Abstract
Importance
Neuroticism is a personality trait that is briefly defined by emotional instability. It is a robust genetic risk factor for Major Depressive Disorder (MDD) and other psychiatric disorders. Hence, neuroticism is an important phenotype for psychiatric genetics. The Genetics of Personality Consortium (GPC) has created a resource for genome-wide association analyses of personality traits in over 63,000 participants (including MDD cases).
Objective
To identify genetic variants associated with neuroticism by performing a meta-analysis of genome-wide association (GWA) results based on 1000Genomes imputation, to evaluate if common genetic variants as assessed by Single Nucleotide Polymorphisms (SNPs) explain variation in neuroticism by estimating SNP-based heritability, and to examine whether SNPs that predict neuroticism also predict MDD.
Setting
30 cohorts with genome-wide genotype, personality and MDD data from the GPC.
Participants
The study included 63,661 participants from 29 discovery cohorts and 9,786 participants from a replication cohort. Participants came from Europe, the United States or Australia.
Main outcome measure(s)
Neuroticism scores harmonized across all cohorts by Item Response Theory (IRT) analysis, and clinically assessed MDD case-control status.
Results
A genome-wide significant SNP was found in the MAGI1 gene (rs35855737; P=9.26 × 10−9 in the discovery meta-analysis, and P=2.38 × 10−8 in the meta-analysis of all 30 cohorts). Common genetic variants explain 15% of the variance in neuroticism. Polygenic scores based on the meta-analysis of neuroticism in 27 of the discovery cohorts significantly predicted neuroticism in 2 independent cohorts. Importantly, polygenic scores also predicted MDD in these cohorts.
Conclusions and relevance
This study identifies a novel locus for neuroticism. The variant is located in a known gene that has been associated with bipolar disorder and schizophrenia in previous studies. In addition, the study shows that neuroticism is influenced by many genetic variants of small effect that are either common or tagged by common variants. These genetic variants also influence MDD. Future studies should confirm the role of the MAGI1 locus for neuroticism, and further investigate the association of MAGI1 and the polygenic association to a range of other psychiatric disorders that are phenotypically correlated with neuroticism.
Dimensions of personality have been linked with the liability to suffer from psychiatric illness.1 Perhaps the strongest link between personality and psychiatric illness is the association of neuroticism with Major Depressive Disorder (MDD)2–5. Neuroticism is also associated with other psychiatric disorders that entail emotional dysregulation, including personality, substance use and anxiety disorders.2,6–8 Furthermore, neuroticism is associated with neurological diseases such as migraine and Alzheimer’s disease.9,10 Hence, neuroticism is a psychological risk factor of profound public health significance.11
Neuroticism refers to the tendency to experience diverse and relatively more intense negative emotions. Neuroticism and similar traits such as harm avoidance and negative emotionality share an affective underpinning12 and are found in all main theories of personality.13–24 Twin studies of neuroticism, harm avoidance or negative emotionality generally find that between 40 and 60% of the trait variance is explained by genomic variation,3,25–28 without large age or sex by genotype effects, modest assortative mating and large genetic and phenotypic stability across the lifespan.28–31 These findings and the fact that neuroticism is strongly related to MDD7,32–35 make neuroticism an important phenotype for psychiatric genetic studies.
Genome-Wide Association (GWA) studies require large sample sizes to have sufficient statistical power, which is often achieved by aggregating results in multiple cohorts in a meta-analysis. This however requires a measurement scale that is comparable across cohorts. We recently showed for neuroticism and extraversion how different personality instruments could be linked through Item Response Theory (IRT) analysis in order to assess the same underlying constructs.36 Personality item data were harmonized in >160,000 participants from the Genetics of Personality Consortium (GPC). A meta-analysis of data from >29,000 twin pairs from six of the participating cohorts showed that the heritability of the harmonized neuroticism scores was 48%.36 This estimate was based on twin correlations that ranged between 0.39 and 0.53 for monozygotic twin pairs, and between 0.11 and 0.26 for dizygotic twin pairs across cohorts and genders. The opposite-sex twin correlations were not significantly lower than the same-sex dizygotic twin correlations, illustrating that the same genetic factors influence neuroticism in men and women.
Gene finding studies for MDD and neuroticism-like personality traits have had limited success to date. There have been two meta-analytic GWA studies for personality traits, including neuroticism and harm-avoidance. The sample sizes were small by current standards (N=11,590,37 and 17,37538) and single nucleotide polymorphisms (SNPs) were imputed using HAPMAP as a reference. The largest GWA39–41 studies for MDD are those from the Psychiatric Genomics Consortium (PGC), with 9,240 MDD cases and 9,519 controls in the discovery phase of the study, and 6,783 MDD cases and 50,695 controls in the replication phase, and imputation based on HAPMAP. These studies did not detect genome-wide significant SNPs.42
To assess if gene-finding efforts are likely to have success, techniques have been developed that test whether common variants that are tagged by genome-wide SNP arrays contribute to variation in the phenotype.43 Two of such studies for neuroticism found effects of common SNPs, explaining about 6% of the phenotypic variance, and another study for MDD found that common SNPs explain 28–32% of the phenotypic variance.44–46 In young children, genome-wide SNPs explained 13% to 43% of the variance in internalizing problems.47
Here we report results of the largest GWA study for neuroticism so far, conducted in 63,661 participants from 29 cohorts. Imputation was performed against the 1000Genomes (1000G) reference panel. The aims of the study were: (1) to identify genetic variants for neuroticism by performing a meta-analysis of GWA results, (2) to estimate SNP-based heritability in the two largest cohorts to establish that the sets of SNPs contain information to detect genetic variants, and (3) to test whether these variants predict MDD status in a large cohort of clinically assessed MDD cases and screened controls.
Materials and methods
Cohorts
The meta-analysis included 29 discovery cohorts, with 21 cohorts from Europe, 6 from the Unites States and 2 from Australia. All participants were of European descent. The total sample size was 63,661 for the GWA meta-analysis. The Generation Scotland cohort (N=9,786) was included for replication of GWA top results. For detailed information on each cohort see the eMaterials and methods.
Phenotyping
After harmonizing all item data on neuroticism from multiple instruments, comparable neuroticism scores were obtained for all cohorts.36 These scores were estimated for all participants after conducting IRT analysis on the available item data for neuroticism from the NEO Personality Inventory, Eysenck Personality Questionnaire, and the International Personality Item Pool inventory, all item data for harm avoidance from the Cloninger’s Tridimensional Personality Questionnaire, and all item data for negative emotionality from the Multidimensional Personality Questionnaire (see eMaterials and methods). For the Generation Scotland cohort phenotypes were summed scores on the neuroticism scale of the EPQ Revised Short Form.
Genotyping and imputation
An overview of SNP genotyping, quality control (QC), and imputation is given in eTable 2. QC of genotype data was performed in each study independently, using comparable but study specific criteria. Basic QC steps included checks for European ancestry, sex inconsistencies, Mendelian errors, and high genome-wide homozygosity. Checks for relatedness were carried out in those samples that aimed to include unrelated individuals only. Genotype data were further checked based on Hardy–Weinberg Equilibrium (HWE), minor allele frequencies (MAF), SNP and sample call rates. Genotype data were imputed using the 1000G phase 1 version 3 (build 37, hg19) reference panel with standard software packages such as IMPUTE, MACH or Minimac (see eTable 2).
Statistical analyses
GWA analysis in each cohort
GWA analyses were conducted in each cohort using linear regression (additive model, with sex and age as covariates) with the aim to identify single common genetic variants that influence neuroticism in both men and women of different ages. Depending on the characteristics of the cohort, additional covariates such as PCs were added. Different software packages were used to run the association analysis (see eTable 2). Uncertainty of the imputed genotypes was taken into account. In those cohorts that included related individuals, the dependency among participants was accounted for. Locations of SNPs are reported on build 37 (hg19).
Meta-analysis of GWA results across cohorts
A meta-analysis of the GWA results of the discovery cohorts was conducted using the weighted inverse variance method in METAL (http://www.sph.umich.edu/csg/abecasis/metal/index.html). This is a fixed effects model in which effect sizes (beta’s) are weighted by the inverse of their variance and then summed over cohorts. This model is appropriate if phenotypes are on a similar scale, which was the case for the harmonized neuroticism scores. Poorly imputed SNPs (r2<0.30 or proper_info<0.40) and SNPs with low MAF (MAF < √(5/N, which corresponds to less than 5 estimated individuals in the least frequent genotype group, under the assumption of HWE) were excluded, resulting in a total number of 1.1M to 6.6M SNPs across cohorts. The number of unique SNPs available for meta-analysis was 7,480,565. For 530,951 SNPs association results were available in one cohort only and were discarded, leading to a final 6,949,614 SNPs for which results are reported. Genomic control inflation factors (lambda) and Manhattan and quantile–quantile plots per cohort are provided in eTable 3 and eFigures 1 and 2. SNPs with a P-value of 5 × 10−8 or smaller were considered genome-wide significant. In the Generation Scotland cohort, all SNPs with a P-value smaller than 1 × 10−5 were tested for replication. For these SNPs, a meta-analysis of all 30 cohorts was conducted. Because in the Generation Scotland cohort, sum scores were available for neuroticism, this meta-analysis was based on combining P-values, taking into account the direction of effect and weighting by sample size, rather than combining effect sizes.
Meta-analysis results at the SNP level were used as the input to compute P-values at the gene level. These analyses were performed with VEGAS2.48,49 A gene with a P-value of 1 × 10−6 or lower is considered genome-wide significant in gene-wide analyses.
Variance explained by common SNPs
In two large cohorts included in the meta-analysis, the Netherlands Twin Register (NTR, N = 3,599 unrelated individuals) cohort and the QIMR Berghofer Medical Research Institute (QIMR, N= 3,369) adult cohort, Genomic-relatedness-matrix Restricted Maximum Likelihood (GREML) analysis in the GCTA software was applied to estimate the proportion of variance in neuroticism that can be explained by common SNPs.43,50 GCTA analysis was based on best guess genotypes obtained in PLINK using a threshold of a maximum genotype probability >0.70, and additionally filtering on r-squared >0.80. Next, in estimating the GRM matrix in the GCTA software, SNPs with MAF <0.05 were excluded. The additive genetic relationship matrix (GRM) for all individuals in the data sets estimated based on SNPs was used to estimate the proportion of phenotypic variance due to additive genetic variance. Sex, age and population-specific principal components (PCs) were included as covariates.
Polygenic risk score analysis
Polygenic risk scores (PGS) analyses were conducted to test the predictive power of the meta-analysis results for neuroticism itself and for MDD. PGS were computed in NTR and Netherlands Study of Depression and Anxiety (NESDA51) and were based on the meta-analysis results excluding the NTR and NESDA cohorts, further referred to as the discovery set. PGS were calculated for all individuals of the NTR and NESDA target set by taking a set of most significant SNPs from the analysis in the discovery set and by multiplying the individual’s genotypic score (0, 1 or 2 for genotyped SNPs, or any value in between 0 and 2 for imputed SNPs) by the effect size of a particular SNP (unstandardized regression coefficient based on the meta-analysis), and summing this over SNPs. PGS were calculated for six different P-value thresholds (P<1 × 10−5, P<1 × 10−4, P<1 × 10−3, P<0.01, P<0.05 and P<0.5). Next, linear/logistic regression was conducted to predict neuroticism from the PGS in 8,648 NTR participants and MDD status in 1,859 unrelated MDD cases and 2,391 unrelated controls from NTR and NESDA. MDD case-control status was defined as a lifetime DSM-IV diagnosis using the Composite International Diagnostic Interview (CIDI). Age, sex and nine PCs were included as covariates. See for more details eMaterials and methods.
Results
Meta-analysis of GWA results for neuroticism
Meta-analysis of GWA results across the 29 cohorts revealed one genome-wide significant SNP (rs35855737; P=9.26 × 10−9). The SNP is located in an intron of the MAGI1 gene (Figure 1). The pooled regression effect was −0.04 with the minor allele C coded as the effect allele (Figure 2). Imputation quality was very high (r-squared or proper_info>0.94) in all cohorts, except in ERF (r2=0.63). MAF of the SNP ranged from 0.13 to 0.22 across cohorts with imputation quality >0.94 and showed a mean of 0.18 (SD=0.02), which corresponds to the MAF for this SNP in the 1000G reference set. MAF in the ERF cohort was 0.07. Eleven other SNPs in the MAGI1 gene showed suggestive genome-wide significance (P<1 × 10−5); all SNPs were intronic; one SNP was in very high LD with rs35855737 (rs1404544; r2>0.80; P=8,59 × 10−6) and three SNPs were in high LD with rs35855737 (rs1524970, rs1880522 and rs6799284; r2>0.60, 3.64 × 10−6 < P < 8.54 × 10−7). The Manhattan and quantile-quantile plots are given in Figures 3 and 4. A list with all 127 suggestively genome-wide significant SNPs is provided in eTable 41. The lowest p-value for the gene-based tests did not reach genome-wide significance (P<1 × 10−6).
Figure 1.
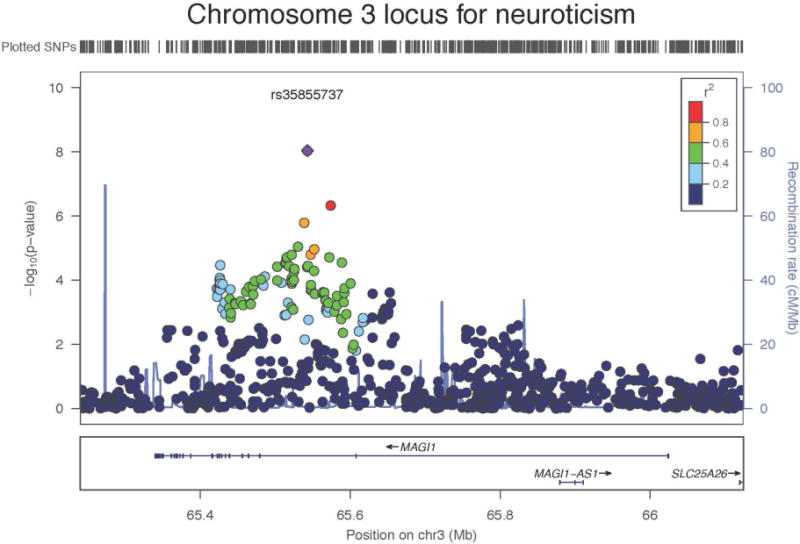
Region plot for genome-wide significant SNP rs35855737 in the MAGI1 gene on chromosome 3 for neuroticism
Figure 2.
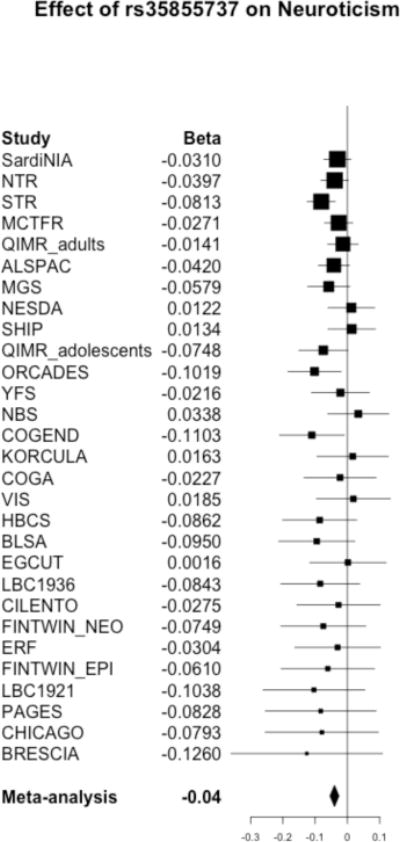
Forest plot for genome-wide significant SNP rs35855737 in the MAGI1 gene on chromosome 3 for neuroticism
Figure 3.
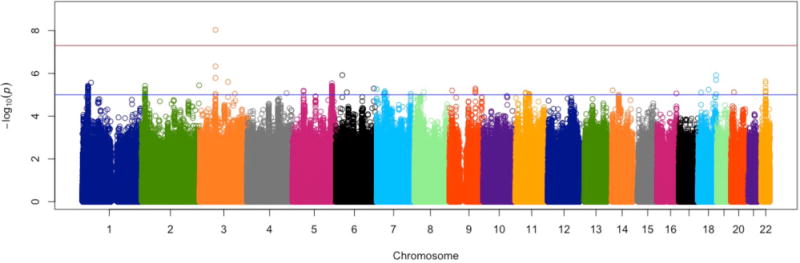
Manhattan plot for meta-analysis results for neuroticism in 29 discovery cohorts
Figure 4.
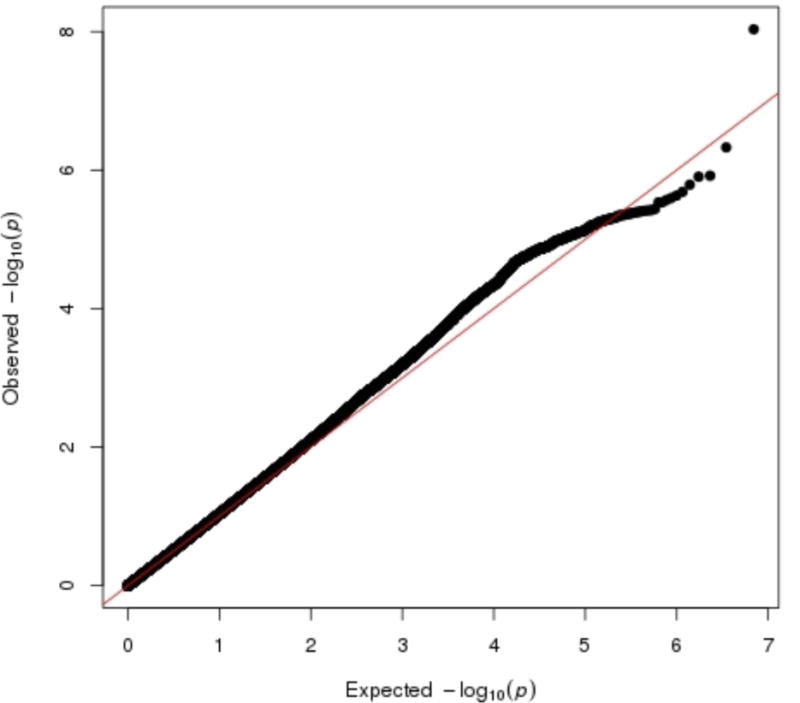
Quantile-quantile plots for meta-analysis results for neuroticism in 29 discovery cohorts
Results of the follow-up analysis for all SNPs with P-value < 1 × 10−5 in the Generation Scotland cohort are displayed in eTable 4. Rs35855737 is not significantly associated with neuroticism in the Generation Scotland cohort, but the direction of the effect is the same (beta=−0.02 for effect allele C; P-value=0.32). A meta-analysis of the results from all 30 cohorts shows that rs35855737 remains genome-wide significant (beta=−0.04; P-value=2.38 × 10−8).
Variance in neuroticism explained by common SNPs
In the NTR cohort, 14.7% of the variance in neuroticism was explained by all SNPs (P=0.02; 95% Confidence Interval (CI) 0.002–0.29). In the QIMR cohort, 15.7% of the variance was explained by SNPs (P=0.18; 95% CI 0–0.47).
Polygenic risk score analysis for Neuroticism and MDD
The results of the polygenic risk score analyses are presented in Figure 5. In the NTR, polygenic risk scores are significantly (P<0.05) associated with neuroticism when polygenic scores are based on SNP sets with P-value thresholds of 1 × 10−3 and lower. The most significant result was found for the SNP set with a P-value threshold of 0.05, with an explained variance of 0.66% and a P-value of 1.09 × 10−12. In the combined NTR/NESDA cohort, polygenic risk scores are significantly (P<0.05) associated with MDD for SNP sets with P-value thresholds of 0.01 and 0.05, with higher neuroticism predicting larger risk for MDD. The most significant result was found for the SNP set with a P-value threshold of 0.05, with an explained variance of 1.05% and a P-value of 4.02 × 10−9.
Figure 5.
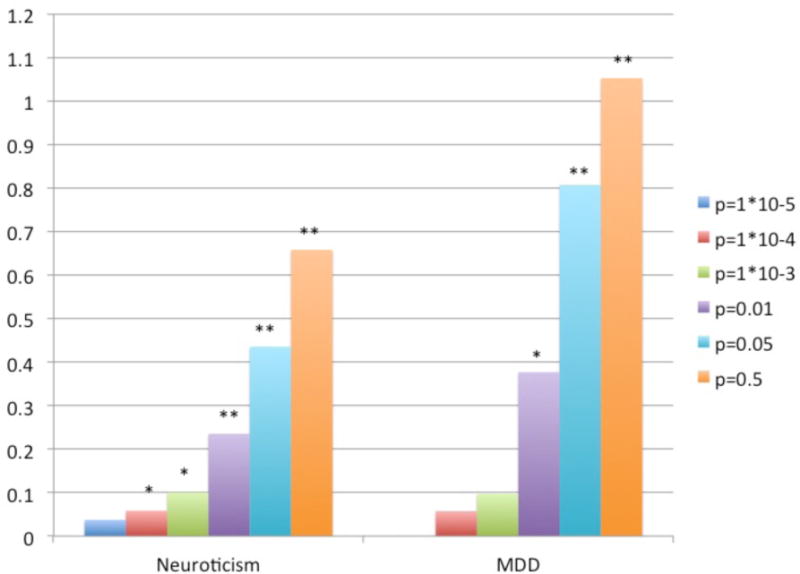
Results of polygenic risk score analyses predicting MDD and neuroticism based on neuroticism polygenic risk scores
Note: Prediction of neuroticism and MDD in NTR/NESDA cohorts based on neuroticism polygenic risk scores from meta-analysis results omitting NTR/NESDA cohorts significant with *P<0.05 or ** P<0.001. Different colored bars refer to different P-value thresholds used to calculate the polygenic risk scores.
Discussion
This study evaluated in 63,661 individuals if common genetic variants explain variation in neuroticism by performing a meta-analysis of GWA results for neuroticism and by estimating SNP-based heritability. In addition, it was examined whether genetic variants that predict neuroticism also predict MDD.
The meta-analysis of GWA results for neuroticism showed a genome-wide significant SNP in an intron of the MAGI1 gene. The MAGI1 gene is expressed in neuronal tissue, in particular the hippocampus, and is found at the synaptic plasma membrane.52 In addition, it has been shown that MAGI1 acts as a scaffolding protein in the neurite growth factor (NGF) receptor-mediated signaling pathway.53 Interestingly, the MAGI1 gene has previously been implicated in bipolar disorder, schizophrenia and episodicity in MDD54–56, disorders that in part share their genetic etiology.57 A genome-wide linkage scan for early onset bipolar disorder type 1 revealed genome-wide significant linkage in the 3p14 region where MAGI1 is located.56 A copy number variation study found evidence for deletions in MAGI1 to be associated with bipolar disorder and duplications to be associated with schizophrenia.54 Further, a genome-wide association study found suggestive association (P=5.1 × 10−7) of a SNP in MAGI1 with episodicity in MDD, a feature of MDD that shows increased risk to shifting to bipolar disorder.55
It was further estimated that SNP-based genetic similarity across individuals accounted for approximately 15% of the variance in neuroticism. This estimate is larger than in earlier reports using the same technique (about 6% explained)44,46. Heritability estimates from twin studies are usually larger and range between 40 and 55%.36
Polygenic risk scores based on the GWA meta-analysis significantly predicted MDD status in a large independent target set consisting of MDD cases and screened controls. The polygenic scores for neuroticism reassuringly also predicted neuroticism in MDD controls of this same independent set. MDD and neuroticism were explained about equally well by neuroticism polygenic scores (up to 1.05% explained variance). These findings are consistent with previous reports that studied the prediction of MDD and bipolar disorder based on polygenic scores derived from Big Five neuroticism GWA results.58,59
This study demonstrates that increasing the number of subjects and SNPs in a meta-analysis is successful in identifying a novel locus for neuroticism. Yet, the effect size of the identified SNP is very small (regression coefficient of −0.04 for the harmonized score with a variance of around 1). Together with our findings of a SNP-based heritability of around 15% and an increase in explained variance in the polygenic risk score analysis when polygenic scores are based on larger sets of SNPs, this suggests that neuroticism is highly polygenic.
Our results further indicate that the heritability of neuroticism likely consists not only of common SNPs with small additive effects. Common SNPs with non-additive effects, rare SNPs and indels may also influence neuroticism, possibly in interaction (epistasis). As a consequence, to further our understanding of the genetic and molecular basis of neuroticism (and associated psychiatric disorders) different routes need to be taken in future studies. One route would be to increase the number of subjects and SNPs to further identify more common variants of additive effect, which has shown to be successful for schizophrenia.60 Also, the study of variants other than common variants of additive effect could be pursued. Alternative routes could include pathway analyses and genetic studies that are informed by results from the animal literature on basic emotions such as fear, sadness and anger.61–64
The current study more than tripled the sample size compared to the previous published meta-analysis on personality38, providing a substantial increase in power to detect variants. The power to detect variants that explain 0.23% of the variance (corresponding to the effect size for the most significant SNP in the previous meta-analysis38) increased from 84% to 100%. In addition, with a sample size of 63,661 individuals there is 80% power to detect variants that explain at least 0.063% of the variance in neuroticism, compared to 1.6% power given the 17,375 subjects that were included in the previous meta-analysis.38 The large increase in sample size was possible because an IRT approach enabled harmonization of personality data obtained from different personality questionnaires, which may serve as an example for gene-finding studies for other psychological, cognitive and psychiatric traits where harmonization is required to increase sample size (e.g. symptoms of depression or ADHD measured by different questionnaires).
The results for neuroticism were predictive for MDD. Future analyses may focus on whether the MAGI1 locus and polygenic variance for neuroticism is also associated with psychiatric disorders that are phenotypically associated with neuroticism, such as Borderline personality disorder, bipolar disorder, schizophrenia, ADHD and substance use disorders. This could be achieved by combining data from with GPC with those available within the PGC65 and the Social Science Genetic Association Consortium66. Novel methodologies will be needed to test whether neuroticism represents a causal risk factor for MDD and other disorders, whether reverse causality is also present, or whether the genetic association between neuroticism and psychiatric disorders reflects an underlying common liability.57,67–69 It is expected that such studies will increase our understanding of the role that emotional instability plays in the occurrence and course of psychiatric disorders and other important health outcomes.
Supplementary Material
Acknowledgments
We would like to thank all participating subjects. Analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Organization for Scientific Research (NWO 480-05-003).
ALSPAC We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council (grant 74882), the Wellcome Trust (grant 076467) and the University of Bristol provide core support for ALSPAC. We thank 23andMe for funding the genotyping of the ALSPAC children’s sample. This publication is the work of the authors, and they will serve as guarantors for the contents of this paper.
BLSA We acknowledge support from the Intramural Research Program of the NIH, National Institute on Aging. We thank Robert McCrae.
BRESCIA We acknowledge support from the Italian Ministry of Health (RC and RF2007 Conv. 42) and Regione Lombardia (ID: 17387 SAL-13). We thank Ilaria Gandin for imputation analysis support.
CHICAGO This work was supported by NIH grants, DA007255 (ABH), HG006265 (to BEE), DA02812 (to HdW), and DA021336 and DA024845 (to AAP). BEE was also funded through the Bioinformatics Research Development Fund, supported by Kathryn and George Gould. We wish to thank Andrew D. Skol for providing advice about genotype calling.
CILENTO We acknowledge Dr Maria Enza Amendola for the test administration and thank the personnel working in the organization of the study in the villages. MC received funding support from the Italian Ministry of Universities (FIRB – RBNE08NKH7, INTEROMICS Flaghip Project), the Assessorato Ricerca Regione Campania, the Fondazione con il SUD (2011-PDR-13), and the Fondazione Banco di Napoli.
SAGE – COGA/CONGEND Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401) and the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease”(HHSN268200782096C). The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Texas Biomedical Research Institute (L. Almasy), Howard University (R. Taylor) and Virginia Commonwealth University (D. Dick). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, S. O’Connor, L. Wetherill, X. Xuei (Indiana University); Grace Chan (University of Iowa); S. Kang, N. Manz, M. Rangaswamy (SUNY Downstate); J. Rohrbaugh, J-C Wang (Washington University in St. Louis); A. Brooks (Rutgers University); and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). The Collaborative Genetic Study of Nicotine Dependence (COGEND) project is a collaborative research group and part of the NIDA Genetics Consortium. Subject collection was supported by NIH grant P01 CA089392 (L.J. Bierut) from the National Cancer Institute. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C (J. Tischfield and J. Rice). Jaime Derringer was supported by NIH T32 MH016880.
EGCUT AM and TE received support from FP7 Grants (201413 ENGAGE, 212111 BBMRI, ECOGENE (No. 205419, EBC)) and OpenGENE. AM and TE also received targeted financing from Estonian Government SF0180142s08 and by EU via the European Regional Development Fund, in the frame of Centre of Excellence in Genomics. The genotyping of the Estonian Genome Project samples were performed in Estonian Biocentre Genotyping Core Facility, AM and TE thank Mari Nelis and Viljo Soo for their contributions. AR and JA were supported by a grant from the Estonian Ministry of Science and Education (SF0180029s08).
ERF The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947) and also received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002-01254). The ERF study was further supported by ENGAGE consortium and CMSB. High-throughput analysis of the ERF data was supported by joint grant from Netherlands Organisation for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). ERF was further supported by the ZonMw grant (project 91111025). We are grateful to all study participants and their relatives, general practitioners and neurologists for their contributions and to P. Veraart for her help in genealogy, J. Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection.
Finnish Twin Cohort (FTC) We acknowledge support from the Academy of Finland Center of Excellence in Complex Disease Genetics (grant numbers: 213506, 129680), the Academy of Finland (grants 100499, 205585, 118555 and 141054 to JK, grant 257075 to EV), Global Research Awards for Nicotine Dependence (GRAND), ENGAGE (European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant agreement number 201413), DA12854 to P A F Madden, and AA-12502, AA-00145, and AA-09203 to RJRose, AA15416 and K02AA018755 to DM Dick
HBCS We thank all study participants as well as everybody involved in the Helsinki Birth Cohort Study. Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation.
KORCULA The CROATIA-Korcula study was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). We would like to acknowledge the invaluable contributions of the recruitment team in Korcula, the administrative teams in Croatia and Edinburgh and the people of Korcula. The SNP genotyping for the CROATIA-Korcula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany.
LBC1921 & LBC1936 For the Lothian Birth Cohorts, we thank Paul Redmond for database management; Alan Gow, Michelle Taylor, Janie Corley, Caroline Brett and Caroline Cameron for data collection and data entry; nurses and staff at the Wellcome Trust Clinical Research Facility, where blood extraction and genotyping was performed; staff at the Lothian Health Board, and the staff at the SCRE Centre, University of Glasgow. The research was supported by a program grant from Research Into Ageing. The research continues with program grants from Age UK (The Disconnected Mind). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1). Funding from the Biotechnology and Biological Sciences Research Council (BBSRC) and Medical Research Council (MRC) is gratefully acknowledged.
MCTFR We would like to thank Rob Kirkpatrick for his help running analyses. Research reported in this publication was supported by the National Institutes of Health under award numbers R37DA005147, R01AA009367, R01AA011886, R01DA013240, R01MH066140, and U01DA024417.
MGS Samples were collected under the following grants: NIMH Schizophrenia Genetics Initiative U01s: MH46276, MH46289, and MH46318; and Molecular Genetics of Schizophrenia Part 1 (MGS1) and Part 2 (MGS2) R01s: MH67257, MH59588, MH59571, MH59565, MH59587, MH60870, MH60879, MH59566, MH59586, and MH61675. Genotyping and analyses were funded under the MGS U01s: MH79469 and MH79470. NBS Principal investigators of the Nijmegen Biomedical Study are L.A.L.M. Kiemeney, M. den Heijer, A.L.M. Verbeek, D.W. Swinkels and B. Franke.
Zweden96
NESDA The Netherlands Study of Depression and Anxiety (NESDA) were funded by the Netherlands Organization for Scientific Research (Geestkracht program grant 10-000-1002); the Center for Medical Systems Biology (CMSB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL), VU University’s EMGO Institute for Health and Care Research and Neuroscience Campus Amsterdam. Genotyping was funded by the US National Institute of Mental Health (RC2MH089951) as part of the American Recovery and Reinvestment Act of 2009. BP is financially supported by NWO-VIDI grant no. 91811602.
NTR We acknowledge financial support from the Netherlands Organization for Scientific Research (NWO): Grants 575-25-006, 480-04-004, 904-61-090; 904-61-193, 400-05-717 and Spinozapremie SPI 56-464-14192. MHMdeM is financially supported by NOW VENI Grant No. 016-115-035. Genotyping was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis was supported by grants from Genetic Association Information Network and the NIMH (MH081802). Genotype data were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/dbgap, accession number phs000020.v1.p1).
ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the MRC Human Genetics Unit, Arthritis Research UK and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney.
PAGES none
QIMR Berghofer adolescents/adults We thank Marlene Grace and Ann Eldridge for sample collection; Megan Campbell, Lisa Bowdler, Steven Crooks and staff of the Molecular Epidemiology Laboratory for sample processing and preparation; Harry Beeby, David Smyth and Daniel Park for IT support. We acknowledge support from the Australian Research Council grants A79600334, A79906588, A79801419, DP0212016, DP0343921, DP0664638, and DP1093900 (to NGM and MJW), Beyond Blue and the Borderline Personality Disorder Research Foundation (to NGM), NIH Grants DA12854 (to PAFM), AA07728, AA07580, AA11998, AA13320, AA13321 (to ACH) and MH66206 (to WSS); and grants from the Australian National Health and Medical Research Council; MLP is supported by DA019951. Genotyping was partly funded by the National Health and Medical Research Council (Medical Bioinformatics Genomics Proteomics Program, 389891) and the 5th Framework Programme (FP-5) GenomEUtwin Project (QLG2-CT-2002-01254). Further genotyping at the Center for Inherited Disease Research was supported by a grant to the late Richard Todd, M.D., Ph.D., former Principal Investigator of Grant AA13320. SEM and GWM are supported by the National Health and Medical Research Council Fellowship Scheme. Further, we gratefully acknowledge Dr Dale R Nyholt for his substantial involvement in the QC and preparation of the QIMR GWA data sets. Dr Nyholt also contributed 8% of the GWAS for the QIMR adult cohort (NHMRC IDs 339462, 442981, 389938, 496739).
SardiNIA We acknowledge support from the Intramural Research Program of the NIH, National Institute on Aging. Funding was provided by the National Institute on Aging, NIH Contract No. NO1-AG-1-2109 to the SardiNIA (‘ProgeNIA’) team.
SHIP SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG. This work was also funded by the German Research Foundation (DFG: GR 1912/5-1).
STR The STR was supported by grants from the Ministry for Higher Education, the Swedish Research Council (M-2005-1112 and 2009-2298), GenomEUtwin (EU/QLRT-2001-01254; QLG2-CT-2002-01254), NIH grant DK U01-066134, The Swedish Foundation for Strategic Research (SSF; ICA08-0047), the Swedish Heart-Lung Foundation, the Royal Swedish Academy of Science, and ENGAGE (within the European Union Seventh Framework Programme, HEALTH-F4-2007-201413).
VIS The CROATIA-Vis study was funded by grants from the Medical Research Council (UK) and Republic of Croatia Ministry of Science, Education and Sports research grants to I.R. (108-1080315-0302). We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to The University of Split and Zagreb Medical Schools, the Institute for Anthropological Research in Zagreb and Croatian Institute for Public Health. The SNP genotyping for the CROATIA-Vis cohort was performed in the core genotyping laboratory of the Wellcome Trust Clinical Research Facility at the Western General Hospital, Edinburgh, Scotland.
YFS The Young Finns Study has been financially supported by the Academy of Finland (grants 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), 41071 (Skidi), and 265869 (Mind)), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9N035 for Dr. Lehtimäki), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (for Dr. Lehtimäki). The expert technical assistance in statistical analysis by Irina Lisinen, Mika Helminen, and Ville Aalto is gratefully acknowledged.
Generation Scotland SFHS is funded by the Scottish Executive Health Department, Chief Scientist Office, grant number CZD/16/6. Exome array genotyping for GS:SFHS was funded by the Medical Research Council UK and performed at the Wellcome Trust Clinical Research Facility Genetics Core at Western General Hospital, Edinburgh, UK. We would like to acknowledge the invaluable contributions of the families who took part in the Generation Scotland: Scottish Family Health Study, the general practitioners and Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes academic researchers, IT staff, laboratory technicians, statisticians and research managers.
Footnotes
Authors’ contributions
Writing group: MHMdeM, SMvdB, KJHV, RFK, ML, AAV, LKM, JD, TE, DIB
Analytic group: MHMdeM, SMvdB, KJHV, ML, AAV, LKM, JDe, TE, NA, SG, NKH, ABH, JH, BK, JL, ML, MM, TT, ATeu, AV, JW, IOF, NT, DME, TL, IS, EP, GRA, JM, JDi, RK, YQ
Study design and project management: LF, LPR, JGE, AAP, GWM, MJW, PAFM, DP, AMin, AP, DR, MC, IG, CH, IR, AMet, JK, IJD, KR, JFW, LKJ, JMH, HJG, BWJHP, CMvD, DME, NLP, PTC, ATer, MMG, NGM, DIB, RFK, AAV, GDS, TL, OTR, PKEM, KH, JMS, DS, GRA, HC, RJR
Sample and phenotype data collection: BWJHP, AMet, AR, JA, IG, AMH, PAFM, ACH, NGM, MJW, KA, MN, LJB, JGE, LF, PTC, ATer, GDS, MGK, HdW, AAP, AKH, WSS, RS, DR, Amin, MJ, LPR, LKJ, OTR, PKEM, EV, KH, AM, FB, OP, LZ
Data preparation: MHMdeM, SMvdB, JW, KGO, JJH, SEM, NKH, YM, TE, AR, GD, ML, RG, AA, JD, EW, BK, GD, BEE, COS, GH, KJHV, SDG, DEA, TBB, JPK, NJT, BP, CM, MJ, AL, ARS, ATer, DCL, HT
Conflict of interest disclosures: there is no conflict of interest.
Full results of the meta-analysis can be downloaded from http://www.tweelingenregister.org/GPC.
References
- 1.Fanous AH, Kendler KS. The genetic relationship of personality to major depression and schizophrenia. Neurotox Res. 2004;6(1):43–50. doi: 10.1007/BF03033295. [DOI] [PubMed] [Google Scholar]
- 2.Kotov R, Gamez W, Schmidt F, Watson D. Linking “Big” Personality Traits to Anxiety, Depressive, and Substance Use Disorders: A Meta-Analysis. Psychol Bull. 2010;136(5):768–821. doi: 10.1037/a0020327. [DOI] [PubMed] [Google Scholar]
- 3.Distel MA, Trull TJ, Willemsen G, et al. The Five Factor Model of personality and borderline personality disorder: A genetic analysis of comorbidity. Biol Psychiatry. 2009;66:1131–1138. doi: 10.1016/j.biopsych.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Middeldorp CM, Cath DC, van den Berg M, Beem AL, Van Dyck R, Boomsma DI. The Association of Personality with Anxious and Depressive Psychopathology. In: Canli T, editor. The Biological Basis of Personality and Individual Differences. New York: Guilford Press; 2006. pp. 251–272. [Google Scholar]
- 5.Xia J, He Q, Li YH, et al. The relationship between neuroticism, major depressive disorder and comorbid disorders in Chinese women. J Affect Disorders. 2011 Dec;135(1–3):100–105. doi: 10.1016/j.jad.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saulsman LM, Page AC. The five-factor model and personality disorder empirical literature: A meta-analytic review. Clinical Psychology Review. 2004;23(8):1055–1085. doi: 10.1016/j.cpr.2002.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Myers J. The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol Med. 2009;40(5):1–6. doi: 10.1017/S0033291709991140. [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Myers J. The boundaries of the internalizing and externalizing genetic spectra in men and women. Psychol Med. 2013 Apr 11;:1–9. doi: 10.1017/S0033291713000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ligthart L, Boomsma DI. Causes of comorbidity: pleiotropy or causality? Shared genetic and environmental influences on migraine and neuroticism. Twin Res Hum Genet. 2012 Apr;15(2):158–165. doi: 10.1375/twin.15.2.158. [DOI] [PubMed] [Google Scholar]
- 10.Terracciano A, S A, An Y, O’Brien RJ, Ferrucci L, Zonderman AB, Resnick SM. Personality and risk of Alzheimer’s disease: New data and meta-analysis. Alzheimer’s & Dementia. 2014 doi: 10.1016/j.jalz.2013.1003.1002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahey BB. Public health significance of neuroticism. Am Psychol. 2009 May-Jun;64(4):241–256. doi: 10.1037/a0015309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson D, Clark LA. On Traits and Temperament – General and Specific Factors of Emotional Experience and Their Relation to the 5-Factor Model. J Pers. 1992 Jun;60(2):441–476. doi: 10.1111/j.1467-6494.1992.tb00980.x. [DOI] [PubMed] [Google Scholar]
- 13.De Fruyt F, Van De Wiele L, Van Heeringen C. Cloninger’s psychobiological model of temperament and character and the five-factor model of personality. Pers Indiv Differ. 2000;29(3):441–452. [Google Scholar]
- 14.Ferrando PJ. The measurement of neuroticism using MMQ, MPI, EPI and EPQ items: a psychometric analysis based on item response theory. Pers Indiv Differ. 2001;30(4):641–656. [Google Scholar]
- 15.Church AT. Relating the Tellegen and 5-Factor Models of Personality Structure. J Pers Soc Psychol. 1994 Nov;67(5):898–909. doi: 10.1037//0022-3514.67.5.898. [DOI] [PubMed] [Google Scholar]
- 16.Markon KE, Krueger RF, Watson D. Delineating the structure of normal and abnormal personality: An integrative hierarchical approach. J Pers Soc Psychol. 2005;88(1):139–157. doi: 10.1037/0022-3514.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa PT, McCrae RR. Professional manual: Revised NEO Personality Inventory (NEO-PI-R) and NEO Five-Factor-Inventory (NEO-FFI) Odessa, FL: Psychological Assessment Resources; 1992. [Google Scholar]
- 18.Eysenck HJ, Eysenck SBG. Eysenck Personality Inventory. San Diego, CA: Educational and Industrial Testing Service; 1964. [Google Scholar]
- 19.Eysenck HJ, Eysenck SBG. Manual of the Eysenck Personality Questionnaire. London: Hodder & Stoughton; 1975. [Google Scholar]
- 20.Eysenck SBG, Eysenck HJ, Barrett P. A Revised Version of the Psychoticism Scale. Pers Indiv Differ. 1985;6(1):21–29. [Google Scholar]
- 21.Cloninger CR. A Systematic Method for Clinical Description and Classification of Personality Variants – A Proposal. Arch Gen Psychiatry. 1987;44(6):573–588. doi: 10.1001/archpsyc.1987.01800180093014. [DOI] [PubMed] [Google Scholar]
- 22.Cloninger CR, Svrakic DM, Przybeck TR. A Psychobiological Model of Temperament and Character. Arch Gen Psychiatry. 1993;50(12):975–990. doi: 10.1001/archpsyc.1993.01820240059008. [DOI] [PubMed] [Google Scholar]
- 23.Patrick CJ, Curtin JJ, Tellegen A. Development and validation of a brief form of the multidimensional personality questionnaire. Psychological Assessment. 2002;14(2):150–163. doi: 10.1037//1040-3590.14.2.150. [DOI] [PubMed] [Google Scholar]
- 24.Tellegen A. Manual of the Multidimensional Personality Questionnaire. Minneapolis: University of Minnesota Press; 2000. [Google Scholar]
- 25.Bouchard TJ, Loehlin JC. Genes, evolution, and personality. Behav Genet. 2001;31(3):243–273. doi: 10.1023/a:1012294324713. [DOI] [PubMed] [Google Scholar]
- 26.Yamagata S, Suzuki A, Ando J, et al. Is the genetic structure of human personality universal? A cross-cultural twin study from North America, Europe, and Asia. J Pers Soc Psychol. 2006;90(6):987–998. doi: 10.1037/0022-3514.90.6.987. [DOI] [PubMed] [Google Scholar]
- 27.Finkel D, McGue M. Sex differences and nonadditivity in heritability of the Multidimensional Personality Questionnaire Scales. J Pers Soc Psychol. 1997 Apr;72(4):929–938. doi: 10.1037//0022-3514.72.4.929. [DOI] [PubMed] [Google Scholar]
- 28.Keller MC, Coventry WL, Heath AC, Martin NG. Widespread evidence for non-additive genetic variation in Cloninger’s and Eysenck’s personality dimensions using a twin plus sibling design. Behav Genet. 2005;35(6):707–721. doi: 10.1007/s10519-005-6041-7. [DOI] [PubMed] [Google Scholar]
- 29.Vink JM, Bartels M, van Beijsterveldt TCEM, et al. Sex Differences in Genetic Architecture of Complex Phenotypes? PLoS One. 2012 Dec 18;7(12) doi: 10.1371/journal.pone.0047371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake RI, Eaves LJ, Maes HH, Heath AC, Martin NG. Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45,850 twins and relatives on two continents. Behav Genet. 2000 May;30(3):223–233. doi: 10.1023/a:1001918408984. [DOI] [PubMed] [Google Scholar]
- 31.Wray NR, Birley AJ, Sullivan PF, Visscher PM, Martin NG. Genetic and phenotypic stability of measures of neuroticism over 22 years. Twin Res Hum Genet. 2007 Oct;10(5):695–702. doi: 10.1375/twin.10.5.695. [DOI] [PubMed] [Google Scholar]
- 32.Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS. Neuroticism, Major Depression and Gender: a Population-Based Twin Study. Psychol Med. 2002;32(4):719–728. doi: 10.1017/s003329170200541x. [DOI] [PubMed] [Google Scholar]
- 33.Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. American Journal of Psychiatry. 2006;163(5):857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 34.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression – A Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63(10):1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 35.Boomsma DI, Beem AL, van den BM, et al. Netherlands twin family study of anxious depression (NETSAD) Twin Res. 2000;3(4):323–334. doi: 10.1375/136905200320565300. [DOI] [PubMed] [Google Scholar]
- 36.van den Berg SM, de Moor MH, McGue M, et al. Harmonization of Neuroticism and Extraversion phenotypes across inventories and cohorts in the Genetics of Personality Consortium: an application of Item Response Theory. Behav Genet. 2014 Jul;44(4):295–313. doi: 10.1007/s10519-014-9654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Service SK, Verweij KJH, Lahti J, et al. A genome-wide meta-analysis of association studies of Cloninger’s Temperament Scales. Translational Psychiatry. 2012 May;2 doi: 10.1038/tp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Moor MH, Costa PT, Terracciano A, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012 Mar;17(3):337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray NR, Pergadia ML, Blackwood DHR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17(1):36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan PF, de Geus EJC, Willemsen G, et al. Genomewide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14(4):359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohli MA, Lucae S, Saemann PG, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011 Apr 28;70(2):252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripke S, Wray NR, Lewis CM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013 Apr;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vinkhuyzen AA, Pedersen NL, Yang J, et al. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl Psychiatry. 2012;2:e102. doi: 10.1038/tp.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lubke GH, Hottenga JJ, Walters R, et al. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol Psychiatry. 2012 Oct 15;72(8):707–709. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verweij KJ, Yang J, Lahti J, et al. Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012 Oct;66(10):3238–3251. doi: 10.1111/j.1558-5646.2012.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benke KS, Nivard MG, Velders FP, et al. A genome-wide association meta-analysis of preschool internalizing problems. Journal of the American Academy of Child and Adolescent Psychiatry. 2014 Jun;53(6):667–676. doi: 10.1016/j.jaac.2013.12.028. e667. [DOI] [PubMed] [Google Scholar]
- 48.Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra AMS. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2014 doi: 10.1017/thg.2014.79. first published online: http://dx.doi.org/10.1017/thg.2014.1079. [DOI] [PubMed]
- 50.Visscher PM, Yang J, Goddard ME. A commentary on ‘Common SNPs explain a large proportion of the heritability for human height’ by Yang et al. (2010) Twin Research and Human Genetics. 2010;13(6):517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- 51.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder 2. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito H, Morishita R, Sudo K, et al. Biochemical and morphological characterization of MAGI-1 in neuronal tissue. Journal of neuroscience research. 2012 Sep;90(9):1776–1781. doi: 10.1002/jnr.23074. [DOI] [PubMed] [Google Scholar]
- 53.Ito H, Morishita R, Iwamoto I, Mizuno M, Nagata K. MAGI-1 acts as a scaffolding molecule for NGF receptor-mediated signaling pathway. Biochimica et biophysica acta. 2013 Oct;1833(10):2302–2310. doi: 10.1016/j.bbamcr.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Karlsson R, Graae L, Lekman M, et al. MAGI1 copy number variation in bipolar affective disorder and schizophrenia. Biol Psychiatry. 2012 May 15;71(10):922–930. doi: 10.1016/j.biopsych.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 55.Ferentinos P, Rivera M, Ising M, et al. Investigating the genetic variation underlying episodicity in major depressive disorder: suggestive evidence for a bipolar contribution. Journal of affective disorders. 2014 Feb;155:81–89. doi: 10.1016/j.jad.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 56.Etain B, Mathieu F, Rietschel M, et al. Genome-wide scan for genes involved in bipolar affective disorder in 70 European families ascertained through a bipolar type I early-onset proband: supportive evidence for linkage at 3p14. Mol Psychiatry. 2006 Jul;11(7):685–694. doi: 10.1038/sj.mp.4001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 Apr 20;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Middeldorp CM, de Moor MH, McGrath LM, et al. The genetic association between personality and major depression or bipolar disorder. A polygenic score analysis using genome-wide association data. Transl Psychiatry. 2011;1:e50. doi: 10.1038/tp.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luciano M, Huffman JE, Arias-Vasquez A, et al. Genome-wide association uncovers shared genetic effects among personality traits and mood states. Am J Med Genet B Neuropsychiatr Genet. 2012 Sep;159B(6):684–695. doi: 10.1002/ajmg.b.32072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014 Jul 24;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Li MY, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montag C, Reuter M. Disentangling the molecular genetic basis of personality: From monoamines to neuropeptides. Neurosci Biobehav R. 2014 Jun;43:228–239. doi: 10.1016/j.neubiorev.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 63.Flint J. The genetic basis of neuroticism. Neurosci Biobehav R. 2004 May;28(3):307–316. doi: 10.1016/j.neubiorev.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Nivard MG, Mbarek H, Hottenga JJ, et al. Further confirmation of the association between anxiety and CTNND2: replication in humans. Genes, brain, and behavior. 2014 Feb;13(2):195–201. doi: 10.1111/gbb.12095. [DOI] [PubMed] [Google Scholar]
- 65.Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010 Oct 21;68(2):182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013 Jun 21;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Moor MHM, Boomsma DI, Stubbe JH, Willemsen G, de Geus EJC. Testing causality in the association between regular exercise and symptoms of anxiety and depression. Arch Gen Psychiatry. 2008;65(8):897–905. doi: 10.1001/archpsyc.65.8.897. [DOI] [PubMed] [Google Scholar]
- 68.Fanous AH, Neale MC, Aggen SH, Kendler KS. A longitudinal study of personality and major depression in a population-based sample of male twins. Psychol Med. 2007;37(8):1163–1172. doi: 10.1017/S0033291707000244. [DOI] [PubMed] [Google Scholar]
- 69.Visscher PM, Hemani G, Vinkhuyzen AA, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014 Apr;10(4):e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


