Abstract
Autosomal-dominant missense mutations in leucine-rich repeat kinase 2 (LRRK2) are a common genetic cause of Parkinson's disease (PD). LRRK2 is a multidomain protein with kinase and GTPase activities. Dominant mutations are found in the domains that have these two enzyme activities including the common G2019S mutation that increases kinase activity by 2-3 fold. However, there is also a genetic variant in some populations, G2385R, that lies in a C-terminal WD40 domain of LRRK2 and acts as a risk factor for PD. In this study we show that the G2385R mutation causes a partial loss of the kinase function of LRRK2 and deletion of the C-terminus completely abolishes kinase activity. This effect is strong enough to overcome the kinase activating effects of the G2019S mutation in the kinase domain. Hsp90 has an increased affinity to G2385R variant compare to wild type LRRK2 and inhibition of the chaperone binding combined with proteasome inhibition leads to association of mutant LRRK2 with high molecular weight native fractions that likely represent proteasome degradation pathways. The loss of function of G2385R correlates with several cellular phenotypes that have been proposed to be kinase dependent. These results suggest that the C-terminus of LRRK2 plays an important role in maintaining enzymatic function of the protein and that G2385R may be associated with PD in a way that is different from kinase activating mutations. These results may be important in understanding the differing mechanism(s) by which mutations in LRRK2 act and may also have implications for therapeutic strategies for PD.
Keywords: Parkinson's disease, WD40 domain, Hsp90, gene mutation, kinase activity, neurite length, inclusion bodies
Introduction
Parkinson's disease (PD) is a neurodegenerative disorder with varied symptoms including tremor, rigidity, and postural instability [1]. The most common monogenic cause of PD are dominant missense mutations in the Leucine-rich repeat kinase 2 (LRRK2) gene [2-4]. Clinically, LRRK2-associated PD is indistinguishable from idiopathic cases [5].
LRRK2 is a large (286 kDa) complex protein that possesses two enzymatic domains, a ROC (Ras of complex proteins) domain that binds GTP and a kinase domain. The COR (C-terminus of ROC) domain is found between ROC and kinase domains and may contribute to the formation of LRRK2 dimers and regulation of GTP binding [6]. Other domains, including LRR (leucine-rich repeat) and WD40 domains are proposed to participate in protein-protein interactions of LRRK2. One mutation, G2019S in the kinase domain, increases kinase activity whereas mutations in the ROC and COR domain decrease GTPase activity [7-10]. It is thought that these two observations are linked in that there is evidence of communication between the GTPase and kinase activities, although the precise molecular basis is poorly understood [11].
In contrast, the function of the WD40 domain of LRRK2 remains relatively unknown. Deletion of the WD40 domain and the extreme C-terminus decreases kinase activity and limits neurotoxicity, possibly due to diminished dimer formation [12]. However, deletion of only the last 7 amino acids at the extreme C-terminus of LRRK2 also completely abolishes kinase activity [13]. Therefore, it is unclear whether the effects of deletions at the C-terminus on toxicity are due to the WD40 domain itself or are related to kinase activity.
Several amino acid variants in the WD40 domain have been found, and one, G2385R, shows a genetic association with PD [14, 15]. The effects of G2385R on LRRK2 kinase activity are controversial. Some reports have shown that G2385R LRRK2 has a decreased kinase activity [13] while others did not find any significant difference compare to WT protein [10, 16]. The G2385R substitution decreases 14-3-3 binding mediated by phosphorylation of S910/S935 residues which are both proposed to be cellular correlates of kinase activity of LRRK2 [16].
In this study we asessed how the G2385R variant affects the functional properties of the LRRK2 protein. We found G2385R decreased kinase activity, phosphorylation of LRRK2 in vivo and the ability to bind 14-3-3 proteins to about half of the WT protein. At the same time, this mutation increased binding to heat shock protein 90 (Hsp90), which plays a role in maintaining of LRRK2 stability. The effect on in vitro kinase activity was seen whether G2385R was present on a WT background or if the constructs also contained the activating G2019S mutation. Additionally, G2385R LRRK2 had a different profile on size-exclusion chromatography compared to WT protein. We speculate that G2385R destabilizes LRRK2 structure, partially misfolds the protein, and thus impairs LRRK2 function.
Materials and Methods
Constructs
2xMyc-LRRK2 constructs were cloned into pCMV plasmid as described previously [7]. 3xFlag-tagged construct of LRRK2 in pCHMWS plasmid was a generous gift of Dr. J.– M. Taymans (De Katholieke Universiteit van Leuven, Belgium). Mutagenesis was performed using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). All plasmids were analyzed with EcoRI and EcoRV restriction enzymes (Fermentas) and fully sequenced.
In Vitro Kinase Assays
HEK293FT cells were transiently transfected with 3xFlag-LRRK2 plasmid using polyethylenimine (Sigma), harvested 48 hours after transfection and lysed with buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% (v/v) Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 μg ml−1 leupeptin (Cell Signalling) supplemented with protease inhibitor cocktail (Roche). Lysates were pre-cleared with EZveiw Red Protein G Affinity Gel for 30 min and immunoprecipitated using EZview Red ANTI-FLAG M2 Affinity Gel (both Sigma) for 2 h. Complexes were then washed 3 times with wash buffer (25 mM Tris-HCl (pH 7.5), 400 mM NaCl, 1% Triton) and once with kinase buffer (25 mM Tris-HCl (pH 7.5), 5 mM beta-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2). LRRK2 was eluted with 200 μg ml−1 3xFlag-peptide (Sigma) in kinase buffer with 0.02% (v/v) Triton for 1 h. To achieve reproducible data in the kinase assays, LRRK2 protein concentrations were estimated before the experiments by running aliquots and BSA standards on 4-20% PAGE-SDS Criterion TGX pre-cast gels with subsequent staining with GelCode Blue Stain Reagent (Thermo Scientific). In some experiments, we treated cells with 1μM Geldanamycin for 2h at 37°C. In preliminary experiments, this treatment was sufficient to displace Hsp90 from LRRK2 (data not shown) but did not decrease the amount of LRRK2 recovered.
For LKKR2 autophosphorylation assays, reactions were initiated by addition of 10 μM of nonradioactive ATP (Sigma) and 10 μCi of γP33-ATP (Perkin Elmer) and incubated at 30°C for 30 min in Thermomixer R (Eppendorf) at 1400 rpm. The samples were run on 4-20% PAGE-SDS Criterion TGX pre-cast gels (Bio-rad), transferred to PVDF membrane and exposed to phophoscreen, which was subsequently imaged on a Storm phosphorimager (Amersham Pharmacia Biosciences). Results of LRRK2 autophosphorylation were normalized to the amount of LRRK2 protein detected by western blot against the FLAG peptide. Each experiment was repeated on 3-4 separate days with independent transfections, purifications and autoradiography measurements.
For estimation of LRRK2 substrate phosphorylation activity, the reaction was additionally supplemented with 200 μM lrrktide (Enzo Life sciences). Equivalent amounts of radioactivity of the γP33-ATP were added to each reaction, taking decay into consideration. At each time point, 20 μl aliquots were removed from the reaction, mixed with 10 μl of 0.5 M EDTA, and applied to P-81 paper (Whatman). The P-81 paper was then dried, washed 4 times with 75 mM phosphoric acid (Sigma), immersed onto Biosafe II (RPI) scintillation cocktail and activity was measured on an LS6500 scintillation counter (Beckman Coulter). The phosphorylation of lrrktide, estimated as CPM incorporated in the first 30 minutes of the reaction, were normalized to the amount of protein detected by western blot and to the overall incorporation of radiolabel at 60 minutes, thereby correcting for small differences in the overall CPM. Each experiment was repeated 3-4 times with independent transfections, protein purifications and time courses.
Metabolic labeling
HEK293FT cells were transfected with 3xFlag-LRRK2 constructs in 6-well plates. Twenty-four hours after transfection, cells were washed twice in phosphate-free DMEM (Invitrogen) and labeled in the same medium containing 40 μCi/well P-32 radionuclide disodium phosphate (Perkin Elmer) for 4 hours. After labeling, cells were collected in PBS, pH 7.4 and lysed in 1× cell signaling lysis buffer (Cell Signaling), containing protease (Roche) and phosphatase inhibitors (Thermo scientific). After incubation on ice for 30 min, lysates were centrifuged at 12,000 ×g for 10 min at 4°C. Supernatants were collected and precleared with 20 μl of EZveiw Red Protein G Affinity Gel (Sigma) for 30 min at 4°C then incubated with 20 μl of EZview Red ANTI-FLAG M2 affinity gel (Sigma) for 1 h at 4 °C. Immunoprecipitated proteins were washed five times in buffer containing 25 mM Tris pH 7.5, 400 mM NaCl, 1% (v/v) Triton and subjected to SDS-PAGE. Results were from three independent transfections that were metabolically labeled and separated.
Co-immunoprecipitation
HEK293FT cells were transfected with 3xFlag-LRRK2. After 48 hours, cells were lysed with buffer containing 20 mM Tris-HCl (pH 7.4), 137 mM NaCl, 3 mM KCl, 10% (v/v) glycerol, 1 mM EDTA, 0.3% (v/v) triton X-100 supplemented with protease inhibitor cocktail (Roche) and HALT phosphatase inhibitor cocktail (Thermo Scientific). Lysates were pre-cleared with EZview Red Protein G Affinity Gel for 1 h and immunoprecipitated with EZview Red ANTI-FLAG M2 Affinity Gel (both Sigma) for 2 h. EZview Red ANTI-FLAG M2 Affinity Gel were then gently washed 6 times with buffer containing 20 mM Tris-HCl (pH 7.4), 137 mM NaCl, 3 mM KCl (all KD Medical), 0.1% Triton. LRRK2 was eluted with Gentle Ag/Ab Elution Buffer, pH 6.6 (Thermo Scientific) with 0.01% (v/v) Triton for 30 min. Eluate was desalted with 7K molecular weight cutoff Zeba Spin Desalting Columns (Thermo Scientific). Each co-immunoprecipitation was repeated in 3-4 independent experiments and quantification was performed by estimating the ratio of immunoprecipitated binding partner to the amount of LRRK2 construct pulled down on the beads.
Cell Fractionation
HEK293FT cells transiently transfected with different 3xFlag-LRRK2 mutants or human lymphoblastoma cell line #00057 (Coriell) were homogenized in 1 ml of sedimentation medium (250 mM sucrose, 3 mM Tris-HCl (pH 7.3), 0.5 mM EGTA, protease inhibitors cocktail) by applying of 20 gentle strokes of a Potter-Elvenhjem homogenizer. Mouse brains (FVB/N strain) were minced into 1–2 mm pieces prior to homogenization. The resulting homogenates were centrifuged at 1,000 ×g for 10 min. The first pellet was discarded, 200 μl of supernatant was used as a total fraction and 800 μl were subsequently centrifuged at 10,000 ×g for 10 min. The second pellet was washed 2 times by gentle resuspention and centrifugation (10,000 ×g, 10 min) and used as a mitochondrial fraction. The second supernatant from cells was centrifuged at 200,000 ×g for an additional 90 min, while the second supernatant form mouse brain tissues was centrifuged at 100,000 ×g for 60 min. The pellet after the third centrifugation was taken as the microsomal fraction and the supernatant as the cytosolic fraction. Volumes of all fractions were adjusted up to 800 μl with sedimentation medium. All fractions were then lysed with lysis buffer, sonicated and cleared at 20,000 ×g for 10 min. All procedures were carry out at 4°C. In each case, experiments are representative of triplicate purifications performed on different days. All mouse work followed the guidelines approved by the Institutional Animal Care and Use Committees of the National Institute of Child Health and Human Development.
Size-Exclusion Chromatography
HEK293FT cells were harvested in a PBS supplemented with Roche protease inhibitors cocktail and lysed using 4 freeze-thaw cycles in liquid nitrogen. In some experiments, we incubated the cells at 37°C with 1μM Geldanamycin for 2h before extraction. To inhibit the proteasome, we exposed cells to 25μM MG132 alone or combined with 1μM Geldanamycin for 4h at 37°C. Insoluble material was removed from the lysates by centrifugation and lysates were then passed through 0.45 μm filters (Nanosep MF, Pall Life Sciences) prior to analysis. Size exclusion chromatography was performed using a BioAssist G4SWXL column (7.8mm × 30.0cm, Tosoh Bioscience) with PBS as the mobile phase (0.8mL/min, 4°C). Samples (0.100 mL, 0.7% of the total geometric column volume compared to a maximum recommended by the manufacturer of <2%) were injected into the column and fractions (0.28 mL) were collected. Aliquots containing LRRK2 (elution volume 5.75–11.75mL) were analyzed by SDS-PAGE followed by Western blot for FLAG-LRRK2. Thyroglobulin (660 kDa), Ferritin (440 kDa) and IgM (~1 MDa) were used as standards (Fig. S1). Distribution of LRRK2 in each fraction was estimated by measuring the integrated optical density of the LRRK2 band, corrected to the total amount of immunoreactivity in all fractions. Each construct or treatment was transfected on at least three separate days and results were compiled for all experiments.
Differential Scanning Fluorimetry
Differential Scanning Fluorimetry (DSF) was performed as specified elsewhere [17, 18]. Briefly, SYPRO Orange (Invitrogen) fluorescent dye was used to obtain thermal denaturation curves with excitation and emission wavelengths of 488 and 520 nm respectively and a thermal gradient from 25 °C to 99 °C in 1 °C increments on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Each 20 μl reaction contained 25 mM HEPES (pH 7.5), 150 mM NaCl, 0.02% Triton, 200 μg/ml 3xFlag-peptide, 80 nM 3xFlag-LRRK2 and 5× SYPRO Orange. The mid-point of thermal denaturation (Tm) was estimated as a temperature with the maximal first derivative of the normalized fluorescence.
Immunostaining
HEK293FT or SH-SY5Y cells were seeded at 7×104 per well on 8-well slides (Millicell EZ slide, Millipore) coated with poly-D-lysine (Sigma). HEK293FT cells were cultured in DMEM with 10% FBS without antibiotics. SH-SY5Y cells were maintained in OPTI-MEM without antibiotics. Two hours later, transfections were performed with 3xFlag-LRRK2 constructs using lipofectamine 2000 (Invitrogen) for HEK293FT cells and effectene (Qiagen) for SH-SY5Y cells. Twenty-four hours (HEK293FT cells) or forty-eight hours (SH-SY5Y cells) after transfection, the cells were treated with 10μM LRRK2-IN-1 in DMSO or DMSO alone for 2h at 37C. Cells were then fixed with 4% paraformaldehyde in PBS, blocked with 5% FBS in PBS and stained with anti-Flag (Sigma) in blocking solution with addition of 0.1% Triton100 for 1h. After several washes with PBS, anti-mouse antibody labeled with AlexFluor-488 (Invitrogen) and TO-PRO-3 nuclear stain (Invitrogen) were applied followed by additional washing steps. LRRK2 localization was imaged and counted by an operator blinded to construct/treatments using a Zeiss LSM510 META Confocal Imaging System with 100X/1.4NA objective. At least triplicate experiments, with separate cultures and transfections, were used for each construct.
ImageStream analysis
HEK293FT cells were transfected with 2xMyc-LRRK2 constructs and 48h later, trypsinized cells were fixed for 20 minutes in 4% PFA/PBS and then washed in PBS to remove the fixative. Cell suspensions were then incubated for 1 hour at RT with mouse anti-myc antibody (Roche) in 0.1% Triton/5% FBS/PBS using 1 μg of antibody per 1×106 cells. Cells were washed with PBS and then incubated with the Alexa Fluor 647-conjugated goat anti-mouse IgG secondary antibody (Invitrogen) for 40 min at RT. After washing, cells were counterstained using 10 μg/ml DAPI (Invitrogen) for 5 min at RT then washed in PBS. The cells were assayed using the Amnis ImageStreamX flow cytometer and imaged using a 40X/0.75NA objective. A far-red diode laser (658 nm) and a violet diode laser (405 nm) were used to excite Alexa Fluor 647 and DAPI respectively, and a standard filter array and a high resolution camera was used to image the cells labeled with each dye. The data were processed using IDEAS image analysis software (Amnis Corporation). Approximately 500 myc (LRRK2)-positive cells per construct were analyzed for the presence/absence of inclusions and the percentage of inclusion positive cells was calculated as the number of cells with inclusions divided by the total number of LRRK2-positive cells multiplied by 100.
Neurite outgrowth measurements
M17 human neuroblastoma cells were seeded on PDL-coated (0.05mg/ml) Lab Tech II 2-well chambers (Nalge Nunc) at 105 cells/well and differentiated for total of 9 days with 10 μM Retinoic Acid (Sigma). After 7 days of differentiation, the cells were transfected with 3xFlag-LRRK2 constructs using Effectene (Qiagen). After 48 hours, the cells were fixed with 4% paraformaldehyde (PFA) in PBS and stained using mouse anti-Flag (1:400, Sigma Clone M2) and chicken anti-ß-III-tubulin (1:400, Novus Biologicals) followed by anti-mouse-Alexa Fluor 488 and anti-chicken-Alexa Fluor 647 (both at 1:500, Invitrogen). LRRK2-positive cells were imaged using a Zeiss LSM510 META Confocal Imaging System with 63X/1.4NA objective and neurite length was determined by tracing the entire length of the longest process from the center of the cell using ImageJ. Each experiment was repeated at least three times with independent batches of cells and transfections.
Statistical Analysis
For comparisons of multiple LRRK2 constructs, one-way ANOVA was used with Tukey's honest significance post hoc tests for individual comparisons. In experiments where different LRRK2 constructs were used in the absence and presence of Geldanamycin, we used two-way ANOVA with mutant and treatment as factors, with Bonferroni post hoc tests to compare the effects of Geldanamycin versus DMSO treatment for each construct.
Results
The G2385R LRRK2 mutant has decreased kinase activity
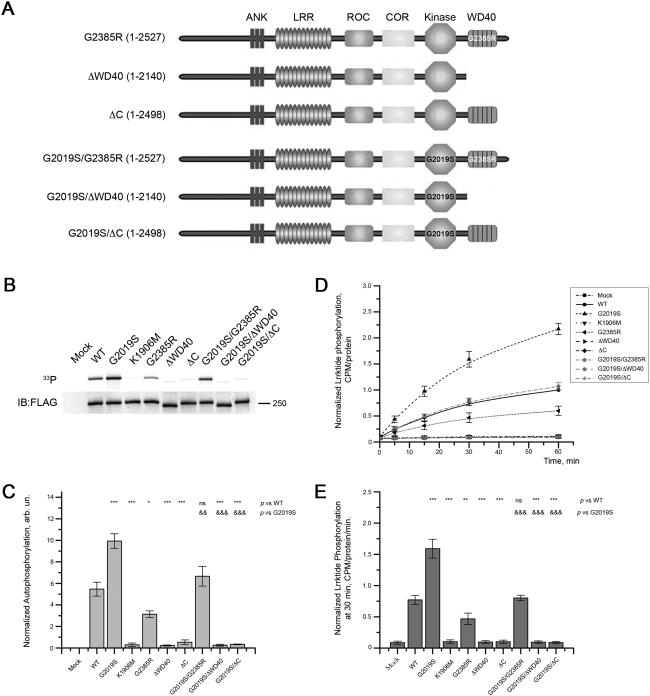
To test the effect of the G2385R variant on kinase activity, we designed a set of mutants with G2385R substitution and C-terminus truncated forms of LRRK2 either on a wild type background or with the G2019S hyperactivating mutant (Fig. 1A). G2385R LRRK2 possessed about 60% of wild-type autophosphorylation activity (Fig. 1B, 1C; p < 0.05). Lrrktide phosphorylation by LRRK2 G2385R was also lower than for wild type protein (Fig. 1D, 1E). The G2029S/G2385R double mutant of LRRK2 had kinase activity intermediate between proteins with either mutation alone and indistinguishable from WT LRRK2 in autophosphorylation (Fig. 1B, 1C) or lrrktide (Fig. 1D, 1E) assays.
Figure 1. The WD40 domain and C-terminus influence LRRK2 kinase activity.
(A) Schematic representation of LRRK2 mutants used in the study. The first panel depicts full-length LRRK2 with G2385R substitution in WD40 domain. Next panel is wild-type LRRK2 with truncation of WD40 domain with the extreme C-terminus (LRRK2 1–2140). The third panel from the top is WT LRRK2 that possesses WD40 domain but lacks the last 29 amino acid at the extreme C-terminus (LRRK2 1–2498). Forth, fifth, and sixth panels represent equivalent mutants with an additional G2019S substitution in the kinase domain. (B) The indicated forms of LRRK2 were subjected to autophosphorylation activity assays. The upper panel is an autoradiogram and the lower panel is a respective western blot showing equal amounts of LRRK2 mutants used in experiment. (C) Quantitation of replicate experiments similar to (B) shows that removal of either WD40 domain or the C-terminal 29 amino acids abolished autophosphorylation activity. Introduction of the G2385R mutation to WT LRRK2 or G2019S mutant reduced their kinase activity by 40%. Autophosphorylation was normalized to the amount of LRRK2 in the gel estimated by western blots. (D) Truncated forms of LRRK2 and G2385R mutations were further assayed for their ability to phosphorylate the model substrate lrrktide at times from 5-60 minutes. (E) Normalized CPM at 30 min of lrrktide phosphorylation, corrected to amount of LRRK2 protein at the end of the experiment, confirmed the inhibitory effects of mutations on kinase activity. For the plots in (C) and (E), one-way ANOVA with Tukey's honest significance test was used to assess statistical significance relative to WT (*, P<0.05; **, P<0.01; ***, P<0.001; ns, not significant) or G2019S (&) LRRK2 depicted in the upper part of the plots. Data for each time-point show mean±SEM (n = 4).
To further delineate the regions of the C-teminus required to influence kinase activity, we introduced stop codons just before (1–2140 or LRRK2ΔWD40) or just after (1–2498 or LRRK2ΔC) the predicted WD40 domain of the protein. Both deletions completely abolished autophosphorylation activity of WT or G2019S protein (Fig. 1B, 1C). Similar results were obtained for 2xMyc-LRRK2 constructs (data not shown). The truncated forms of LRRK2 also showed no lrrktide phosphorylation activity, similar to the kinase dead version (K1906M) of the protein (Fig. 1D, 1E).
These data show that the extreme C-terminus is essential for LRRK2 kinase activity and this effect is sufficient to overcome the activating effect of G2019S. Additionally, the G2385R mutation decreases kinase activity by approximately 40% and does not change the ratio in decrease of phosphorylation activity between WT and G2019S LRRK2 backgrounds. In turn, this indicates that the G2385R variant has a partial effect on LRRK2 kinase activity and supports the concept that the WD40 domain and C-terminus influence activity of the kinase domain of LRRK2.
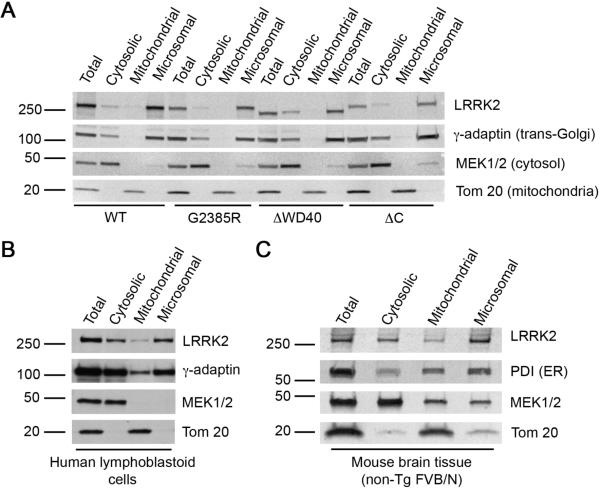
LRRK2 predominantly localizes to microsomes
It has been suggested that the subcellular localization of LRRK2 could affect its biochemical properties, with membrane associated protein forming kinase active dimers [19]. We considered whether the WD40 domain and the extreme C-terminus could influence intracellular distribution of the protein and therefore explain the effects on activity. To test this hypothesis, we performed fractionation of HEK293FT cells transiently transfected with WT and G2385R LRRK2 as well as WD40 and the extreme C-terminus deleted mutants. All LRRK2 isoforms were predominantly associated with the microsomal-enriched fraction of HEK293FT cells. A smaller portion of 3xFlag-LRRK2 was found in the cytosol with only small amounts in the mitochondria-enriched fraction (Fig. 2A). Because it has been suggested that LRRK2 is found in the mitochondria, we also performed immunocytochemistry for LRRK2 with Mitotracker Orange staining. Again, the amount of LRRK2 costaining with mitochondria was modest (Fig. S2). Endogenous LRRK2 had similar pattern of subcellular fractionation in human lymphoblastoid cells (Fig. 2B) and mouse brain tissue (Fig. 2C), although the quality of fractionations for these types of cells/tissue was not as good as for HEK293FT cells, seen as contamination of cytosolic and mitochondrial fractions with microsomal markers (γ-adaptin or PDI).
Figure 2. Distribution of LRRK2 in cellular compartements.
(A) Full-length WT and G2385R 3xFlag-LRRK2 as well as C-terminus truncated forms of the protein are predominantly associated with microsomal-enriched fraction when transiently expressed in HEK293FT cells. (B, C) Endogenous LRRK2 was mostly found in microsomal-enriched fraction of lymphoblastoid cells or whole brain homogenates of 2 month old mouse (FVB/N) using MJF2 antibody. The following subcellular markers were used for fraction identification: PDI for endoplasmic reticulum or γ-adaptin for trans-Golgi (both second panels), MEK1/2 for cytosol (third panels), and Tom 20 for mitochondria (bottom panels). Each fractionation is representative of triplicate experiments performed at different times.
These results suggest that LRRK2 is found predominantly in microsomal fractions and allowed us to test if the G2385R mutation altered this. Deletion of WD40 domain together with the extreme C-terminus slightly shifted the 3xFlag-LRRK2 fractionation pattern to the cytosol, but the majority remained microsomal. G2385R substitution or deletion of the extreme C-terminus alone did not change the fractionation profile of 3xFlag-LRRK2 (Fig. 2A). Similar results were obtained with 2xMyc-LRRK2 mutants (data not shown). Therefore, the absence of kinase activity in C-terminus truncated LRRK2 mutants and reduced autophosphorylation and lrrktide phosphorylation found in G2385R variant is not due to a change of subcellular distribution.
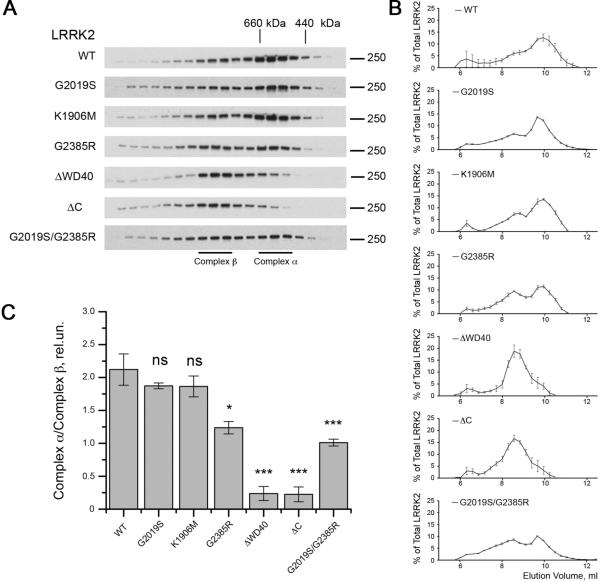
The G2385R mutation changes the behavior of LRRK2 under native separation techniques
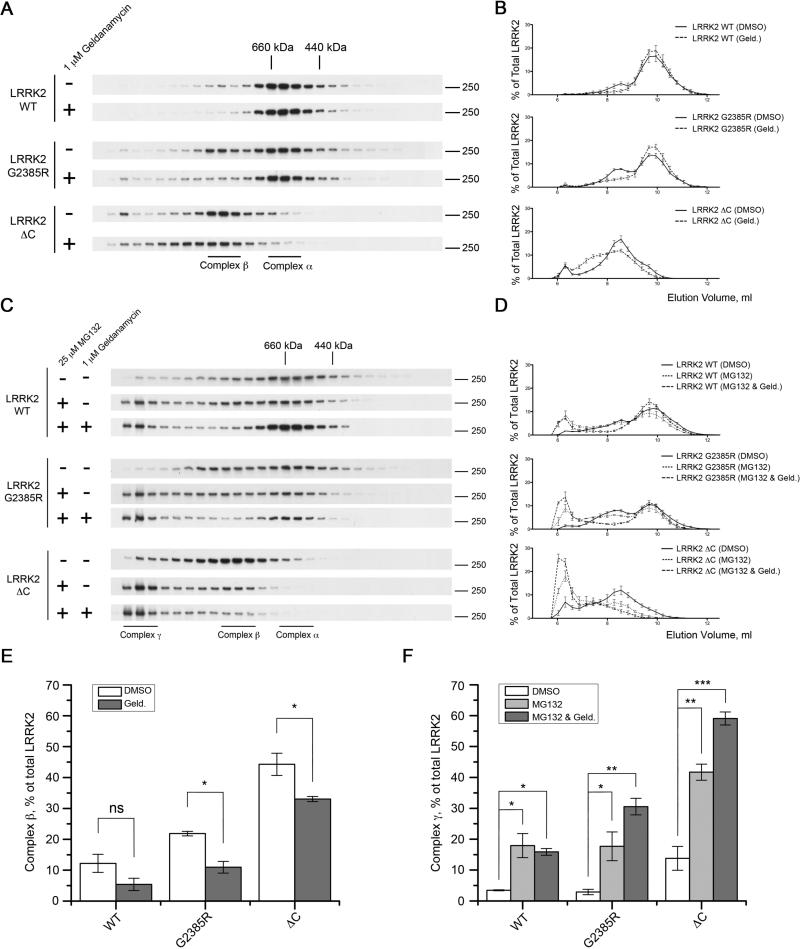
For many kinases, proper folding is crucial for the normal phosphorylation activity and structural stability [20, 21]. To examine the effects of mutations on the behavior of LRRK2 under native conditions, we used size exclusion chromatography and found that WT, G2019S and K1906M 3xFLAG-LRRK2 eluted over a broad range of molecular weights from ~400-1000 kDa but before the void volume of the column (Fig. S1). Within this range, there was a higher signal for LRRK2 immunoreactivity centered at 10.0mL and a shoulder of the separation at ~8.5mL (Fig. 3A, 3B), which are estimated to be between 440 and 660 kDa and between 660 kDa and 800 kDa, respectively. A small amount of LRRK2 eluted at very high molecular weights (>1 MDa) but before the void volume of the column. Surprisingly, C-terminus truncated forms of LRRK2 (ΔWD40 and ΔC) were eluted from the column at lower volumes, with highest immunoreactivity found at ~8.5mL, despite the fact that they have lower molecular weights compare to the intact protein. Furthermore, proteins with the G2385R substitution tended to elute more in the ~8.5mL fraction compared to wild type protein (Fig. 3A, 3B). To capture this behavior, we considered that the 440-600kDa and 660-800kDa regions of the separation as potentially distinct protein complexes, which we labeled α and β for clarity (Fig. 3A). Under these separation conditions, these two regions are not completely distinct and we defined complex α representing elution volume 9.67–10.23 ml while complex β represents 8.27–8.83 ml. Calculating the ratio of the amount of protein in the lower molecular weight to the higher molecular weight complexes, we found a striking effect of the deletion mutations and an intermediate effect of the G2385R mutation either on a WT or G2019S background (Fig. 3C). These results show that the apparent molecular weight of WT LRRK2 is approximately 600 kDa, as previously reported [12, 22, 23], while the two truncated proteins exist as apparently higher molecular weight species, also consistent with previous results [12]. Surprisingly, the G2385R mutant exhibits behavior intermediate between these two extremes.
Figure 3. C-terminal region is essential for the maintenance of LRRK2 native conformation.
(A) Size exclusion chromatography was used to demonstrate that WT, G2019S and K1906M 3xFlag-LRRK2 have elution volumes between 440 and 660 kDa (complex α), a second region of ~600-800 kDa (complex β) and a minor amount eluting at a higher molecular weight. C-terminus truncated LRRK2 mutants elute at relatively higher apparent molecular weights compared to wild type protein and the G2385R variants elute broadly. Markers indicate elution volumes of the standards ferritin (440 kDa), thyroglobulin (660 kDa) and IgM (~1 MDa). (B) Quantification of amounts of LRRK2 in each fraction compared to total immunoreactivity for all fractions for constructs as indicated (n = 3, mean±SEM). (C) Summary of the ratio of complex α to complex β. The percentage of total LRRK2 was calculated for each fraction collected, and the sums of the three fractions representing complexes α and β were used to calculate the ratios shown. Statistical significance relative to WT was assessed by one-way ANOVA with Tukey's honest significance test (*, P<0.05; **, P<0.01; ***, P<0.001; ns, not significant; n = 3).
The shift towards apparently higher molecular weight caused by the deletion of the C-terminus, despite the smaller size of the protein, or to a lesser extent the G2385R substitution could be explained by conformational shifts that increase its hydrodynamic volume. To test whether the G2385R mutation destabilized LRRK2, we used differential scanning fluorimetry [17]. DSF did not revealed any difference in thermal stability between WT and G2385R LRRK2 (Fig. S3). These results indicate that the G2385R substitution does not grossly perturb the secondary structure of LRRK2 to the level at which we can detect with DSF. We therefore next considered whether the apparent increase in molecular weight of some of these variants might be due to enhanced associations with protein binding partners.
The G2385R mutation affects protein-protein interactions of LRRK2
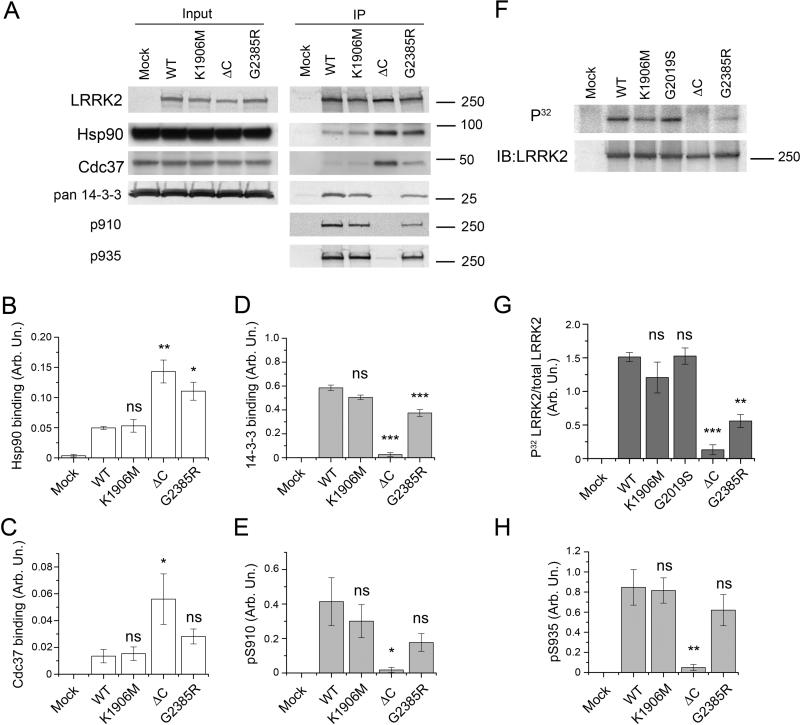
LRRK2 has a number of known protein-protein interaction partners and, as well as being a kinase, LRRK2 is also a phosphoprotein. 14-3-3 proteins bind LRRK2 at S910 and S935 in a manner that depends on an unknown kinase that phosphorylates these sites [16, 24, 25]. This effect is also sensitive to overall conformation of LRRK2, as mutations in the ROC and COR domains that do not alter kinase activity can also disrupt phosphorylation and recruitment of 14-3-3. Furthermore, LRRK2 is also known to bind Hsp90 cell division cycle 37 homolog (Cdc37), probably via the kinase domain [26].
In order to estimate binding, we performed immunoprecipitation experiments against endogenous Hsp90, cdc37 and 14-3-3 proteins (Fig. 4A). Strikingly, both deletion of the extreme C-terminus and G2385R mutation caused an increase in Hsp90 binding to LRRK2, which correlated with higher binding of Cdc37 (Fig. 4A, 4B, 4C). Interestingly, that interaction with Hsp90 was independent of kinase activity of LRRK2 as a kinase dead (K1906M) mutant bound Hsp90 similarly to WT protein.
Figure 4. Intact C-terminus is crutial for LRRK2 interaction with other proteins and for its phosphorylation status in vivo.
(A) Co-immunoprecipitation of endogenous 14-3-3 proteins and transiently transfected 3xFlag-LRRK2 overexpressed in HEK293FT cells with Flag affinity gel. Input (left panel) and immunoprecipitates (right panel) were probed with anti-pan-14-3-3 (Santa-Cruz), anti-Hsp90 (Cell signaling), anti-Cdc37 (BD Biosciences), and MJF2 anti-LRRK2 antibody (Epitomics) as well as with antibody to S910/S935 phosphosites (University of Dundee). Representative experiments are shown. Normalized quantifications of Hsp90 (B), Cdc37 (C) and 14-3-3 (D) binding relative to immunoprecipitated LRRK2 and intensity of signals from pS910, pS935 phosphosites (E and H respectively) for different LRRK2 mutants. (F) Phosphorylation of LRRK2 immunoprecipitated after metabolic labeling with [P32]orthophosphate (top panel). The bottom panel demonstrates loading of LRRK2. (G) LRRK2 phosphorylation in vivo represented as autoradiography results normalized to the amount of protein detected by western blotting. Statistical significance relative to WT was assessed by one-way ANOVA with Tukey's honest significance test (*, P<0.05; **, P<0.01; ***, P<0.001; ns – not significant). All experiments were replicated at least 3 times. Data presented as mean±SEM.
Although 14-3-3 proteins were bound by WT and kinase dead (K1906M) 3xFlag-LRRK2 in simlar amounts, the 14-3-3 binding ability of G2385R mutant was significantly decreased compared to WT protein and the extreme C-terminus deficient mutant did not bind any detectable amount of 14-3-3 proteins (Fig. 4A, 4D). It has been suggested that the binding of 14-3-3 to LRRK2 requires phosphorylation at S910/S935 [24]. To check phosphorylation of S910/S935 in f the LRRK2 variants, we performed metabolic labeling with [P32]orthophosphate. Although the kinase dead (K1906M) and kinase activating (G2019S) LRRK2 mutants had phosphorylation similar to wild-type protein, G2385R substitution significantly decreased LRRK2 phosphorylation while deletion of the extreme C-terminus completely abolished it (Fig. 4F, 4G). These results indicate that in vivo LRRK2 is phosphorylated by other kinases in a manner that depends on the WD40 and C-terminal regions. In line with these experiments, we found that decreased 14-3-3 binding ability of different LRRK2 mutants correlated with phosphorylation status of S910/935 residues (Fig. 4E, 4H). This suggests that the C-terminus of LRRK2 influences phosphorylation of Ser910 and Ser935 by other kinases and the G2385R variant is intermediate to WT LRRK2.
Collectively, these results suggest that one of the effects of mutations at the C-terminus of LRRK2 is to change binding to several protein interaction partners. Given this, we next asked whether binding of one of these was sufficient to explain the behavior of LRRK2 under native conditions noted in our earlier experiments.
Hsp90 contributes to the behavior of G2385R LRRK2 under native conditions
Taking into consideration the stronger association of Hsp90 with LRRK2 mutants that eluted at higher apparent molecular weights compared to the wild type protein, we hypothesized that the binding of Hsp90 itself could be important in mobility under native conditions. In order to test this, we treated 3xFlag-LRRK2 transfected HEK293FT cells with the Hsp90 inhibitor geldanamycin (1 μM) for 2 h before lysing the cells and separating proteins using size-exclusion chromatography. Inhibition of Hsp90 binding changed the elution profiles of G2385R and the extreme C-terminus truncated LRRK2 (Fig. 5A, 5B). Pretreatment with 1 μM geldanamycin for 2 hours selectively diminished the relative amount of the G2385R protein that eluted in the slightly higher apparent molecular weight fractions, labeled here as complex β, while the elution profile of C-terminus truncated protein was shifted to higher molecular weights (Fig. 5A, 5B). A smaller effect was seen with wild type LRRK2. These results suggest that disruption of a single binding partner, Hsp90, is sufficient to change the native elution profile of LRRK2 suggests that the two regions of the native separation represent distinct protein complexes.
Figure 5. Inhibition of Hsp90 binding facilitates proteosomal degradation of G2385R and C-terminus truncated LRRK2.
(A) Mobility of WT, G2385R and C-terminus truncated 3xFlag-LRRK2 on size-exclusion chromatography changes after 2 h pretreatment of transiently transfected HEK293FT cells with the Hsp90 inhibitor geldanamycin (1 μM). (B) Respective chromatograms displaying the elution profiles of different 3xFlag-LRRK2 mutants after pretreatment of HEK293FT cells with either DMSO alone or 1 μM geldanamycin. (C) Pretreatment of the HEK293FT cells with geldanamycin (1 μM) and proteasomal inhibitor MG132 (25 μM) for 4 h promotes accumulation of very high molecular weight forms of WT, G2385R and C-terminus truncated LRRK2 (complex γ). (D) Chromatograms depicting the elution profiles of WT, G2385R and C-terminus truncated 3xFlag-LRRK2. (E) Histogram showing the change in relative amount of complex β for WT, G2385R and ΔC LRRK2 after inhibition of Hsp90 binding with geldanamycin (1 μM) for 2 h (filled bars) compared to controls (open bars) and (F) representing relative amount of complex γ after pretreatment of HEK293FT cells with DMSO (open bars), 25 μM MG132 alone (grey bars) or 25 μM MG132 and 1 μM geldanamycin (filled bars). Data shown as mean±SEM (n = 3).
Hsp90 can help fold and stabilize LRRK2, and prevent degradation by the proteasome [27]. Under native conditions, we saw accumulation of a small amount of very high molecular weight species that eluted before the void volume of the column for wild type, G2385R, and C-terminus truncated LRRK2, particularly for the truncated construct (Fig. 5A, 5B). These fractions were enhanced when we treated cells with the proteasome inhibitor MG132 (25 μM) alone for 4 hours (Fig. 5C, 5D). Importantly, the accumulation of the high molecular weight species of the G2385R and C-terminus truncated LRRK2 mutants was facilitated by additional inhibition of Hsp90 binding with geldanamycin (1 μM), while wild type protein was less affected.
Because the behavior of the high molecular weight native species was distinct, and responded to MG132, we infer that these >1MDa regions represent LRRK2 associated with the proteasome but not yet degraded. We therefore labeled these as complex γ, assuming that they represent LRRK2 bound to different partners than the other two regions of the separation. We used these three complexes to summarize the effects of Geldanamycin and MG132 treatment on the behavior of LRRK2 under native conditions. In the Geldanamycin experiment, we see that there is a decrease in complex β, and that this reaches statistical significance for G2385R and the C-terminal deletion construct (Fig. 5E). Conversely, incubation with MG132 alone increases the amount of the high molecular weight species labeled here as complex γ significantly for all constructs (Fig 5F). This effect is increased further for G2385R and the C-terminally deleted LRRK2 in the presence of Geldanamycin. Collectively, these results suggest that Hsp90 is essential for maintaining stability of LRRK2 and inhibition of this activity promotes proteasomal degradation of LRRK2 [27], although it should be noted that the timing of exposure in the current experiments was deliberately short to minimize loss of LRRK2 due to proteasome turnover. Furthermore, these results suggest that the chaperone activity of Hsp90 is more important for the G2385R mutant compared to the wild type LRRK2.
Hsp90 does not control the kinase activity of LRRK2
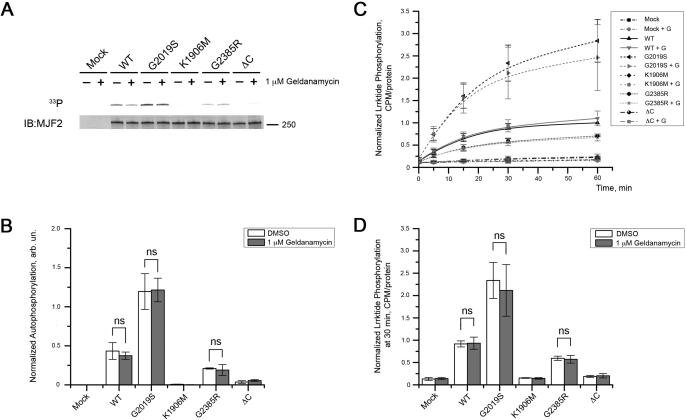
Given the increased association of Hsp90 to G2385R and, to a greater extent, delta C-terminus LRRK2 compared to other variants, we next asked whether this enhanced binding was responsible for the lower observed kinase activity. To test this, we exposed cells to 1μM Geldanamycin for 2 h then extracted LRRK2 and measured kinase activity using autophosphorylation (Fig. 6A, 6B) or lrrktide (Fig. 6C, 6D) assays. In both sets of assays we replicated the results in figure 1 that G2019S had higher (~250%), and G2385R and delta-C had lower (~60%) of wild type protein. However, we found that Geldanamycin did not alter the activity of any of the variants. Using two-way ANOVA, the difference between mutations was highly significant (P<0.0001) whereas the effect of geldanamycin was not and there was no significant interaction term. These results show that Hsp90 is not important for kinase activity of LRRK2 and more likely influences protein folding and/or turnover by the proteasome.
Figure 6. Inhibition of Hsp90 binding by geldanamycin does not affect kinase activity of LRRK2.
HEK239FT cells were transiently transfected with 3xFlag-LRRK2 and treated with 1 μM geldanamycin or DMSO alone for 2 hours prior lysis. Proteins were further purified and subjected to autophosphorylation (A) and lrrktide (B) assays. (A) The upper panel depicts an autoradiogram and the lower panel represents respective western blot. (B) Autophosphorylation activity of different LRRK2 mutants was normalized to the amount of protein with pretreatment with 1 μM geldanamycin (grey bars) or DMSO alone (open bars). (C) The same proteins were analyzed for their ability to phosphorylated lrrktide. Data normalized to CPM at 60 min point detected for wild type LRRK2 pretreated with the vehicle (DMSO) compared to 1 μM geldanamycin for 2 hours (“+ G”). (E) Normalized CPM at 30 min of lrrktide phosphorylation showed similar effects of mutations on kinase activity, but no difference was detected when HEK293FT cells were pretreated with 1 μM geldanamycin compare to DMSO. For the histograms in (B) and (D), two-way ANOVA with Bonferroni post-hoc test was used to assess statistical significance between pretreatment of cells with 1 μM geldanamycin or DMSO alone (mean±SEM, n = 3).
The G2385R substitution prevents formation of skein-like structures in HEK293FT cells
The above data suggest that C-terminal deletions and G2385R mutation decrease kinase activity. However, it remains possible that these results are true only in vitro and might not be reflective of the effects of mutations within living cells. Recognizing that there is no well-established direct measure of LRRK2 kinase activity in vivo, we used several phenotypes that have been proposed to depend on the kinase activity of LRRK2.
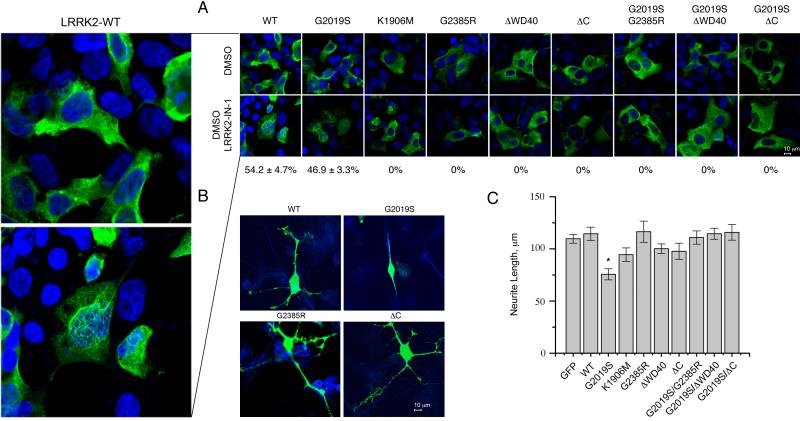
It has been shown previously that dephosphorylation of S910 and S935, associated with loss of 14-3-3 binding, causes the formation of skein-like inclusions in HEK293FT cells [24]. These effects can be triggered by application of the LRRK2 selective inhibitor LRRK2-IN-1 [28]. Using this approach, we confirmed that LRRK2-IN-1 caused redistribution of kinase active WT and G2019S LRRK2 to skein-like structures but did not change localization of kinase dead K1906M LRRK2 mutant. In contrast, neither C-terminus truncated LRRK2 mutants or proteins with G2385R substitution revealed formation of skein-like inclusions before or after the treatment with LRRK2-IN-1 (Fig. 7A). Similar results were seen in SH-SY5Y cells (Fig. S4). These results suggest that the C-terminal deletions and the G2385R mutations are insensitive to inhibition by LRRK2-IN1 in a cellular context and that the loss of 14-3-3 binding by itself is not sufficient to cause inclusion body formation.
Figure 7. LRRK2 G2385R behaves similarly to kinase dead mutant in cellular assays.
(A) HEK293FT cells were transfected with the indicated constructs and treated with DMSO (upper panels) or the selective LRRK2 inhibitor, LRRK2-IN1 (lower panels), then stained for LRRK2 (green) and nuclei (blue). Skein-like inclusions were identified as cytoplasmic fibrillar structures formed after application of LRRK2-IN-1. The percentage of LRRK2-positive cells with skein-like cellular structures are shown below the LRRK2-IN1 treated cells and were calculated based on 3 independent experiments (mean±SEM). Panel on the left is a magnification of HEK293FT cells transfected with 3xFlag-LRRK2-WT and treated with DMSO (upper plot) or LRRK2-IN-1 (lower plot). (B) M17 neuroblastoma cells were transfected with indicated constructs and stained for LRRK2 (green) and the neuronal marker β-III-tubulin (blue). Expression of G2019S LRRK2 has a dramatic effect on neurite length compared to wild type (left) or Δ-C LRRK2 (right). (C) Quantitation of neurite length from LRRK2-positive cells transfected with the full range of constructs used in this study or 3x-Flag-GFP shows that only G2019S LRRK2 decreases neurite length relative to wild-type protein (P<0.05, one-way ANOVA with Tukey's HSD post-hoc test, n = 15 cells measured and representative of duplicate or more experiments per construct).
We supplemented these assays with an automated unbiased ImageStream analysis. The percentage of HEK293FT cells with inclusions correlated with kinase activity of LRRK2 mutants in that G2019S mutant demonstrated increased proportion of cells with inclusion bodies while D1994A kinase dead LRRK2 revealed decrease in formation of these structures (Fig. S5). G2385R and G2019S/G2385R double LRRK2 mutant did not show significant difference in percentage of cells with inclusions compared to the wild type protein. Additionally, deletion of C-terminus completely abolished inclusion body formation, again consistent with the hypothesis that these mutations are kinase dead in a cellular context.
Finally, we measured neurite length in M17 neuroblastoma cell line, which has also been shown to be dependent on LRRK2 kinase activity [29]. Replicating previous work, we found that the activating G2019S mutation decreased the average length of neurites and, at these levels of overexpression, we did not see an effect of WT LRRK2 compared to a control protein, GFP (Fig. 7B, 7C). Importantly, deletions of the C-terminus or inclusion of the G2385R mutation placed on a G2019S background prevented the neurite shortening effect. Overall, these data show that in independent assays of LRRK2 in intact cells, the C-terminus is required for kinase activity and that the G2385R mutant behaves as a kinase dead protein in a cellular context.
Discussion
In the present study we investigated the effects of the G2385R mutation on the function of LRRK2. Because G2385R is a risk factor for PD, we predicted that it might share some properties with mutations that are inherited in a Mendelian manner, although perhaps with a quantitatively smaller effect. Surprisingly, we found that in many assays G2385R behaves in the opposite way to pathogenic mutations and can in some assays overcome the effects of the G2019S variant that has attracted much attention because of its ability to cause kinase activation.
In our studies, the G2385R mutant of LRRK2 has a decreased kinase activity, reduced phosphorylation of the S910/S935 sites and a concomitant diminished ability to bind 14-3-3 proteins. In contrast, there was higher binding to Hsp90 which correlates with an accumulation of slightly higher molecular weight forms under native separation conditions. The cellular effects of the protein with G2385R substitution were also different from those of WT LRRK2 and, again, were sufficient to overcome the effects of the G2019S mutation when the two variants were placed on the same molecule (Table 1).
Table 1.
Comparison of functional properties of different LRRK2 mutants.
| Functional properties | LRRK2 isoforms | |||
|---|---|---|---|---|
| WT | K1906M | ΔC | G2385R | |
| Kinase activity | ++ | – | – | + |
| Elution peak | 440–660 kDa | 440–660 kDa | Above 660 kDa | 440–660 kDa, above 660 kDa |
| 14-3-3 binding ability | ++ | ++ | – | + |
| Phosphorylation of S910/S935 | ++ | ++ | – | + |
| Hsp90 binding | + | + | +++ | ++ |
| Metabolic labeling | ++ | ++ | – | + |
| Skein-like cellular structures | ++ | – | – | – |
| Neurite length | ++ | ++ | ++ | ++ |
Binding of Hsp90 to LRRK2 has been reported previously by several independent groups [27, 30-32]. Our current data confirms this and suggests that Hsp90 plays an important role in formation of the proportion of LRRK2 that separates at apparently higher molecular weight using native techniques such as SEC. Mutations such as G2385R and, more dramatically, deletions of the C-terminus that bind Hsp90 more tightly than WT LRRK2 cause a shift towards these higher apparent molecular weight regions of the separation. Because brief treatment of cells with the Hsp90 inhibitor geldanamycin decreases the amount of LRRK2 separating between ~660-800 kDa, we infer that this species is represents LRRK2 bound to chaperone complexes. Due to the accumulation of LRRK2 at very high molecular weight fractions (>1 MDa, but below the void volume of the column used here) after treatment with the proteasome inhibitor MG132, we further infer that these fractions represent LRRK2 that is associated with, and would normally be degraded by, the proteasome. The proteasome is at least one pathway by which LRRK2 is degraded [27] and our proposal is that Hsp90 binding occurs to maintain properly folded LRRK2 protein and prevent such degradation.
The behavior of LRRK2 under SEC in this paper is largely consistent with previous studies using native separation techniques that identify a prominent region of immunoreactivity at ~500-600 kDa. Given the limitations of precision of molecular weight estimates of SEC, this has been previously interpreted to represent dimeric LRRK2 [22, 23], but this interpretation has been challenged using alternative techniques that suggest it may represent LRRK2 monomer [33]. In the absence of a source of LRRK2 purified to homogeneity, this interpretation will probably remain uncertain, but the overall behavior of LRRK2 is consistent between laboratories, as is the presence of a range of species at higher apparent molecular weights discussed above. One discrepancy is that in our previous work [22], a triple kinase dead mutant of LRRK2 (K1906A, D1994A, D2017A) eluted above 600 kDa and into the void volume of the column used there while in our current experiments the single kinase mutant (K1906M) eluted predominately between 660 and 440 kDa. Although we cannot directly compare these experiments performed under different conditions, we have since found the triple kinase dead to be partly unstable in cells as are some mutations that lack the ability of LRRK2 to bind GTP/GDP such as T1348N (Ruth Chia, David Hauser and Mark Cookson, unpublished observations). The K1906M variant is less prone to degradation and, as shown here, does not demonstrate enhanced Hsp90 binding so is likely more properly folded and therefore may be a more useful kinase dead construct than less conservative mutations.
The lack of effect of K1906M on the behavior of LRRK2 in terms of formation of different protein complexes is important to the interpretation of our current results. Because the C-terminal deletion constructs are kinase inactive, and G2385R is less kinase active, the behavior of the protein might be due either to diminished kinase activity or related more directly to the role of the C-terminus in LRRK2 behavior. Wild type, K1906M, and G2019S LRRK2 all demonstrated similar in vivo phosphorylation patterns and were phosphorylated specifically at S910 and S935, consistent with previously published results [16, 24] which is likely because LRRK2 is predominantly phosphorylated in vivo by other kinase(s). Consequently, kinase dead LRRK2 (K1906M) maintained its ability to bind 14-3-3 proteins and has similar to the WT protein elution profile on size-exclusion chromatography considering the specific behavior of the protein around 500-800 kDa. The strong effect of C-terminal deletions and the moderate effect of G2385R on phosphorylation of LRRK2 leads us to propose that the WD40 domain and C-terminus are important in either recruiting other kinases to LRRK2 and/or influence the availability of the S910/935 to other kinases or phosphatases.
Although the three dimensional structure of LRRK2 is not currently solved, several pieces of indirect evidence suggest that deletion of the extreme C-terminus of LRRK2 leads to a change in protein conformation of LRRK2. Because deletions or point mutations in the WD40 domain of LRRK2 lead to abolished or decreased kinase activity, this implies that the overall fold of the protein is important for maintaining the kinase domain in its active state. Given that these regions are separated by several hundred amino acids in the primary sequence, it seems unreasonable to infer a very local event in the kinase domain itself, although this cannot be excluded without a structure that includes both regions. We have shown previously that the WD40 domain and C-terminus binds to the ROC domain of LRRK2 and speculate that the WD40-Cterminal region plays a role in tethering of the kinase domain, maintaining the relative orientation of the two lobes of the kinase domain within the multidomain protein [22]. Furthermore, that the G2385R variant or C-terminal deletions can overcome the hyperactivating effect of G2019S when placed on the same molecule might imply that this is likely an intra-rather than inter- molecular event, i.e. self-interaction within a monomer rather than between monomers within a dimer. Again, both of these hypotheses are speculative without detailed structures against which to test them.
What is clear is that the binding of Hsp90 is not responsible for the decrement in kinase activity of C-terminal mutations in LRRK2. Incorporating all the available data into one model, it is therefore likely that the effects of mutations in the C-terminal region that influence kinase activity are also recognized independently by chaperones as being imperfectly folded. If the C-terminus does indeed play a role in maintaining the overall multidomain fold of LRRK2, then loss of the C-terminus may lead to unfolding of the molecule, exposing new regions for Hsp90 interaction.
An outstanding question is why G2385R has a relatively strong effect in these assays. It is possible that the neutral and flexible glycine residue, which is predicted to be on the outer surface of the WD40 domain [34], also plays a role in overall folding of the molecule. Substitution of Gly2385 with a positively charged arginine might have a locally destabilizing effect. Interestingly, it has been already shown in transferrin and collagen related disorders that glycine to arginine substitutions can lead to formation and existence of several forms of proteins simultaneously [35, 36]. Whether this occurs for mutant, or wild type, LRRK2 is not clear at this time.
Several groups have proposed that selective inhibition of LRRK2 kinase activity is a promising therapeutic strategy for treatment of PD caused be single mutation in LRRK2 gene [7, 28, 37]. One surprise from our current data, and against our initial hypothesis, was that G2385R, a very well established genetic risk factor for sporadic PD, has robustly decreased kinase activity. This contrasts to the increase in kinase activity for G2019S and to the reports that kinase activity is important for neurotoxicity in model systems. The implication of this result for pathogenesis is uncertain. One possibility is that kinase activity of LRRK2 can be tolerated over only a narrow range, i.e. that both loss or gain of function variants are pathogenic, which would then imply that LRRK2 inhibitors would probably have very narrow therapeutic index. However, it is also possible that the G2385R substitution changes scaffolding properties of the protein and that this can lead to pathogenic effects. If this is true, then blocking kinase activity would not be the only way in which to interfere with LRRK2 dysfunction, raising important additional therapeutic avenues for PD [38].
In summary the present study shows that G2385R LRRK2 mutant is a partial loss of function mutation. G2385R has lower kinase activity of LRRK2 and altered binding to several currently validated protein interactions. Future studies will be important to understand how G2385R act as a risk factor for PD and whether kinase activity is important for the actions of this mutation.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging and of the National Institute of Neurological diseases and Stroke. We are very thankful for Dr. Kazutoshi Nakazawa (National Institute of Mental Health, NIH, USA) for usage of the scintillation counter, for Dr. Jean-Marc Taymans (De Katholieke Universiteit van Leuven, Belgium) for generous sharing with 3xFlag-tagged construct of LRRK2 in pCHMWS plasmid.
Abbreviations
- Cdc37
cell division cycle 37 homolog
- CPM
counts per minute
- COR
C-terminus of ROC
- DSF
differential scanning fluorimetry
- Hsp90
heat shock protein 90
- LRRK2
leucine rich repeat kinase 2
- PD
Parkinson's disease
- ROC
Ras of complex proteins
- WT
wild type
References
- 1.Halliday G, Lees A, Stern M. Milestones in Parkinson's disease--clinical and pathologic features. Mov Disord. 2011;26(6):1015–1021. doi: 10.1002/mds.23669. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 5.Haugarvoll K, Wszolek ZK. Clinical features of LRRK2 parkinsonism. Parkinsonism Relat Disord. 2009;15(Suppl 3):S205–208. doi: 10.1016/S1353-8020(09)70815-6. [DOI] [PubMed] [Google Scholar]
- 6.Daniels V, Vancraenenbroeck R, Law BM, Greggio E, Lobbestael E, Gao F, De Maeyer M, Cookson MR, Harvey K, Baekelandt V, et al. Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. J Neurochem. 2011;116(2):304–315. doi: 10.1111/j.1471-4159.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23(2):329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun. 2007;357(3):668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007;103(1):238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16(2):223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 11.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11(12):791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorgensen ND, Peng Y, Ho CC, Rideout HJ, Petrey D, Liu P, Dauer WT. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One. 2009;4(12):e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405(2):307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung HC, Chen CM, Hardy J, Singleton AB, Wu YR. A common genetic factor for Parkinson disease in ethnic Chinese population in Taiwan. BMC Neurol. 2006;6:47. doi: 10.1186/1471-2377-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funayama M, Li Y, Tomiyama H, Yoshino H, Imamichi Y, Yamamoto M, Murata M, Toda T, Mizuno Y, Hattori N. Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian population. Neuroreport. 2007;18(3):273–275. doi: 10.1097/WNR.0b013e32801254b6. [DOI] [PubMed] [Google Scholar]
- 16.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430(3):393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Dunn L, Greggio E, Krumm B, Jackson GS, Cookson MR, Lewis PA, Deng J. The R1441C mutation alters the folding properties of the ROC domain of LRRK2. Biochim Biophys Acta. 2009;1792(12):1194–1197. doi: 10.1016/j.bbadis.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger Z, Smith KA, Lavoie MJ. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49(26):5511–5523. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu QY, Li F, Wang XY, Chen ZJ. Impact of inter-subunit interactions on the dimeric arginine kinase activity and structural stability. Arch Biochem Biophys. 2011;512(1):61–68. doi: 10.1016/j.abb.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Lu S, Jiang Y, Lv J, Zou J, Wu T. Mechanism of kinase inactivation and nonbinding of FRATide to GSK3beta due to K85M mutation: molecular dynamics simulation and normal mode analysis. Biopolymers. 2011;95(10):669–681. doi: 10.1002/bip.21629. [DOI] [PubMed] [Google Scholar]
- 22.Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283(24):16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J Neurochem. 2009;111(3):703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 24.Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J. 2010;430(3):405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Wang QJ, Pan N, Lee S, Zhao Y, Chait BT, Yue Z. Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson's disease. PLoS One. 2011;6(3):e17153. doi: 10.1371/journal.pone.0017153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15(2):223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28(13):3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee JD, Patricelli MP, Nomanbhoy TK, Alessi DR, et al. Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2. Nat Chem Biol. 2011;7(4):203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52(4):587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Ding X, Goldberg MS. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One. 2009;4(6):e5949. doi: 10.1371/journal.pone.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waxman EA, Covy JP, Bukh I, Li X, Dawson TM, Giasson BI. Leucine-rich repeat kinase 2 expression leads to aggresome formation that is not associated with alpha-synuclein inclusions. J Neuropathol Exp Neurol. 2009;68(7):785–796. doi: 10.1097/NEN.0b013e3181aaf4fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, Zhang YJ, Jiang H, Ross CA, Moore DJ, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106(8):2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito G, Iwatsubo T. Re-examination of the dimerization state of leucine-rich repeat kinase 2: predominance of the monomeric form. Biochem J. 2012;441(3):987–994. doi: 10.1042/BJ20111215. [DOI] [PubMed] [Google Scholar]
- 34.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 2006;29(5):286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Mason AB, Halbrooks PJ, James NG, Byrne SL, Grady JK, Chasteen ND, Bobst CE, Kaltashov IA, Smith VC, MacGillivray RT, et al. Structural and functional consequences of the substitution of glycine 65 with arginine in the N-lobe of human transferrin. Biochemistry. 2009;48(9):1945–1953. doi: 10.1021/bi802254x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards A, Narcisi P, Lloyd J, Ferguson C, Pope FM. The substitution of glycine 661 by arginine in type III collagen produces mutant molecules with different thermal stabilities and causes Ehlers-Danlos syndrome type IV. J Med Genet. 1993;30(8):690–693. doi: 10.1136/jmg.30.8.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat Med. 2010;16(9):998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudenko IN, Chia R, Cookson MR. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson's disease? BMC Med. 2012;10:20. doi: 10.1186/1741-7015-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.