Center) and were cultured in DMEM/F12 media containing 10% FBS. Resistant cells (SKBR3-R) were cultured with the addition of 4 µg/mL trastuzumab [39]. TMD-231 cells were a gift from Dr. Harikrishna Nakshatri (Indiana University School of Medicine) [40] and were cultured in DMEM media containing 10% FBS.
Treatment with Imetelstat and/or Trastuzumab
Cells were allowed to attach overnight prior to drug treatment. To refresh imetelstat, media was spiked with imetelstat every 3 days and all cells were counted and passaged every 6-7 days. For the CSC marker expression studies, cells were treated with 2.5 µM imetelstat and/or trastuzumab [0.625 µM (1 trastuzumab: 4 imetelstat) for HCC1954 and SKBR3 cells and 0.3125 µM (1 trastuzumab: 8 imetelstat) for HCC1569 cells]. For the cell sorting experiments, cells were treated with 2.5 µM imetelstat or sense oligonucleotide for 3 days for telomerase activity and 6 weeks for telomere length.
Flow Cytometry and Fluorescence Activated Cell Sorting (FACS)
Flow cytometric analysis was performed using an LSRII 407 nm laser cytometer (BD Biosciences) and cell sorting using a Special Order Research Product FACSAria sorter (BD Biosciences). Cells were stained with APC-H7-conjugated CD44, PE-Cy7- conjugated CD24, and violet LIVE/DEAD fixable cell stain (all antibodies from BD Biosciences and viability stain from Life Technologies). FMO (fluorescence minus one) controls were used to determine appropriate gates. The Aldefluor assay was used to measure and separate cells based on ALDH activity according to manufacturer’s guidelines (StemCell Technologies). Control samples treated with DEAB (diethylaminobenzaldehyde) were used for gating the negative population. FlowJo software was used for all analyses.
Methods for telomerase activity and telomere length determination, methylene blue cell proliferation for combination studies, mammosphere cultures, invasion assays, xenograft animal studies, and statistical analyses are available in supplemental information.
RESULTS
Long term treatment with imetelstat inhibits cell growth
To study the effects of imetelstat treatment in HER2+ breast cancer cell lines, we first measured cumulative population doublings in two HER2+ cell lines not previously studied for telomerase inhibition, HCC1569 and HCC1954. Cumulative population doublings of the HCC1569 and HCC1954 cell lines statistically significantly differed between imetelstat and untreated cells at the time point indicated by the arrow and remained different throughout the remainder of the experiment (Fig, S1, two-way repeated measures ANOVA, p < 0.05). HCC1569 cells stopped doubling and reached the stationary phase or plateau of the population doubling graph after 17 weeks of treatment. HCC1954 reached the stationary phase after 10 weeks of treatment. The sense oligonucleotide control had a minor effect on cell proliferation, but it occurred much later than imetelstat and did not inhibit cells from continuing to proliferate. Although cumulative population doublings began to differ sooner in the HCC1569 cells, it took longer to reach the plateau than in HCC1954 cells. The HCC1569 cells were slower growing, doubling about 4 times per week versus 5 times in the HCC1954, and have longer baseline telomere length, which may explain why a longer treatment regimen is required to reach the stationary phase in this cell line.
HER2+ CSCs have active telomerase and are sensitive to telomerase inhibition via imetelstat
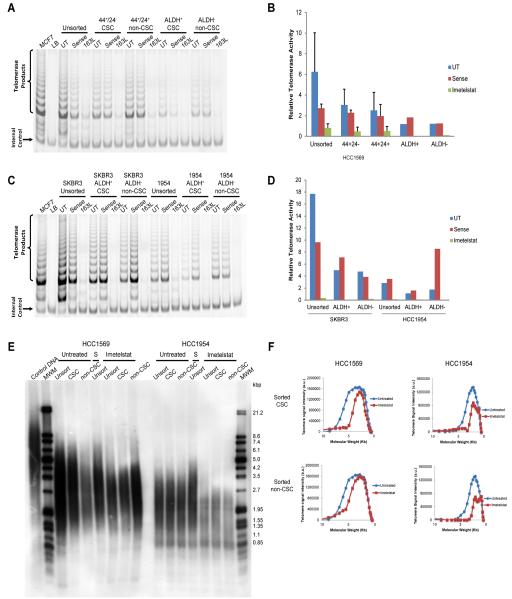
Previous studies have reported similar telomerase activity between CSCs and their bulk tumor cell counterparts, as well as similarities in telomere length [35,6]. We determined whether our HER2+ cell lines also had similar telomerase activity and telomere length in the CSC, non-CSC (bulk tumor cells), and unsorted populations. HCC1569 cell line was flow sorted based on either CD44/CD24 expression or ALDH enzymatic activity. Unsorted, non-CSC, and CSC populations all had active telomerase (Fig. 1a-b). There was no difference in relative telomerase activity between CSC, non-CSC, and unsorted cells in the HCC1569 cell line based on CD44/CD24 expression (Fig. 1b, one-way ANOVA, p > 0.05 for untreated, sense, and imetelstat groups). We also tested whether imetelstat can inhibit telomerase activity in the different subpopulations. Imetelstat (163L) was able to abolish telomerase activity in both the CSC and non-CSC populations; however, as expected, the sense oligonucleotide control did not affect telomerase activity (Fig. 1a-b). ALDH enzymatic activity was used to sort the HCC1954 cell line, as well as another HER2+ breast cancer cell line we have previously used for telomerase inhibition studies, the SKBR3 cell line [27]. Again, we found unsorted, non-CSC, and CSC populations had active telomerase that imetelstat was able to inhibit in both subpopulations (Fig. 1c-d). In line with the previous reports, we found similar average telomere length between the CSC and non-CSC populations in our HER2+ breast cancer cell lines (Fig. 1e, S2). Six weeks of imetelstat treatment decreased average telomere length in the HCC1954 cell line, but not the HCC1569 cell line (Fig. 1e, S2). HCC1569 cells had a longer baseline telomere length than HCC1954, possibly explaining why we did not see differences in average telomere length at this time point of treatment. However, there appeared to be a shortening of longer telomeres in both cell lines with imetelstat treatment, shown by a shift downward in the telomere smear (Fig. 1e). Indeed, examination of the densitometry of the telomere smears revealed shortening of longer telomeres with imetelstat treatment, shown by a shift in the curve toward a smaller molecular weight (Fig. 1f).
Fig. 1.
CSCs have active telomerase that can be inhibited by imetelstat leading to telomere shortening. A) Detection of telomerase activity by TRAP assay of HCC1569 flow sorted CSCs and non-CSCs by marker expression following 3 day treatment. UT= untreated cells, 163L= imetelstat treated cells B) Average quantification of relative telomerase activity from A) using ratio of telomerase products to internal standard, n = 3 for CD44/CD24 sorting and n = 2 for ALDH sorting, p = 0.2344 for UT, p = 0.4720 for sense, p = 0.5872 for 163L (one-way ANOVA comparing unsorted, CD44+/CD24−, and CD44+/CD24+) C) TRAP assay of SKBR3 and HCC1954 flow sorted CSCs and non-CSCs following 4 day treatment. D) Average quantification of relative telomerase activity from C), n = 2. E) Telomere length determination by Terminal restriction fragment analysis of HCC1569 and HCC1954 flow sorted cells following 6 weeks of treatment. MWM= molecular weight marker, Unsort= unsorted cells, S= sense oligonucleotide control, HCC1569 CSC= CD44+/CD24−, HCC1569 non-CSC= CD44+/CD24+, HCC1954 CSC= ALDH+, HCC1954 non-CSC= ALDH− . F) Telomere length quantification using TELORUN of sorted HCC1569 (left panels) and HCC1954 (right panels) cells pretreated with imetelstat or untreated for 6 weeks prior to sorting CSC (top panels) and non-CSC (bottom panels) populations. Shift in Molecular Weight (telomere length in Kb) after imetelstat treatment (red lines) shows telomere shortening of the longer telomeres within each sample. a.u. = arbitrary units.
Telomerase inhibition can decrease CSCs and limit mammosphere formation
The use of breast cancer cell lines to study CSCs has been validated [41,42]. SKBR3, HCC1954, and HCC1569 cell lines were subjected to flow cytometry analysis of CD44/CD24 marker expression and ALDH enzymatic activity. The HCC1569 cell line was positive for CD44 expression and contained positive and negative CD24 expressing cells, allowing us to use this cell line to measure the CD44+/CD24− CSC population (Fig. S3a). Both HCC1954 and SKBR3 cell lines did not have a measureable CD44+/CD24− population (Fig. S3b-c). As predicted by the correlation of HER2 and ALDH, all three cell lines had ALDH+ CSC subpopulations [9].
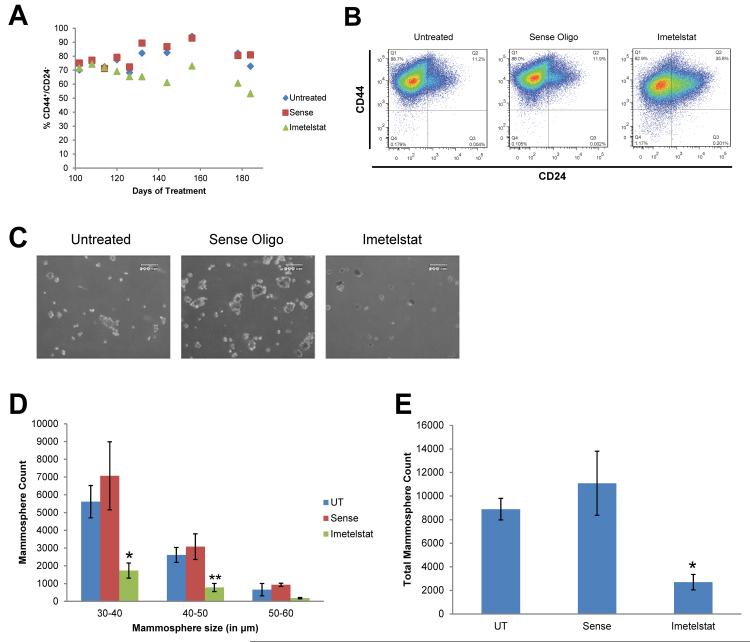
To determine the effect of telomerase inhibition on the CSC population, we measured CD44+/CD24− expression in the HCC1569 cell line. Long-term treatment with imetelstat decreased the CSC population by more than twenty percent, whereas the sense oligonucleotide control had no effect on the CSC population (Fig. 2a-b). Furthermore, imetelstat pretreatment significantly decreased mammosphere count, an in vitro assessment of stem cell function, compared to untreated and sense controls (Fig. 2c-e, one-way ANOVA, p < 0.05).
Fig. 2.
Imetelstat but not the sense oligonucleotide control decreases the CSC population and mammosphere counts. A) Scatter plot of CSC marker expression following treatment. B) Flow cytometry analysis of CSC marker expression. C) Representative images of mammosphere cultures following pretreatment. D) Primary mammosphere count grouped by mammosphere size (n=3), average ± SD, one-way ANOVA, * p<0.05, ** p< 0.01 compared to untreated. E) Sum of mammosphere size groups as total mammosphere count, average ± SD, ANOVA, * p< 0.05 compared to untreated.
Imetelstat augments the effects of trastuzumab in HER2+ breast cancer cell lines
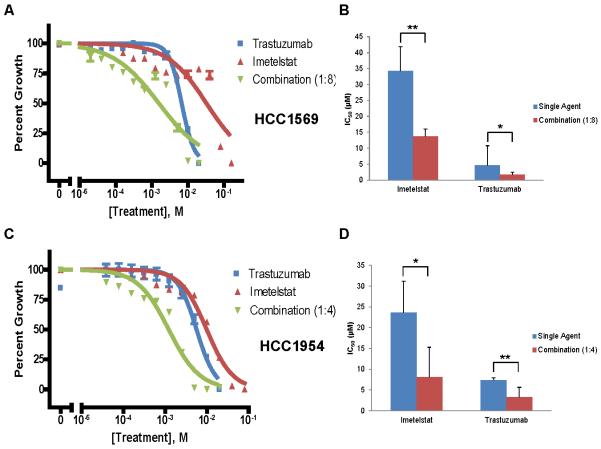
Our lab has previously reported a synergistic effect of imetelstat and trastuzumab combination therapy in vitro [27]. We next verified this effect applied to the HCC1569 and HCC1954 cell lines, which have previously been classified as having a de novo resistance to trastuzumab [43]. IC50 values of trastuzumab and imetelstat were determined for both cell lines and used to select drug ratios for combination treatments. Trastuzumab and imetelstat combination shifted the dose-response curve and significantly decreased the concentration of both drugs needed to achieve the IC50 (Fig. 3). Moreover, the combination index showed a synergistic effect (CI < 1) at most concentrations tested (Table 1). Although these cells are reported to be innately resistant to trastuzumab and we did notice little effect on cell proliferation at lower concentrations, we were able to determine IC50 values and showed combination treatment decreased the IC50 value for both trastuzumab and imetelstat. These combination studies suggest imetelstat can augment the effects of trastuzumab.
Fig. 3.
Imetelstat augments the effects of trastuzumab in HER2+ breast cancer cells. A) Representative dose-response curve for HCC1569 cells treated with imetelstat, trastuzumab, or 1:8 combination for 5 days. B) Average IC50 values of each drug as a single agent or in combination, Average + SD, n = 4, p = 0.0055 for imetelstat, p = 0.0470 for trastuzumab (Student’s t-test). C) Representative dose-response curve for HCC1954 cells treated with imetelstat, trastuzumab, or 1:4 combination for 5 days. D) Average IC50 values of each drug as a single agent or in combination, Average + SD, n = 5, p = 0.0107 for imetelstat, p = 0.0058 for trastuzumab (Student’s t-test). * = p < 0.05, ** = p < 0.01.
Table 1.
Combination Index Values for Trastuzumab and Imetelstat Combination Treatment
| Cell Line | [Imetelstat] (μM) |
[Trastuzumab] (μM) |
Combination Index |
|---|---|---|---|
| HCC1569 | 0.156 | 0.020 | 0.466 |
| 0.313 | 0.039 | 0.908 | |
| 0.625 | 0.078 | 0.554 | |
| 1.25 | 0.156 | 0.334 | |
| 2.5 | 0.313 | 0.295 | |
| 5 | 0.625 | 0.313 | |
| 10 | 1.25 | 1.148 | |
| 20 | 2.5 | 9.013 | |
| 40 | 5 | 0.327 | |
| 80 | 10 | 0.060 | |
|
| |||
| HCC1954 | 0.156 | 0.039 | 0.137 |
| 0.313 | 0.078 | 0.228 | |
| 0.625 | 0.156 | 0.322 | |
| 1.25 | 0.313 | 0.654 | |
| 2.5 | 0.625 | 1.136 | |
| 5 | 1.25 | 1.343 | |
| 10 | 2.5 | 1.301 | |
| 20 | 5 | 0.823 | |
| 40 | 10 | 0.723 | |
| 80 | 20 | 0.917 | |
Imetelstat in combination with trastuzumab decreases the CSC population
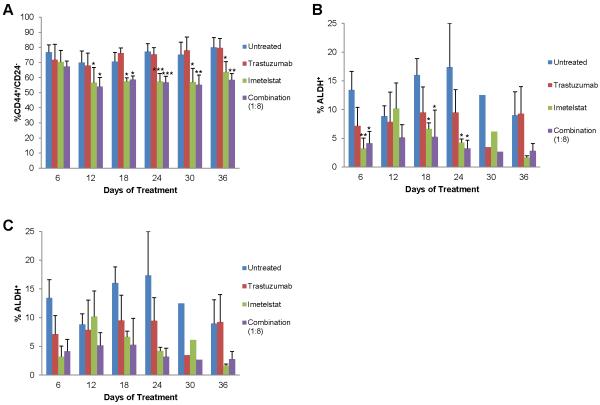
We next wanted to determine the effect of imetelstat and trastuzumab combination treatment on the CSC population. In accordance with Fig. 2, we found imetelstat alone was able to decrease the CSC population (Fig. 4a-c, green bars). Although trastuzumab alone did not affect the percentage of CD44+/CD24− CSCs (Fig. 4a, red bars), we found trastuzumab decreased the ALDH+ CSC population (Fig. 4b-c, red bars) as previously published [10,11]. Combination treatment significantly decreased the CD44+/CD24− CSC population in the HCC1569 cell line (p < 0.05 for 12-54 days of treatment) (Fig. 4a, purple bars). Moreover, combination treatment decreased the ALDH+ CSC population in all cell lines tested, although statistical analyses is hindered due to high variability of enzymatic activity (Fig. 4b-c, purple bars).
Fig. 4.
Imetelstat and trastuzumab combination treatment decreases CSCs. A) CSC marker expression of HCC1569 cells analyzed by flow cytometry. Average percent CD44+/CD24− + SD, n=3, one-way ANOVA, * p < 0.05, ** p < 0.01, *** p < 0.001 compared to untreated B) ALDH enzymatic activity of HCC1569 cells measured by flow cytometry. Average percent ALDH+ + SD when error bars are shown, n=3, one-way ANOVA, * p < 0.05, ** p < 0.01 compared to untreated C) ALDH enzymatic activity of HCC1954, SKBR3, and SKBR3-R cells measured by flow cytometry after 6 days of treatment. Average percent ALDH+ + SD, n=3
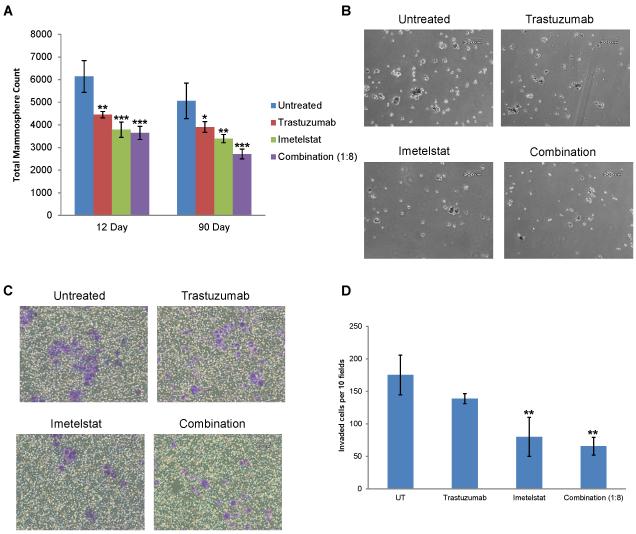
We next wanted to determine if the decrease in the CSC population after combination treatment resulted in decreased functional ability of the CSCs. We observed a significant decrease in mammosphere count and smaller spheroids following trastuzumab, imetelstat, and combination treatment after short term (12 days) and long term (90 days) pretreatment, suggesting CSC self-renewal is continually inhibited by these agents (Fig. 5a-b). Moreover, short term treatment with imetelstat may not lead to telomere shortening, but long term treatment certainly does, suggesting imetelstat may inhibit the self-renewal potential of CSCs in telomere length dependent and independent mechanisms. It has been reported that the CSC population has an increased invasive potential, an early step required for metastasis [44,41]. We performed invasion assays to determine if the decrease in CSCs following imetelstat and/or trastuzumab treatment correlated with decreased invasive potential. Twelve day imetelstat and combination treatments significantly decreased the invasive potential compared to untreated samples, with less than half as many cells invading (Fig. 5c-d).
Fig. 5.
Imetelstat and trastuzumab combination treatment inhibits mammosphere formation and invasive potential. A) Total mammosphere counts from 12 and 90 day pretreatment HCC1569 mammosphere cultures, Average ± SD, n=3, one-way ANOVA, * p < 0.05, ** p < 0.01, *** p < 0.001 compared to untreated. B) Representative Images of Primary Mammospheres cultured from HCC1569 cells, 4X Magnification, 200 µm scale bar. C) Representative images of invaded cells in purple, 10X magnification, 60 µm scale bar. D) Invaded cells in 10 random fields per well, Average ± SD, n=3, one-way ANOVA, ** p < 0.01 compared to untreated
Imetelstat and trastuzumab combination treatment decreases primary tumor growth in vivo
We and others have shown imetelstat treatment can decrease primary tumor growth in xenograft mouse models [24,28,30,31]. However, the effect of imetelstat and trastuzumab combination treatment on xenograft tumor growth has not previously been studied. We found a significant decrease in primary tumor growth of HCC1954 cells implanted into the mammary fat pad of NSG mice and then treated with combination of imetelstat and trastuzumab (Fig. 6a). However, the study was concluded, due to burdensome tumor volume in the vehicle control group, before differences in tumor volume between the trastuzumab alone and combination group could be observed. Tumor growth rates, calculated as the slope of the line of tumor volume versus days post inoculation, appeared to be different between combination and trastuzumab alone groups, suggesting addition of imetelstat can enhance the effects of trastuzumab on tumor growth (Fig. 6b). HCC1954 cells did not reliably metastasize following tertiary passaging in mice (data not shown); therefore, we studied the effect of imetelstat alone in a triple negative breast cancer cell line, TMD-231 [45]. The primary tumor tissue from this study was digested to generate a single cell suspension, which were then grown in monolayer culture or subjected to flow cytometry analysis of CSC marker expression. The PBS treated xenograft cells have a more mesenchymal-like phenotype with elongated morphology and reduced cell-cell contact, a phenotype typical of breast CSCs [46], whereas the imetelstat treated xenograft cells have more epithelial morphologic features, namely rounder with more cell-cell contact (Fig. S4c). Furthermore, there is a trend of decreased CSC marker expression in imetelstat treated xenograft cells compared to vehicle control, but this decrease was not statistically significant due to higher variability in the PBS group and small sample size (Fig. S4d). These data suggest imetelstat can target the CSC population in vivo.
Fig. 6.
Imetelstat, trastuzumab, and combination treatment decrease primary tumor growth in vivo. A) Tumor volume of HCC1954 cells by caliper measurements in PBS control (thrice weekly, n=9), trastuzumab (20 mg/kg twice weekly, n=10), imetelstat (30 mg/kg, thrice weekly, n=10), and combination (trastuzumab twice weekly and imetelstat thrice weekly, n=9) treated NSG mice. Average ± SEM B) Rate of tumor growth calculated from A) is the slope of line of the average tumor volume over time for each treatment group
DISCUSSION
Here, we show HER2+ CSCs have active telomerase that can be inhibited by imetelstat treatment, leading to telomere shortening. Imetelstat treatment alone, and in combination with trastuzumab, decreased the number of CSCs, as well as their functional ability, as shown by decreased mammosphere count and invasive potential. We report the first in vivo study of imetelstat and trastuzumab combination, in which the combination treatment has a slower tumor growth rate than either drug alone. Additionally, we found a trend toward lower CSC marker expression in imetelstat treated xenograft cells compared to PBS control using a Triple Negative breast cancer cell line. This study is the first to propose the decreased metastases and invasion following imetelstat treatment are due to decreased CSCs. Moreover, we are the first to investigate the effect of imetelstat and trastuzumab combination therapy on the CSC population.
Interestingly, we observed a decrease in the CSC population following imetelstat treatment both prior to shortening of average telomere length and after telomere shortening, suggesting the effect of imetelstat on the CSC population may occur in telomere length dependent and independent mechanisms, as also reported in multiple myeloma [47]. This study suggested imetelstat can target the CSC population by impacting essential stem cell pathways during stem cell-fate decisions independent of telomere length and that imetelstat modulates CSC growth and self-renewal through decreasing telomere length [47]. In our study, the CSC and non-CSC populations have similar telomerase activity and average telomere length, discounting the possibility that CSCs have shorter baseline telomeres and thus require less time to reach critically short lengths. While we did not observe shortening of average telomere length after 42 days of treatment in the HCC1569 cells (Fig. S2), imetelstat did shorten the longer telomeres at this time point (Fig. 1f). Moreover, it has recently been published that 19 day treatment of imetelstat affected telomere size distribution [32]; therefore, we cannot distinguish whether the decrease in CSCs, mammospheres, and invasive potential after 12 day treatment is due to telomere length dependent or independent mechanisms.
A potential telomere length independent mechanism to explain the effect of imetelstat on CSCs could be due to reversal of the epithelial mesenchymal transition (EMT). EMT activation is associated with maintenance of stem cell properties and acquisition of invasive and metastatic properties; EMT is able to generate CSCs [48,46,49]. We observed a change in cellular phenotype of imetelstat treated xenograft cells compared to PBS treated (Fig. S4c). EMT is crucial for the initial steps of metastasis; however, cells must then undergo a reversion of EMT termed MET to colonize a metastatic lesion [50]. Interestingly, a recent study investigated the off-target effects of imetelstat and found imetelstat disrupted the cytoskeleton through changes in actin, tubulin, and intermediate filament organization; furthermore, imetelstat decreased MMP2 expression and subsequently invasive ability of lung cancer cells [51]. Of note, this study also found imetelstat treatment resulted in a loss of E-cadherin, which could suggest cells have undergone EMT, but are unable to subsequently undergo MET and cannot adhere to colonize a metastatic lesion.
While EMT has been closely associated with CSCs, other studies suggest cells that undergo EMT are not the cells responsible for metastases [52]. Studies were carried out by Liu and collaborators to better understand the relationship between EMT, MET, and CSCs and furthermore distinguish between CD44+/CD24− CSCs and ALDH+ CSCs [53]. The researchers found many genes displayed reciprocal expression patterns between CD44+/CD24− and ALDH+ cell populations. However, a set of transcripts expressed in CD44+/CD24− and ALDH+ cell populations overlapped with spheroid-forming cells, likely representing genes involved in stem cell function. These results suggest two stem cell compartments in human breast cancers are identified by CSC markers. Moreover, CD44+/CD24− CSCs were significantly enriched in EMT- associated genes and ALDH+ CSCs were elevated in genes associated with the epithelial-like state [53]. Furthermore, purified CD44+/CD24− or ALDH+ cells were able to generate heterogeneous populations and recapitulate the populations present in the original cell line, suggesting CSCs display plasticity and are able to reversibly transition between the mesenchymal-like CD44+/CD24− CSC state and the epithelial-like ALDH+ CSC state [53]. This switching between two distinct EMT and non-EMT CSC populations is supported by work in squamous cell carcinoma in which CSCs behaved likewise [54]. Of note, it may be important to target both CSC populations as they can alternate between the two states [53]. Importantly, we show imetelstat alone and in combination with trastuzumab decreased CD44+/CD24− and ALDH+ CSCs in a number of cell lines and most notably both CSC populations in the HCC1569 cell line.
In summary, our results show imetelstat treatment of HER2+ breast cancer cells leads to telomerase inhibition and decreases the CSC population alone and in combination with trastuzumab. Markedly, imetelstat and trastuzumab combination is able to decrease the percentage of CD44+/CD24− and ALDH+ CSCs in the same cell line, suggesting this combination would be effective in targeting CSCs because cells would not benefit from transitioning to the other CSC state, which could potentially occur if we were only able to target one CSC state. Imetelstat decreased the self-renewal potential as well as invasive potential of the CSC population alone and in combination with trastuzumab. CSCs are thought to be responsible for recurrent and metastatic disease and our study suggests adding imetelstat to trastuzumab treatment may provide a more durable clinical response for HER2+ breast cancer patients.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank members of the Herbert Lab for helpful discussion preparing this manuscript. We thank Geron Corporation for generously providing the imetelstat and sense oligonucleotides, Dr. Francisco Esteva for kindly sharing the SKBR3 and SKBR3-R cell lines, Dr. Harikrishna Nakshatri for kindly sharing the TMD-231 cell line, Dr. Harlan Shannon for help with the combination studies, Dr. George Sandusky for help with histology, Dr. Hiromi Tanaka for help with TeloTAGGG analysis, the Indiana University Simon Cancer Center (IUSCC) flow cytometry core facility for their services and expertise, and the IUSCC infusion pharmacy for generously providing the trastuzumab. This investigator was supported, in part, by the National Institutes of Health, National Research Service Award Number T32 HL007910- Basic Science Studies on Gene Therapy of Blood Diseases. This work was also supported in part by a grant from Susan G. Komen for the Cure®, an IUSCC Cancer Biology Training Program Predoctoral Fellowship, a grant from the Mary Kay Ash Charitable Foundation, and the Indiana Genomics Initiative (INGEN; supported in part by the Lilly Endowment, Inc). We are also grateful for the philanthropic support made to the Herbert laboratory through IUSCC in memory of Carol Herbert.
Footnotes
CONFLICT OF INTEREST
Imetelstat and sense oligonucleotides were generously provided by the Geron Corporation. The authors declare they have no other conflicts of interest.
ETHICAL STANDARDS
The authors declare that the experiments performed comply with the current laws.
REFERENCES
- 1.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. doi:10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho RW, Clarke MF. Recent advances in cancer stem cells. Current opinion in genetics & development. 2008;18(1):48–53. doi: 10.1016/j.gde.2008.01.017. doi:10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea--a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895-1886. doi:10.1158/0008-5472.can-05-3153. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. doi:10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, Datar RH, Cote RJ. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12(19):5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. doi:10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 6.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. doi:10.1158/0008-5472.can-05-0626. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. doi:10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 8.Phillips TM, McBride WH, Pajonk F. The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98(24):1777–1785. doi: 10.1093/jnci/djj495. doi:10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 9.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. doi:10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnifico A, Albano L, Campaner S, Delia D, Castiglioni F, Gasparini P, Sozzi G, Fontanella E, Menard S, Tagliabue E. Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab. Clin Cancer Res. 2009;15(6):2010–2021. doi: 10.1158/1078-0432.CCR-08-1327. doi:10.1158/1078-0432.ccr-08-1327. [DOI] [PubMed] [Google Scholar]
- 11.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27(47):6120–6130. doi: 10.1038/onc.2008.207. doi:10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buzdar AU, Ibrahim NK, Francis D, Booser DJ, Thomas ES, Theriault RL, Pusztai L, Green MC, Arun BK, Giordano SH, Cristofanilli M, Frye DK, Smith TL, Hunt KK, Singletary SE, Sahin AA, Ewer MS, Buchholz TA, Berry D, Hortobagyi GN. Significantly higher pathologic complete remission rate after neoadjuvant therapy with trastuzumab, paclitaxel, and epirubicin chemotherapy: results of a randomized trial in human epidermal growth factor receptor 2-positive operable breast cancer. J Clin Oncol. 2005;23(16):3676–3685. doi: 10.1200/JCO.2005.07.032. doi:10.1200/JCO.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Korkaya H, Wicha MS. HER2 and breast cancer stem cells: more than meets the eye. Cancer Res. 2013;73(12):3489–3493. doi: 10.1158/0008-5472.CAN-13-0260. doi:10.1158/0008-5472.can-13-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Science. 5193. Vol. 266. New York, NY: 1994. Specific association of human telomerase activity with immortal cells and cancer; pp. 2011–2015. [DOI] [PubMed] [Google Scholar]
- 16.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Science. 5349. Vol. 279. New York, NY: 1998. Extension of life-span by introduction of telomerase into normal human cells; pp. 349–352. [DOI] [PubMed] [Google Scholar]
- 17.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Molecular cell. 2004;14(4):501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13(18):2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nature reviews Drug discovery. 2006;5(7):577–584. doi: 10.1038/nrd2081. doi:10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 20.Gryaznov SM, Jackson S, Dikmen G, Harley C, Herbert BS, Wright WE, Shay JW. Oligonucleotide conjugate GRN163L targeting human telomerase as potential anticancer and antimetastatic agent. Nucleosides Nucleotides Nucleic Acids. 2007;26(10-12):1577–1579. doi: 10.1080/15257770701547271. doi:10.1080/15257770701547271. [DOI] [PubMed] [Google Scholar]
- 21.Herbert BS, Pongracz K, Shay JW, Gryaznov SM. Oligonucleotide N3'-->P5' phosphoramidates as efficient telomerase inhibitors. Oncogene. 2002;21(4):638–642. doi: 10.1038/sj.onc.1205064. doi:10.1038/sj.onc.1205064. [DOI] [PubMed] [Google Scholar]
- 22.Asai A, Oshima Y, Yamamoto Y, Uochi TA, Kusaka H, Akinaga S, Yamashita Y, Pongracz K, Pruzan R, Wunder E, Piatyszek M, Li S, Chin AC, Harley CB, Gryaznov S. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer research. 2003;63(14):3931–3939. [PubMed] [Google Scholar]
- 23.Herbert BS, Gellert GC, Hochreiter A, Pongracz K, Wright WE, Zielinska D, Chin AC, Harley CB, Shay JW, Gryaznov SM. Lipid modification of GRN163, an N3'-->P5' thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene. 2005;24(33):5262–5268. doi: 10.1038/sj.onc.1208760. doi:10.1038/sj.onc.1208760. [DOI] [PubMed] [Google Scholar]
- 24.Dikmen ZG, Gellert GC, Jackson S, Gryaznov S, Tressler R, Dogan P, Wright WE, Shay JW. In vivo inhibition of lung cancer by GRN163L: a novel human telomerase inhibitor. Cancer Res. 2005;65(17):7866–7873. doi: 10.1158/0008-5472.CAN-05-1215. doi:10.1158/0008-5472.can-05-1215. [DOI] [PubMed] [Google Scholar]
- 25.Dikmen ZG, Wright WE, Shay JW, Gryaznov SM. Telomerase targeted oligonucleotide thio-phosphoramidates in T24-luc bladder cancer cells. J Cell Biochem. 2008;104(2):444–452. doi: 10.1002/jcb.21635. doi:10.1002/jcb.21635. [DOI] [PubMed] [Google Scholar]
- 26.Djojosubroto MW, Chin AC, Go N, Schaetzlein S, Manns MP, Gryaznov S, Harley CB, Rudolph KL. Hepatology. 5. Vol. 42. Baltimore, Md: 2005. Telomerase antagonists GRN163 and GRN163L inhibit tumor growth and increase chemosensitivity of human hepatoma; pp. 1127–1136. doi:10.1002/hep.20822. [DOI] [PubMed] [Google Scholar]
- 27.Goldblatt EM, Erickson PA, Gentry ER, Gryaznov SM, Herbert BS. Lipid-conjugated telomerase template antagonists sensitize resistant HER2-positive breast cancer cells to trastuzumab. Breast Cancer Res Treat. 2009;118(1):21–32. doi: 10.1007/s10549-008-0201-4. doi:10.1007/s10549-008-0201-4. [DOI] [PubMed] [Google Scholar]
- 28.Goldblatt EM, Gentry ER, Fox MJ, Gryaznov SM, Shen C, Herbert BS. The telomerase template antagonist GRN163L alters MDA-MB-231 breast cancer cell morphology, inhibits growth, and augments the effects of paclitaxel. Mol Cancer Ther. 2009;8(7):2027–2035. doi: 10.1158/1535-7163.MCT-08-1188. doi:10.1158/1535-7163.mct-08-1188. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Millan J, Goldblatt EM, Gryaznov SM, Mendonca MS, Herbert BS. Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int J Radiat Oncol Biol Phys. 2007;67(3):897–905. doi: 10.1016/j.ijrobp.2006.09.038. doi:10.1016/j.ijrobp.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 30.Hochreiter AE, Xiao H, Goldblatt EM, Gryaznov SM, Miller KD, Badve S, Sledge GW, Herbert BS. Telomerase template antagonist GRN163L disrupts telomere maintenance, tumor growth, and metastasis of breast cancer. Clin Cancer Res. 2006;12(10):3184–3192. doi: 10.1158/1078-0432.CCR-05-2760. doi:10.1158/1078-0432.ccr-05-2760. [DOI] [PubMed] [Google Scholar]
- 31.Shammas MA, Koley H, Bertheau RC, Neri P, Fulciniti M, Tassone P, Blotta S, Protopopov A, Mitsiades C, Batchu RB, Anderson KC, Chin A, Gryaznov S, Munshi NC. Telomerase inhibitor GRN163L inhibits myeloma cell growth in vitro and in vivo. Leukemia. 2008;22(7):1410–1418. doi: 10.1038/leu.2008.81. doi:10.1038/leu.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burchett KM, Yan Y, Ouellette MM. Telomerase inhibitor Imetelstat (GRN163L) limits the lifespan of human pancreatic cancer cells. PloS one. 2014;9(1):e85155. doi: 10.1371/journal.pone.0085155. doi:10.1371/journal.pone.0085155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju Z, Rudolph KL. European journal of cancer. 9. Vol. 42. Oxford; England: 2006. Telomeres and telomerase in cancer stem cells; pp. 1197–1203. 1990. doi:10.1016/j.ejca.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 34.Castelo-Branco P, Zhang C, Lipman T, Fujitani M, Hansford L, Clarke I, Harley CB, Tressler R, Malkin D, Walker E, Kaplan DR, Dirks P, Tabori U. Neural tumor-initiating cells have distinct telomere maintenance and can be safely targeted for telomerase inhibition. Clin Cancer Res. 2011;17(1):111–121. doi: 10.1158/1078-0432.CCR-10-2075. doi:10.1158/1078-0432.CCR-10-2075. [DOI] [PubMed] [Google Scholar]
- 35.Joseph I, Tressler R, Bassett E, Harley C, Buseman CM, Pattamatta P, Wright WE, Shay JW, Go NF. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010;70(22):9494–9504. doi: 10.1158/0008-5472.CAN-10-0233. doi:10.1158/0008-5472.can-10-0233. [DOI] [PubMed] [Google Scholar]
- 36.Marian CO, Wright WE, Shay JW. The effects of telomerase inhibition on prostate tumor-initiating cells. Int J Cancer. 2010;127(2):321–331. doi: 10.1002/ijc.25043. doi:10.1002/ijc.25043. [DOI] [PubMed] [Google Scholar]
- 37.Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Mickey BE, Wright WE, Shay JW, Bachoo RM. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16(1):154–163. doi: 10.1158/1078-0432.CCR-09-2850. doi:10.1158/1078-0432.ccr-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer treatment reviews. 2013;39(5):444–456. doi: 10.1016/j.ctrv.2012.06.007. doi:10.1016/j.ctrv.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Nahta R, Esteva FJ. In vitro effects of trastuzumab and vinorelbine in trastuzumab-resistant breast cancer cells. Cancer chemotherapy and pharmacology. 2004;53(2):186–190. doi: 10.1007/s00280-003-0728-3. doi:10.1007/s00280-003-0728-3. [DOI] [PubMed] [Google Scholar]
- 40.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, Broxmeyer HE, Nakshatri H. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278(24):21631–21638. doi: 10.1074/jbc.M300609200. doi:10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 41.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer research. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. doi:10.1158/0008-5472.can-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. doi:10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brien NA, Browne BC, Chow L, Wang Y, Ginther C, Arboleda J, Duffy MJ, Crown J, O'Donovan N, Slamon DJ. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9(6):1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. doi:10.1158/1535-7163.mct-09-1171. [DOI] [PubMed] [Google Scholar]
- 44.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr., Badve S, Nakshatri H. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. doi:10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, Bhat-Nakshatri P, Goswami C, Badve S, Nakshatri H. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res. 2013;73(18):5821–5833. doi: 10.1158/0008-5472.CAN-13-1080. doi:10.1158/0008-5472.CAN-13-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. doi:10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan SK, Wang Q, Tressler R, Harley C, Go N, Bassett E, Huff CA, Jones RJ, Matsui W. Telomerase inhibition targets clonogenic multiple myeloma cells through telomere length-dependent and independent mechanisms. PloS one. 2010;5(9) doi: 10.1371/journal.pone.0012487. doi:10.1371/journal.pone.0012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. doi:10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PloS one. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. doi:10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27(20):2192–2206. doi: 10.1101/gad.225334.113. doi:10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mender I, Senturk S, Ozgunes N, Akcali KC, Kletsas D, Gryaznov S, Can A, Shay JW, Dikmen ZG. Imetelstat (a telomerase antagonist) exerts offtarget effects on the cytoskeleton. International journal of oncology. 2013;42(5):1709–1715. doi: 10.3892/ijo.2013.1865. doi:10.3892/ijo.2013.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, Hu GF. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer Res. 2008;68(24):10377–10386. doi: 10.1158/0008-5472.CAN-08-1444. doi:10.1158/0008-5472.can-08-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D'Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC, Wicha MS. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem cell reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. doi:10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71(15):5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. doi:10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 55.Wright WE, Shay JW, Piatyszek MA. Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic acids research. 1995;23(18):3794–3795. doi: 10.1093/nar/23.18.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat Protoc. 2006;1(3):1583–1590. doi: 10.1038/nprot.2006.239. doi:10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- 57.Herbert BS, Shay JW, Wright WE. Analysis of telomeres and telomerase. Current protocols in cell biology. 2003 doi: 10.1002/0471143030.cb1806s20. editorial board, Juan S Bonifacino [et al] Chapter 18:Unit 18 16. doi:10.1002/0471143030.cb1806s20. [DOI] [PubMed] [Google Scholar]
- 58.Oliver MH, Harrison NK, Bishop JE, Cole PJ, Laurent GJ. A rapid and convenient assay for counting cells cultured in microwell plates: application for assessment of growth factors. Journal of cell science. 1989;92(Pt 3):513–518. doi: 10.1242/jcs.92.3.513. [DOI] [PubMed] [Google Scholar]
- 59.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.