Abstract
Motile cells navigate through tissue by relying on tactile cues from gradients provided by extracellular matrix (ECM) such as ligand density or stiffness. Mesenchymal stem cells (MSCs) and fibroblasts encounter adhesive or ‘haptotactic’ gradients at the interface between healthy and fibrotic tissue as they migrate towards an injury site. Mimicking this phenomenon, we developed tunable RGD and collagen gradients in polyacrylamide hydrogels of physiologically relevant stiffness using density gradient multilayer polymerization (DGMP) to better understand how such ligand gradients regulate migratory behaviors. Independent of ligand composition and fiber deformation, haptotaxis was observed in mouse 3T3 fibroblasts. Human MSCs however, haptotaxed only when cell-substrate adhesion was indirectly reduced via addition of free soluble matrix ligand mimetic peptides. Under basal conditions, MSCs were more contractile than fibroblasts. However, the presence of soluble adhesive peptides reduced MSC-induced substrate deformations; increased contractility may contribute to limited migration, but modulating cytoskeletal assembly was ineffective at promoting MSC haptotaxis. When introduced to gradients of increased absolute ligand concentrations, 3T3s displayed increased contractility and no longer haptotaxed. These data suggest that haptotactic behaviors are limited by adhesion and that although both cell types may home to tissue to aid in repair, fibroblasts may be more responsive to ligand gradients than MSCs.
Keywords: migration, mesenchymal stem cell, fibroblast, ligand gradient, surface modification, elasticity
Introduction
Anchorage dependent cells sense and respond to properties of the extracellular matrix (ECM), e.g. stiffness1–3 and ligand density.4–6 Gradients of these cues are often found together in vivo via normal tissue variation or pathological conditions, e.g. the post-infarct myocardial scar that is stiffer than, and compositionally different from, healthy cardiac muscle.7–9 These gradients are thought to aid in directional migration through tissue; for example, mesenchymal stem cells (MSCs) whose fate can be regulated by ECM stiffness10–12 also durotax or migrate in the direction of an increasing stiffness gradient.2 Dermal fibroblast migration into cutaneous wound spaces is also well known and critical for assembly of new ECM.13, 14 However, spatial variation in tissue stiffness in vivo is often accompanied by variation in adhesive ligand density. During fibrosis, ECM accumulates and may present several ligand gradients at the interface of healthy and pathological tissue.15 Because in vivo spatial distributions of ECM proteins15 and stiffness7 have been shown to change with fibrosis and animal models have documented directed migration,13, 14 it is of interest to develop systems to investigate cell migratory behaviors in response to spatial gradients in vitro. Although we and others have developed simplified systems to probe durotaxis1–3, 16, few systems with the ability to separately modulate directed migration due to adhesive ligand gradients,17 i.e., haptotaxis, and migration due to substrate stiffness have been developed.
Adhesive ligand gradients have been shown to induce directed cell migration in human dermal fibroblasts in vitro.18 However, cell type-specific differences in the degree of haptotaxis could be simultaneously influenced by physiologically-relevant substrate stiffness, which is a major regulator of stem cell fate.10, 12 Moreover, haptotactic mechanisms remain unclear; this is in part due to the fact that systems commonly used to study haptotaxis are often limited in gradient resolution, and typically utilize substrates of non-physiologically relevant stiffness. Furthermore, haptotaxis studies often lack direct comparisons between cell and adhesive ligand types. For example, human melanoma cells undergo haptotaxis when exposed to gradients of collagen type IV, fibronectin, and laminin using Boyden chambers; these chambers however, can create only step gradients.19 Myoblast haptotaxis has been observed with more gradual nanodot adhesive gradients; however, these gradients were printed on rigid glass.20 Literature suggests that cell forces exerted on adhesive matrix proteins contribute to cellular mechanical signaling12, 21 and that in vivo gradients are gradual.7, 11 Reductionist systems employing compliant hydrogels deformable by cells, e.g. polyacrylamide22, 23 and polyethylene glycol,18 with gradual haptotactic gradients are thus needed to determine which signaling networks within a variety of cell types regulate haptotaxis. Although not yet studied in detail, mechanisms regulating haptotaxis likely involve adhesive and/or cytoskeletal proteins given their roles in other migratory control mechanisms in processes such as chemotaxis,24 and the establishment of cell polarity and shape.25, 26
Gradients of protein attachment to compliant hydrogels have been achieved via microcontact protein printing, self-assembled monolayers, and microfluidic mixing devices.18, 27, 28 Despite the use of compliant hydrogels, these methods are often difficult to scale down, tune, and require significant quantities of protein or peptide, much of which does not bind. Here, we used density gradient multilayer polymerization (DGMP), to construct compliant culture substrates with spatial gradients of composition.23, 29 DGMP, a simple method that does not require specialized equipment, ingredients, or expertise, relies on density-mediated phase separation to construct layers of distinct composition. By using this method to fabricate gradients of adhesive peptide RGD or collagen type I on a polyacrylamide (PA) hydrogel platform, we demonstrate that while fibroblasts undergo haptotaxis in response to both gradient types under basal conditions, MSC haptotaxis appears to be limited by cell-substrate adhesiveness, and is observed only when cell affinity for the substrate is reduced.
Materials and Methods
Functionalized Coverslips
To covalently attach hydrogel substrates to glass, 25 mm square glass coverslips were cleaned and oxidized by exposing both sides to UV/ozone (BioForce Nanoscience) for 3 minutes. Samples were subsequently functionalized with 20 mM 3-(trimethoxysilyl)propyl methacrylate in 10% acetic acid (AA) in ethanol for 3 minutes, washed with ethanol, and dried. All chemicals were obtained from Sigma-Aldrich and other products from ThermoFisher unless otherwise indicated.
DGMP Hydrogel Fabrication
DGMP involves layering pre-polymer hydrogel solutions into rubber molds cut to fit between two glass slides, followed by UV polymerization. 5 × 5 mm models were cut from super-soft 35A silicone rubber sheets (McMaster-Carr) were clamped between a glass slide, a functionalized glass coverslip to facilitate gel attachment, and a dimethylchlorosilane- (DCDMS-) treated glass slide to facilitate gel detachment. Pre-polymer solutions composed of acrylamide, bis-acrylamide, acrylated poly(ethylene) glycol conjugated to RGD (aPEG-RGD) or acrylic acid, 2,2′-Azobis(2-methylpropion0amidine) dihydrochloride photoinitiator (Sigma), and serial dilutions of iodixanol (Sigma), an inert density modifier, were layered into the silicone molds in order of decreasing density. Hydrogels with gradients of adhesive ligands were fabricated by allowing two 12 μL layers of pre-polymer solution of distinct composition (Fig. 1A) to diffuse and settle for 10 seconds creating a gradient at the mixing interface (Fig. 1B). Layered hydrogels were subsequently polymerized via exposure to 350 nm UV light for 4.5 minutes.
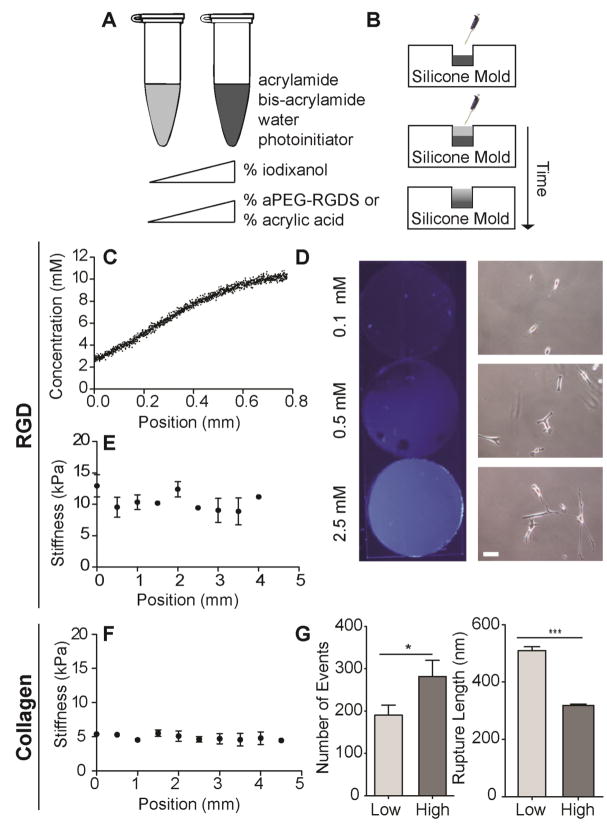
Fig. 1. Fabrication of Polyacrylamide Hydrogels with RGD and Collagen Gradients.
A. Pre-polymer solutions of acrylamide, bis-acrylamide, water, photoinitiator, acrylic acid or acrylated-PEG-RGD, are dissolved with serial dilutions of iodixanol. B. Two pre-polymer solutions are layered in order of decreasing density into a silicone rubber mold that is clamped with a methacrylated glass coverslip in between an untreated glass slide, and a DCDMS treated glass slide. After a settling time of 10 seconds, layered hydrogels are polymerized with UV light for 4.5 minutes. The iodixanol is rinsed away subsequent to polymerization. C. A hydroxycoumarin dye (excitation 350 nm) pre-conjugated to aPEG-RGD allows for gradient characterization; incorporating 0.1 and 10 mM aPEG-RGD into two layers yield a gradient of approximately 10 mM/mm. D. Images of static polyacrylamide gels with 0.1, 0.5, and 2.5 mM aPEG-RGD-dye under UV light (left); brightfield images indicate that MSC spreading correlates with RGD concentration (scale bar, 100 μm). E. Elastic modulus measured every 500 μm via AFM across the RGD gradient hydrogel substrate (n > 3; mean ± S.D.). F. Elastic modulus measured every 500 μm via AFM across the acrylic acid/collagen gradient hydrogel substrate (n > 3; mean ± S.D.). G. Antibody-protein rupture events detected via AFS (left) and corresponding measured rupture lengths (right) on low (0.005%) and high (0.1%) acrylic acid hydrogels activated with collagen I. (n > 500; mean ± S.E.M.; *p < 0.05, ***p < 0.005).
10%/0.15% acrylamide/bis-acrylamide solutions composed of 0.1 or 10 mM aPEG-RGD with 1% or 4% iodixanol respectively, were used to obtain PA-PEG-RGD hydrogels with adhesive RGDS ligand gradients of approximately 10 mM/mm that span an 800 μm region across the center of the hydrogel (Fig. 1C). A hydroxycoumarin dye (excitation 350 nm) pre-conjuated to aPEG-RGD allowed us to visualize serial dilutions of adhesive peptide; differences in cell spreading and fluorescent intensity confirm that substrate adhesiveness scales with aPEG-RGD concentration (Fig. 1D). aPEG-RGD, or acryl-PEG3400-GRGD-amide and acryl-PEG3400-[K-7-hydroxycoumarin-3-OH]GGGRGD-amide were custom ordered (21st Century Biochemicals).
To investigate the migration behavior of cells in response to solely a ligand gradient on a substrate of physiologically relevant stiffness, AFM was used to characterize the hydrogel stiffness in order to ensure that no simultaneous stiffness gradients were created as a consequence of the haptotactic gradient fabrication process (Fig. 1E). Iodixanol and aPEG-RGD did not significantly alter hydrogel stiffness at the concentrations used in this study, as stiffness was maintained at 10 kPa across the substrate.
To obtain collagen gradients, 10%/0.1% acrylamide/bis-acrylamide, acrylic acid, and serial dilutions of iodixanol were used. Solutions of 0.005% and 0.1% acrylic acid, with 1% and 4% iodixanol respectively, were used to fabricate PA-AA hydrogels with gradients of acrylic acid that span a similar 800 μm region across the hydrogel at the mixing interface. Hydrogels were then subsequently functionalized with type I collagen using EDC/NHS crosslinking to create a gradient of collagen on the substrate surface. Hydrogels were incubated in 5 mg/ml EDC (Alfa Aesar) and 9 mg/ml NHS (Alfa Aesar) dissolved in water for 10 minutes at room temperature, rinsed in water, then incubated in 100 μg/ml rat tail collagen type I (BD Biosciences) in DI water for 30 minutes.
AFM indentation indicates that hydrogel stiffness of 5 kPa remained constant across the substrate (Fig. 1F). Force spectrograms from microindentations on both low and high collagen density sides of the substrate were obtained via atomic force spectroscopy (AFS) using a probe functionalized with an anti-collagen type I antibody as described below. When the tip makes contact with the substrate, protein-antibody bonds form. As the tip is retracted from the surface, segments of collagen fibers unfold and stretch until antibody-collagen bonds rupture. These rupture events can be visualized within each spectrogram. A greater number of events (Fig. 1G, left) detected on the high collagen density side of the substrate indicate that collagen density scales with acrylic acid concentration, and shorter rupture lengths on this side of the substrate indicate a greater degree of collagen fiber-EDC/NHS tethering to the surface (Fig. 1G, right). All hydrogels were soaked in sterile PBS, stored at 4°C, and UV sterilized for 10 minutes prior to use in cell culture.
Atomic Force Microscopy
Substrate stiffness was measured via indentation using an MFP-3D-Bio (Asylum Research) atomic force microscope (AFM). Chromium/gold-coated, silicone nitride (SiN) cantilevers with pyramid tips (PNP-TR; NanoWorld) with ~30 pN/nm spring constants as determined from the MFP-3Ds built-in calibration function were used. Samples were mounted on glass slides using vacuum grease and immersed in PBS. The probe was indented into the sample with approach and retraction velocities of 2 μm/s and 20 μm/s and a force trigger of 2 nN. AFM data was analyzed using custom code in Igor Pro (Wavemetrics); the substrate spring constant, i.e., Young’s Moduli, was determined using a linearized Sneddon model.30
To detect fibrous protein on the substrate surfaces, cantilevers were functionalized with the C2456, a mouse monoclonoal anti-collagen type I antibody (Sigma) using a previously established method.31, 32 Briefly, cantilevers were cleaned with chloroform for 30 seconds and immersed in 5 M ethanolamine-HCl in dimethylsulfoxide (DMSO) at room temperature for 3 hours, resulting in the functionalization of an amine group to the surface of the probe. Tips were rinsed in PBS, incubated in 25 mM bis[sulfosuccinimidyl] suberate crosslinker (BS3; Pierce) for 30 minutes, rinsed again in PBS, then immersed in 200 μg/ml C2456 for 30 minutes to crosslink the antibody to the tip. Functionalized cantilevers were rinsed, dried, and stored at 4°C until use. Force spectrograms were taken in 10×10 arrays, with indentations spaced 10 μm apart. The functionalized tips were indented and retracted at 2 μm/s. Force curves were converted to force vs. tip Z-position and analyzed for rupture events using a previously described algorithm.33 Rupture event count along with corresponding event lengths and forces were recorded.
Cell Culture
Human MSCs (Lonza) were cultured between passages 4 to 7 in low glucose Dulbecco’s modified eagle medium (DMEM) (Gibco) supplemented with 5% fetal bovine serum (Gemini Bio-Products) and 1% antibiotic/antimycotic (Corning). NIH-3T3s were cultured in DMEM supplemented with 5% fetal bovine serum (Gemini Bio-Products), 4 mM L-glutamine, 1 mM sodium pyruvate, and 1% penicillin-streptomyocin (Corning). For migration experiments, MSCs and 3T3s were seeded on hydrogels at densities of 1,000 cells/cm2 and 5,000 cells/cm2, respectively. Cells were allowed to attach and spread for 3 hours prior to migration tracking.
Migration Studies
Migration experiments were conducted using Brightfield timelapse video microscopy over a period of 48 hours. Cells were placed in a temperature, CO2, and humidity controlled LiveCell chamber (Pathology Devices). Images were taken every 15 minutes, and cell positions were tracked in FIJI. Absolute migration velocities, velocities in the gradient direction, tactic indices, and angle histograms were calculated from raw position values using custom MATLAB scripts. To minimize error in tracking, a running average was taken of every four velocities. Tactic index, or persistence, is calculated by taking the ratio of displacement (from initial to final position) to total distance traveled. Where indicated, cells were chosen from three regions of the gradient substrates: the low adhesive ligand density region, the gradient region, and the high adhesive ligand density region.
Inhibitors and Drugs and RGD Blocking
Lysophosphatidic acid (Enzo Life Sciences #BML-LP100), bradykinin (Sigma #B3259), nocodazole (Sigma #M1404), cytochalasin D (Sigma #C2618), and Rac inhibitor (Millipore #553502) were included in culture media at concentrations of 20 μM, 10 nM, 0.5 μM, 0.5 μM, and 50 μM respectively. Drugs were stored at 100x in DMSO at −20°C until use. To reduce MSC adhesion to the hydrogel substrate surface by blocking RGD peptide-binding integrins, 0.02, 0.05, 0.1 and 0.2 mM free RGD peptide was included in the culture media throughout the duration of the timelapse.
Immunofluorescence
Cells were fixed in 3.7% formaldehyde for 15 minutes at room temperature, rinsed in PBS, and permeabilized with 1% triton-X in PBS for 15 minutes at room temperature. Cells were incubated in 1:400 rhodamine phalloidin (Life Technologies) in 2% bovine serum albumin (BSA) in PBS supplemented with MgCl2 for 30 minutes, then incubated in 1:2000 Hoescht in DI water for 10 minutes. Samples were washed with DI water and mounted with fluoromount-G (SouthernBiotech). Imaging was performed using a Nikon Eclipse TI microscope equipped with a CARV II confocal system (BD Biosciences), motorized stage and MS-2000 controller (Applied Scientific Instrumentation), and a Cool-Snap HQ camera (Photometrics) and controlled by Metamorph (Molecular Devices).
Förster Resonance Energy Transfer (FRET)
Fibronectin was isolated from human plasma using gelatin-sepharose binding and eluted with 6 M urea. Isolated fibronectin was concentrated to approximately 3 mg/ml using an Amicon Ultra Centrifugal Filter (10 kDa NMWL) (Millipore) and denatured for 15 minutes in 4 M guanidine hydrochloride (GdnHCl). Denatured fibronectin was dual-labeled with Alexa Fluor 488 (donor) and Alexa Fluor 546 (acceptor) fluorophores, as previous described.34, 35 Briefly, denatured fibronectin was incubated with a 30-fold molar excess of Alexa Fluor 546 C5 Maleimide (Life Technologies) for 2 hours to label cysteine residues within the III7 and III15 domains of fibronectin. The single-labeled fibronectin was buffer exchanged into 0.1 M sodium bicarbonate pH 8.3 and separated from unreacted acceptor fluorophores using a spin desalting column (Thermo Scientific). The single-labeled fibronectin was then incubated with a 40-fold molar excess of Alexa Fluor 488 succinimidyl ester (Life Technologies) for 1 hour to label amine residues throughout fibronectin. Unreacted donor fluorophores were removed using a spin desalting column and dual-labeled fibronectin was stored with 10% glycerol at −20°C. The average number of acceptor and donor fluorophores per fibronectin dimer was 3.6 and 8.8, respectively, and was determined using published extinction coefficients and absorbances of the dual-labeled fibronectin at 280, 498, and 556 nm.
Images of the dual-labeled fibronectin were acquired on a Zeiss LSM 780 Confocal Microscope and analyzed using a custom MATLAB script, as previously described 36. Briefly, images were averaged with a 3 × 3 pixel-sliding block and perinuclear regions of DAPI stained cells were manually selected for analysis. The FRET ratio for each pixel within a selected region was calculated by dividing the pixel intensity in the acceptor image by the corresponding pixel intensity in the donor image. FRET ratios less than 0.05 and greater than 1.0 were excluded from analysis. The mean FRET ratio within the selected regions was calculated for each cell and then averaged over all the cells in each condition.
Traction Force Microscopy
Traction force microscopy (TFM) was performed as previously described.37 Briefly, fluorescent 0.2 μm microspheres (Invitrogen) were added to the pre-polymer solution to a concentration of 1% v/v. 0.1% v/v TWEEN was added to the solution to prevent microspheres from forming aggregates. Hydrogel substrates used in TFM experiments were cast in 35 mm glass bottom dishes (MatTek) and layered with an 18 mm DCDMS-treated circular coverslip, then UV polymerized for 4.5 minutes at 350 nm as described above. The microspheres directly underneath selected live cells were imaged with a 60x confocal objective (Zeiss) using the Nikon Eclipse TI microscope. Cells were released with 2.5% trypsin and the same microspheres were imaged again. Embedded microsphere displacement from prior to subsequent to trypsin treatment was determined using a particle image velocimetry script in Matlab (MathWorks). Mean displacements were calculated for each cell, and averaged over all cells within one condition.
Statistical Analyses
All data are expressed as mean ± standard error of the mean. Student’s t-tests were used to determine statistical significance in Figures 1G, 2, 3, 5C, and Supplementary Figures 3 and 5. 1- or 2-way ANOVAs were used in Figures 3, 4, and 5B as indicated. Significant differences were considered for p < 0.05 as indicated; n.s. denotes not significant. Multiple comparisons Tukey’s post-hoc tests were performed where appropriate.
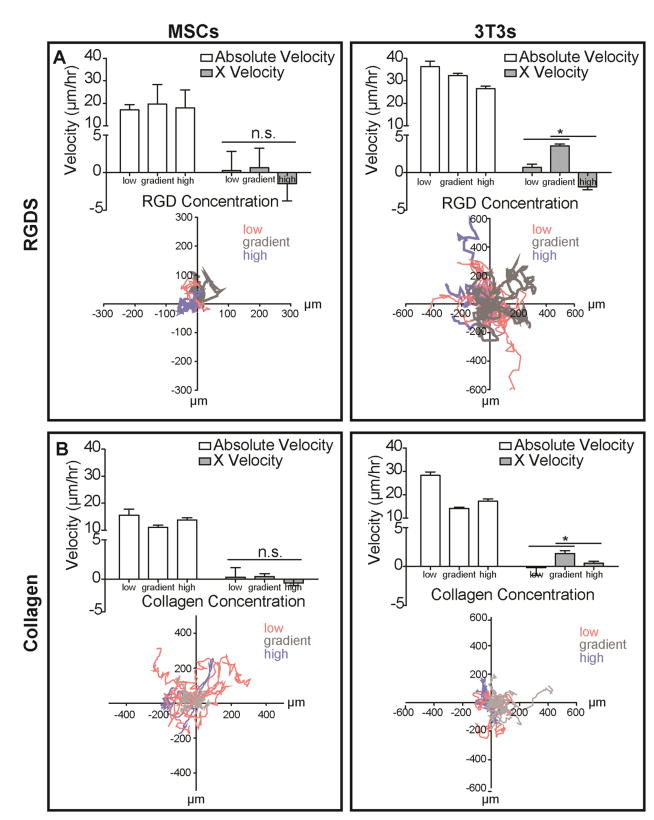
Fig. 2. Migratory Response of Stem Cells and Fibroblasts on Adhesive Ligand Gradients.
Absolute velocities and X velocities of MSCs (left) and 3T3s (right) on indicated regions of hydrogels with A. RGD concentration gradients and B. surface collagen density gradients. Low and high indicates cells found on the low and high static concentration regions of the substrate; gradient indicates cells found at the mixing interface of the substrate. Positive X velocity indicates migration towards regions of increasing adhesive ligand density. Representative rose plots show migratory paths of cells beginning at the origin. (n > 20; mean ± S.E.M.; *p < 0.05; n.s., not significant).
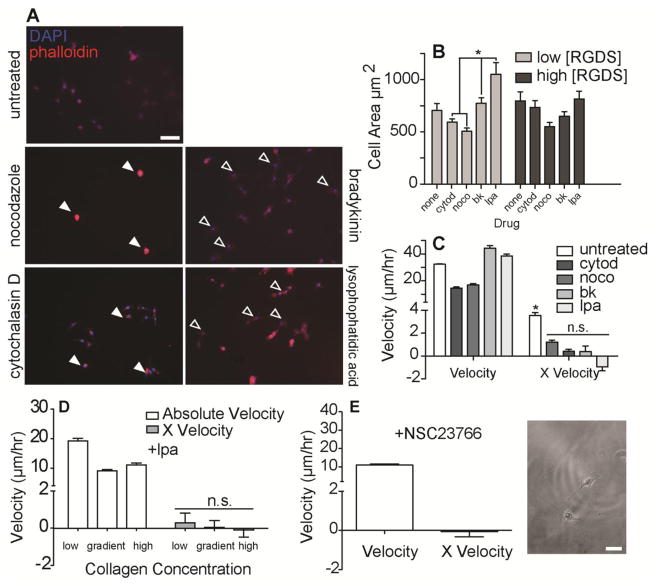
Fig. 3. Influence of Drug Treatment on Fibroblast Morphology and Migration.
A. 3T3 fibroblasts spread on PA-PEG-RGD hydrogels 24 hours after seeding and treating by the indicated drugs (actin, red; nuclei staining, blue). Filled white arrows indicate cells that are smaller and less spread than untreated control cells. Open white arrows indicate cells that are larger and more spread than untreated control cells. (scale bar, 100 μm). B. Spread area and C. absolute migratory velocities and X velocities of cells treated with the indicated drugs on low, gradient, and high regions of RGD gradient hydrogel substrates. (n > 20; mean ± S.E.M.; *p < 0.05; n.s., not significant). D. Absolute velocities and X velocities of 3T3s treated with LPA on indicated regions of acrylic acid hydrogels activated with surface gradients of collagen type I. (n > 20; mean ± S.E.M; n.s., not significant). E. Absolute velocities and X velocities of 3T3s treated with NSC23766 Rac inhibitor on gradient regions of acrylic acid hydrogels activated with surface gradients of collagen type I. Positive X velocity indicates migration towards regions of increasing adhesive ligand density. (n > 20; mean ± S.E.M.; scale bar, 100 μm).
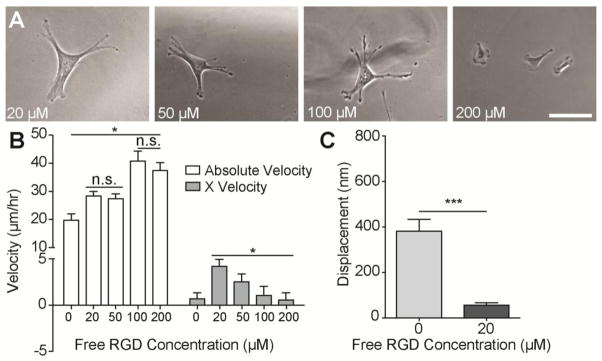
Fig. 5. Reducing MSC-Substrate Adhesion by Blocking Surface Integrins with Free Peptide.
A. MSCs spread on 10 mM PA-PEG-RGD hydrogels with indicated concentrations of free RGD peptide in the culture media. (scale bar, 100 μm). B. Absolute and X velocities of MSCs on the gradient region of RGD gradient PA hydrogels with indicated concentrations of free RGD peptide in the culture media. (n > 40; mean ± S.E.M.; *p < 0.05). C. Mean displacement of particles embedded in PA hydrogels with 10 mM aPEG-RGD resulting from tractions of MSCs in normal media, and media supplemented with 0.02 mM free RGD peptide. (n > 10; mean ± S.E.M.; ***p < 0.005).
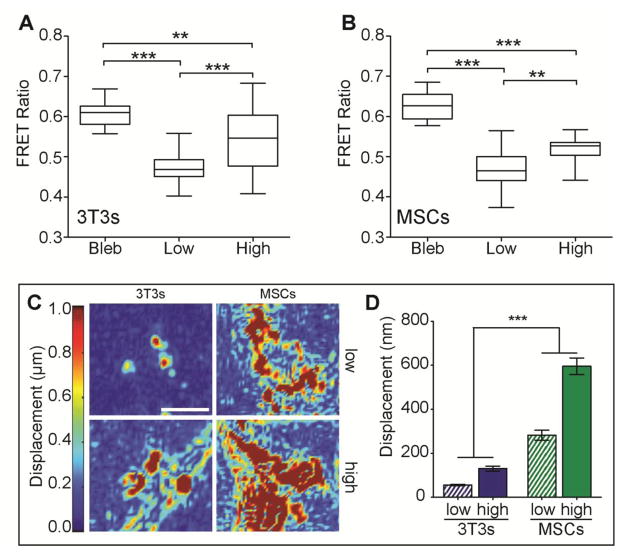
Fig. 4. Measuring Cell-Induced Deformations of the Underlying Substrate.
Mean FRET intensity ratio and ratio maps of fibronectin underneath A. spread 3T3s (left) and B. MSCs (right) on low and high acrylic acid PA hydrogels activated with EDC/NHS and coated with dual-labeled fibronectin. (n > 16; mean ± S.E.M.; **p < 0.01; ***p < 0.005). C. Displacement maps of embedded fluorescent particles (left) and D. mean particle displacement (right) resulting from 3T3 and MSC tractions exerted on low and high acrylic acid PA hydrogels activated with EDC/NHS and coated with collagen type I. (n = 25; mean ± S.E.M.; ***p < 0.005).
Results
Fibroblasts Exhibit Haptotaxis
Murine 3T3 fibroblasts and human mesenchymal stem cells (MSCs) attached to and spread on RGD and collagen ligand gradients within 4 hours of seeding. Quantifying cell density and spread area and across collagen gradient substrates (Fig. S1) 15 hours post-seeding demonstrated that density and spread area both scale with surface collagen density, and further confirmed the presence of a surface adhesive gradient. Migration velocity and haptotactic velocity, i.e. migration in the gradient direction, were measured. MSCs did not display any preferential migration within 48 hours of spreading on gradients of RGD peptide. Absolute MSC migration velocity was also insensitive to ligand density (Fig. 2A, left). However fibroblasts exhibited haptotactic migration in the direction of higher RGD concentration. Moreover, absolute fibroblast migration velocity inversely scaled with RGD concentration, i.e. cells moved faster at low RGD concentration (Fig. 2A, right). To determine whether migration was affected by the presence of a full-length, fibrous protein compared to the short adhesive RGD peptide, MSC and fibroblast migration was assessed on collagen density gradients. Consistent with RGD, MSCs on collagen substrates displayed random migration that lacked preference for either low or high ligand density (Fig. 2B, left) and was not processive (Fig. S2) as measured by a tactic ratio of displacement to total distance travelled over a period of 48 hours. Fibroblasts, on the other hand, displayed biased net migration towards higher collagen density (Fig. 2B, right) that was more processive compared to cells found on static regions of the substrate (Fig. S3). Taken together, these data indicate that fibroblasts undergo haptotaxis on both ligand gradients of RGD and collagen while MSCs do not.
Fibroblast Haptotaxis is Regulated by a Cytoskeletal-mediated Mechanism
To provide insight into how the cytoskeleton potentially regulates migration and haptotaxis, we treated fibroblasts with contractile agonists lysophosphatidic acid (LPA) and bradykinin (BK) as well as with cytoskeleton-perturbing drugs cytochalasin D (CytoD) to prevent actin polymerization and nocodazole (Noco) to prevent microtubule polymerization. Cells treated with nocodazole and cytochalasin D exhibited less-spread and rounded morphology compared to untreated cells (Fig. 3A), while cells treated with LPA and bradykinin were more spread compared to untreated cells, especially at low RGDS concentration (Fig. 3B). While LPA and bradykinin vs. nocodazole and cytochalasin D treatment increased and decreased absolute migration velocity respectively, all drug treatments inhibited haptotaxis (Fig. 3C and S4A). LPA was similarly effective at reducing haptotaxis on collagen gradients (Fig. 3D and S4B). These data suggest that while depolymerizing the cytoskeleton prohibits both spreading and migration, promoting its assembly and contraction produces overly spread, faster moving cells that lose their sensing capabilities. Rac1, a GTP-binding protein that regulates membrane ruffling in migrating cells38 and facilitates labile adhesion,39 may be involved in the sensing capabilities that cause biased migration that which LPA and bradykinin treatment reduced. While Rac inhibited fibroblasts still adhered to and spread on collagen gradient hydrogels, cells were no longer capable of haptotaxis (Fig. 3E). These data overall suggest that appropriate protrusive capabilities, i.e. membrane ruffling, followed by modest cytoskeletal assembly and contractions appear to be crucial for haptotaxis.
Biased migration resulting from cell protrusions produces substrate deformations that are small and transient2 and may be reflected in deformation of matrix proteins and/or the substrate.12, 21 Förster resonance energy transfer (FRET) and traction force microscopy (TFM) were utilized to investigate the effect of adhesive ligand density on cell-induced deformations of the ligand coating and substrate. The FRET ratio decreased underneath both stem cells and fibroblasts, suggesting that the cells exerted traction forces on the ligand coating thus deforming protein fibers, effectively decreasing the FRET ratio. Consequentially, the protein underneath blebbistatin-treated cells that lack the ability to exert forces due to perturbed myosin contractility exhibited increased FRET ratios (Fig. S5). Although fibronectin FRET ratios underneath both cell types scaled with fibronectin surface density, absolute FRET ratio values were similar between the two cell types on both low and high ligand density substrates (Fig. 4A and 4B). These observations suggest that although a greater degree of attachment effectively “ties” down fibronectin to the substrate more tightly and hinders fiber unfolding to a greater extent at higher ligand concentrations, protein tethering is perhaps unrelated to haptotaxis due to differences in migratory behaviors between MSCs and fibroblasts. Conversely, although both fibroblasts and MSCs exhibited adhesive ligand density-dependent differences in mean substrate displacements observed by TFM (Fig. 4C), MSCs displaced the substrate 5-fold more than fibroblasts, regardless of ligand density and degree of tethering (Fig. 4D). Together these data along with the observation that fibroblasts but not MSCs haptotaxed suggest that although the ability of a cell to exert traction forces may be necessary for motility and migration, MSCs tend to surpass traction or adhesiveness thresholds which results in extremely large cell-induced deformations of the substrate. This perhaps occurs to an extent where cells no longer can sense a directional cue such as increasing adhesive ligand density.
Modulating Cell-Substrate Adhesion Affects Haptotactic Behavior in Both Stem Cells and Fibroblasts
To determine whether or not exceeding a traction or adhesiveness threshold can explain the absence of haptotaxis in MSCs, we reduced MSCs tractions by adding free RGD peptide to the culture media to block RGD binding sites on integrins to effectively reduce cell-substrate adhesiveness. MSCs had slightly smaller but still spread morphology (Fig. 5A). Including RGD in the culture media increased absolute migration velocities (Fig. 5B), consistent with the observation of higher migration velocities on low vs. high ligand densities (Fig. 2). Additionally, MSCs displayed haptotaxis in an RGD dose-dependent manner, where 20 μM RGD peptide resulted in the most significant haptotactic response. The presence of free peptide also decreased mean substrate displacement compared to untreated MSCs cultured on high concentration RGD substrates (Fig. 5C). This 7-fold reduction yields deformations within range of fibroblast-induced hydrogel deformations at surface ligand densities where directed migration is observed (Fig. 2). These data also imply that while haptotactic migratory behaviors may be achieved by reducing cell-substrate binding by blocking a subset of relevant integrins, if too many integrins are blocked, cells potentially lose their ability to sufficiently sense ligand density gradients. This further suggests a biphasic phenomenon where cells lose the ability to haptotax if they are too strongly or too weakly adhered to the surface, and the existence of a mid-range of adhesive ligand density where haptotaxis is most profound.
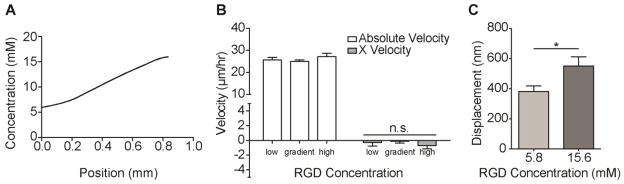
To further support this theory, 3T3 migration was assessed on RGD gradients of similar steepness, but of higher absolute peptide concentrations. Low and high RGD concentrations were increased from 0.1 and 10 mM, to 5.6 and 15.8 mM respectively (Fig. 6A). Total migration velocity and haptotactic velocity were measured. In this system, 3T3s did not display preferential migration towards either end of the gradient substrate (Fig. 6B). Furthermore, absolute fibroblast migration velocity no longer scaled with RGD concentration; cells moved at similar speeds regardless of their position relative to the gradient (Fig. 6B). Mean substrate displacements induced by fibroblasts seeded on these more-adhesive substrates increased by several-fold compared to fibroblasts that haptotaxed on gradients of lower absolute ligand concentrations; these displacements are on par with non-haptotaxing MSCs (Fig. 4D). These data provide additional evidence suggesting that haptotactic behaviors are limited by cell-substrate adhesions, and that these adhesion limits are cell-type specific.
Fig. 6. Increasing Fibroblast-Substrate Adhesion by Increasing Surface Adhesive Ligand Density.
A. A hydroxycoumarin dye (excitation 350 nm) pre-conjugated to aPEG-RGD allows for gradient characterization; incorporating 5.8 and 15.6 mM aPEG-RGD into two layers yield a gradient of approximately 10 mM/mm. B. Absolute velocities and X velocities of 3T3s on indicated regions of hydrogels with RGD concentration gradients spanning a 5.8–15.6 mM range. Low and high indicates cells found on the low and high static concentration regions of the substrate; gradient indicates cells found at the mixing interface of the substrate. Positive X velocity indicates migration towards regions of increasing adhesive ligand density. (n > 50; mean ± S.E.M.; n.s., not significant). C. Mean displacement of particles embedded in PA hydrogels with 5.8 and 15.6 mM aPEG-RGD resulting from tractions of 3T3s in normal media. (n > 10; mean ± S.E.M.; *p < 0.05).
Discussion
By directly comparing migratory behaviors of MSCs and fibroblasts in two systems using density gradient multilayer polymerization (DGMP), these data demonstrate that when introduced to an adhesive ECM ligand gradient–irrespective of whether the ligand is a short adhesive peptide or full length fibrous protein–fibroblasts underwent haptotaxis, while under the same basal culture conditions, MSCs did not. We note two observations contrasting the motility of the two cell types under the same culture conditions and seeded on the same gradient substrates: (1) fibroblasts migrate at faster absolute velocities compared to MSCs, and perhaps more importantly, (2) fibroblasts display a significant inverse relationship between surface ligand density and absolute velocity, similar to classic experiments on uniformly adhesive substrates,40 while MSCs do not. First, the ability of fibroblasts to move faster overall may suggest that they “feel” their surroundings at a faster rate; this perhaps heightens their sensitivity to small changes in adhesive ligand density in their microenvironment, allowing them to respond more quickly relative to slower moving, well-spread MSCs. Second, the inverse scaling of absolute velocities and both RGD and collagen density for fibroblasts confirms that although the exact mechanism is unknown, fibroblasts sense and respond to differences in absolute adhesive ligand density more acutely. Similar MSC motility on all regions of both RGD and collagen coated substrates suggests that MSCs may not be as sensitive as the fibroblasts to these density differences at the ligand concentrations and culture conditions used here. While haptotaxis may play a significant role in directing fibroblasts towards a site of injury, these data suggest that MSCs in vivo may be less sensitive to adhesive ligand gradients. Given that they have been reported to home to sites of injury,41 these data would imply that the MSC migratory process could be regulated by varying concentrations of free ligands in areas of active matrix remodeling, or may perhaps be instead guided by durotactic or other gradient cues.
Intact Cytoskeleton is Necessary for Migration, but Cell-induced Deformations Dictate Haptotaxis
To provide insight into intracellular mechanisms of haptotaxis, fibroblasts were treated with various cytoskeleton inhibitors and activators. Drugs that prevented polymerization of actin and the formation of stable microtubules eliminated haptotaxis and decreased overall motility, as expected. Fibroblasts remained mainly stationary on PA-PEG-RGD hydrogel substrates after treatment with nocodazole and cytochalasin D, reinforcing the well established principle that a stable and intact cytoskeleton is necessary for cell motility;25, 26, 42 similar effects have been shown with MSCs treated with the same inhibitors on durotactic gradients 2. One might expect contractile agonists such as LPA and bradykinin however, to enhance haptotactic migratory behaviors by heightening contractility and thus amplifying the sensitivity or aggregating signal from sensing mechanisms. In fact in fibrotic lungs, LPA treatment induces MSC differentiation into more contractile phenotypes such as myofibroblasts;43 LPA is also known to increase cell motility, and enhance chemotaxis 44. Although cells treated with both bradykinin and LPA in this model of fibrosis displayed increased cell motility and spread area, they no longer underwent directed migration on RGD gradient substrates despite their enhanced ability to “feel” their surroundings, supporting our observations. Lack of tactic enhancement subsequent to LPA treatment is also consistent with previous observations where LPA treatment decreased durotaxis of MSCs on stiffness gradients.2 While chemo- and haptotaxis have been reported to have different signaling mechanisms,19 they would appear to be more complex than simply tuning contractility.
One observation that perhaps relates these results to migrating MSCs is spread area. MSCs are much larger than fibroblasts, regardless of what adhesive ligand was used. Here we found that when fibroblasts are treated with LPA, cells are 1.5-fold larger in spread area, but lose their ability to undergo haptotaxis, i.e. their migratory behavior becomes more similar to MSCs subsequent to LPA treatment. These observations, perhaps reflective of decade-old observations in correlations between cell spread area and both substrate stiffness and adhesive ligand density,4, 13, 45 suggest that extracellular biophysical cues that dictate cell spread area may play a role in limiting taxis. However, closer inspection of the deformations imparted by cells was more suggestive of a mechanism. Deformations within the adhesive ligand coating and in underlying hydrogel substrate revealed that stem cells and fibroblasts could sense differences in low and high surface densities of fibronectin but absolute protein deformations were similar between cell types. These results are consistent with previous findings that although cells may potentially feel differences in fiber tethering, the degree of tethering does not seem to dictate cell behaviors such as differentiation12 and in this case, migration. However, significant differences in hydrogel deformations between MSCs and fibroblasts suggests that greater MSC tractions may impair their ability to sense differences in adhesive ligand density and thus hinder their ability to undergo preferential migration.
Optimal Cell-Substrate Affinity Maximizes Haptotaxis
By including free RGD peptide in the culture media, we were able to effectively diminish MSC-substrate adhesiveness, reduce MSC tractions thus decreasing substrate deformations, and induce haptotactic migratory behaviors in MSCs. By blocking integrin-substrate binding sites, we observed haptotactic velocities in MSCs that were on par with fibroblasts undergoing haptotaxis. However, once the free RGD peptide exceeds an optimal threshold, haptotactic migration is weakened, as MSCs are no longer sufficiently adhered and spread to the substrate. The inhibition of haptotaxis via blocking integrin receptors using free RGD peptide has previously been shown for melanoma cells on fibronectin gradients;19 our results extend these conclusions to MSCs for free RGD peptide beyond a concentration threshold to suggest that MSC haptotactic capabilities are biphasic with respect to adhesiveness.
Furthermore, by increasing the absolute values of RGD surface concentration, we were able to effectively increase fibroblast-substrate adhesiveness and increase fibroblast tractions, thus increasing substrate deformations; this consequently eliminated haptotactic behaviors in fibroblasts. These increased substrate deformations are similar to deformations induced by non-haptotaxing MSCs, suggesting that the ability of a cell to significantly deform the underlying substrate may block haptotactic migratory behaviors. Our results indicate that the ligand concentration range at which haptotaxis is observed differs by cell type, and further emphasize that haptotactic capabilities are dependent on cell-substrate affinity and adhesiveness.
Thresholds for adhesive ligand density that induce haptotaxis are likely adhesive ligand type dependent, as different ligands yield different degrees of cell-substrate affinity.46 Maximal migration of smooth muscle cells has been shown to occur on substrates coated with different densities of collagen and fibronectin that yield similar cell-substrate attachment strength.46 If cyclic RGD or full-length fibronectin bound to the substrate in place of linear RGD, maximal haptotactic migratory behaviors might be displayed at higher concentrations of free peptide or protein in the culture media, as these ligand types have been shown to better facilitate cell-substrate adhesion compared to linear RGD. However as RGD-binding integrins have not been implicated in haptotactic responses to collagen gradients,19 free RGD peptide is unlikely to affect haptotaxis on collagen gradients. Conversely, alternate integrin binding peptides, e.g., laminin-binding YIGSR, would likely not affect MSC migratory behaviors on substrates with bound RGD.
Conclusions
Using DGMP, we presented a gradient fabrication system that is tunable for adhesive ligand type, gradient range, gradient steepness, as well as substrate stiffness. We demonstrate significant differences in the ability of fibroblasts and stem cells to undergo haptotaxis. Our results indicate that haptotaxis mechanisms are dependent on cell-induced tractions, which are largely regulated by substrate adhesiveness, and are cell-type specific.
Supplementary Material
Dots indicate the distribution of cell spread areas, and grey bars indicate the number of cells found across the gradient region of a surface collagen density gradient substrate 15 hours post-seeding.
A. Rose plots of MSC migratory paths beginning at the origin on low, gradient, and high collagen density regions of PA-AA hydrogels. B. Tactic index, or the ratio of displacement to total distance traveled by cells on low, gradient, and high collagen density regions of PA-AA hydrogels. Each data point indicates one cell. (mean ± S.E.M.). C. Histograms depicting initial to final position displacement vector angles of MSCs on low, gradient, and high collagen density regions of PA-AA hydrogels. 0 angle indicates migration directly up the ligand gradient.
A. Rose plots of 3T3 migratory paths beginning at the origin on low, gradient, and high collagen density regions of PA-AA hydrogels. B. Tactic index, or the ratio of displacement to total distance traveled by cells on low, gradient, and high collagen density regions of PA-AA hydrogels. Each data point indicates one cell. (mean ± S.E.M.; ***p < 0.005). C. Histograms depicting initial to final position displacement vector angles of 3T3s on low, gradient, and high collagen density regions of PA-AA hydrogels. 0 angle indicates migration directly up the ligand gradient.
A. Rose plots of migratory paths beginning at the origin on the gradient region of PA-PEG-RGD hydrogels with RGD gradients of 3T3s treated with lysophosphatidic acid, bradykinin, nocodazole, and cytochalasin D. B. Rose plots of migratory paths beginning at the origin on the gradient region of PA-AA hydrogels with collagen gradients of 3T3s treated with lysophosphatidic acid.
FRET intensity ratio map of a blebbistatin treated cell (left) and an untreated cell (center), and measured FRET intensity ratio of undeformed protein underneath a blebbistatin treated cell and an untreated cell (right) seeded on a polyacryalmide hydrogels (right). (n = 8; ***p<0.005).
Acknowledgments
This work was supported by grants from the National Institutes of Health (DP2OD006460 to A.J.E.), National Science Foundation (1463689 to A.J.E.), and the National Science Foundation Graduate Research Fellowship Program (to J.H.W. and H.T.-W.).
Footnotes
Author Contributions
J.H.W. and A.J.E. designed experiments and conceived of the project; J.K. and A.A. provided hydrogel fabrication methods; H.T.-W. fabricated and performed experiments with the FRET probe; A.F. analyzed force spectrograms; J.H.W. and O.C. conducted all other experiments. J.H.W. and A.J.E. analyzed experimental data; J.H.W. performed statistical analyses; J.H.W., A.A., and A.J.E. wrote and edited the manuscript.
Conflict of Interest
Jessica H. Wen, Onkiu Choi, Hermes Taylor-Weiner, Alexander Fuhrmann, Jerome V. Karpaik, Adah Almutairi, and Adam J. Engler declare that they have no conflicts of interest related to this work.
Ethical Standards
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
The authors declare no commercial affiliations or conflicts of interest.
References
- 1.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent LG, Choi YS, Alonso-Latorre B, del Alamo JC, Engler AJ. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol J. 2013;8(4):472–84. doi: 10.1002/biot.201200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Photopolymerization in microfluidic gradient generators: Microscale control of substrate compliance to manipulate cell response. Advanced Materials. 2004;16(23–24):2133–2137. [Google Scholar]
- 4.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86(1 Pt 1):617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudet C, Marganski WA, Kim S, Brown CT, Gunderia V, Dembo M, et al. Influence of type I collagen surface density on fibroblast spreading, motility, and contractility. Biophys J. 2003;85(5):3329–35. doi: 10.1016/S0006-3495(03)74752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89(1):676–89. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MF, Engler AJ, Woo YJ, Pirolli TJ, Bish LT, Jayasankar V, et al. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am J Physiol Heart Circ Physiol. 2006;290(6):H2196–203. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan KE, Quinn KP, Tang KM, Georgakoudi I, Black LD., 3rd Extracellular matrix remodeling following myocardial infarction influences the therapeutic potential of mesenchymal stem cells. Stem Cell Res Ther. 2014;5(1):14. doi: 10.1186/scrt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singelyn JM, DeQuach JA, Seif-Naraghi SB, Littlefield RB, Schup-Magoffin PJ, Christman KL. Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials. 2009;30(29):5409–16. doi: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6(1):e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13(10):979–87. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauffenburger DA, Linderman JJ. Receptors: Models for Binding, Trafficking, and Signaling. New York, New York: Oxford University Press; 1993. [Google Scholar]
- 14.Li W, Fan J, Chen M, Guan S, Sawcer D, Bokoch GM, et al. Mechanism of human dermal fibroblast migration driven by type I collagen and platelet-derived growth factor-BB. Mol Biol Cell. 2004;15(1):294–309. doi: 10.1091/mbc.E03-05-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, et al. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol. 2007;293(6):G1147–54. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 16.Marklein RA, Burdick JA. Spatially controlled hydrogel mechanics to modulate stem cell interactions. Soft Matter. 2010;6(1):136–143. [Google Scholar]
- 17.Burdick JA, Khademhosseini A, Langer R. Fabrication of gradient hydrogels using a microfluidics/photopolymerization process. Langmuir. 2004;20(13):5153–6. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 18.DeLong SA, Gobin AS, West JL. Covalent immobilization of RGDS on hydrogel surfaces to direct cell alignment and migration. J Control Release. 2005;109(1–3):139–48. doi: 10.1016/j.jconrel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Aznavoorian S, Stracke ML, Krutzsch H, Schiffmann E, Liotta LA. Signal transduction for chemotaxis and haptotaxis by matrix molecules in tumor cells. J Cell Biol. 1990;110(4):1427–38. doi: 10.1083/jcb.110.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricoult SG, Pla-Roca M, Safavieh R, Lopez-Ayon GM, Grutter P, Kennedy TE, et al. Large dynamic range digital nanodot gradients of biomolecules made by low-cost nanocontact printing for cell haptotaxis. Small. 2013;9(19):3308–13. doi: 10.1002/smll.201202915. [DOI] [PubMed] [Google Scholar]
- 21.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11(7):642–9. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 22.Grevesse T, Versaevel M, Gabriele S. Preparation of hydroxy-PAAm hydrogels for decoupling the effects of mechanotransduction cues. J Vis Exp. 2014;(90) doi: 10.3791/51010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi-Barr S, Karpiak JV, Ner Y, Wen JH, Engler AJ, Almutairi A. Density gradient multilayered polymerization (DGMP): a novel technique for creating multi-compartment, customizable scaffolds for tissue engineering. J Vis Exp. 2013;(72) doi: 10.3791/50018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Affolter M, Weijer CJ. Signaling to cytoskeletal dynamics during chemotaxis. Dev Cell. 2005;9(1):19–34. doi: 10.1016/j.devcel.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–75. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475(7356):316–23. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HD, Peyton SR. Bio-inspired materials for parsing matrix physicochemical control of cell migration: a review. Integr Biol (Camb) 2012;4(1):37–52. doi: 10.1039/c1ib00069a. [DOI] [PubMed] [Google Scholar]
- 28.Pulsipher A, Yousaf MN. Surface chemistry and cell biological tools for the analysis of cell adhesion and migration. Chembiochem. 2010;11(6):745–53. 730. doi: 10.1002/cbic.200900787. [DOI] [PubMed] [Google Scholar]
- 29.Karpiak JV, Ner Y, Almutairi A. Density gradient multilayer polymerization for creating complex tissue. Adv Mater. 2012;24(11):1466–70. doi: 10.1002/adma.201103501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik G, Fuhrmann A, Cammarato A, Engler AJ. Biophys J. United States: 2011 Biophysical Society. Elsevier Inc; 2011. In situ mechanical analysis of myofibrillar perturbation and aging on soft, bilayered Drosophila myocardium; pp. 2629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonanni B, Kamruzzahan AS, Bizzarri AR, Rankl C, Gruber HJ, Hinterdorfer P, et al. Single molecule recognition between cytochrome C 551 and gold-immobilized azurin by force spectroscopy. Biophys J. 2005;89(4):2783–91. doi: 10.1529/biophysj.105.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chirasatitsin S, Engler AJ. Detecting cell-adhesive sites in extracellular matrix using force spectroscopy mapping. J Phys Condens Matter. 2010;22(19):194102. doi: 10.1088/0953-8984/22/19/194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuhrmann A, Anselmetti D, Ros R, Getfert S, Reimann P. Refined procedure of evaluating experimental single-molecule force spectroscopy data. Physical Review E. 2008;77(3):031912. doi: 10.1103/PhysRevE.77.031912. [DOI] [PubMed] [Google Scholar]
- 34.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A. 2002;99(8):5139–43. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor-Weiner H, Ravi N, Engler AJ. Traction forces via integrin signaling are necessary for definitive endoderm specification. J Cell Sci. 2015 doi: 10.1242/jcs.166157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, et al. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5(10):e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.del Alamo JC, Meili R, Alvarez-Gonzalez B, Alonso-Latorre B, Bastounis E, Firtel R, et al. Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by fourier traction force microscopy. PLoS One. 2013;8(9):e69850. doi: 10.1371/journal.pone.0069850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116(3):431–43. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich JS, Hansen MD, Nelson WJ. Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev Cell. 2002;3(2):259–70. doi: 10.1016/s1534-5807(02)00216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauffenburger DA, Lindermann JJ. Receptors: Models for binding, trafficking, and signaling. 2. London: Oxford University Press; 1996. [Google Scholar]
- 41.Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124(2):407–21. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 42.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–92. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang N, Zhao Y, Feng R, Liu Y, Wang S, Wei W, et al. Lysophosphatidic acid accelerates lung fibrosis by inducing differentiation of mesenchymal stem cells into myofibroblasts. J Cell Mol Med. 2014;18(1):156–69. doi: 10.1111/jcmm.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cencetti F, Bruno G, Blescia S, Bernacchioni C, Bruni P, Donati C. Lysophosphatidic acid stimulates cell migration of satellite cells. A role for the sphingosine kinase/sphingosine 1-phosphate axis. FEBS J. 2014;281(19):4467–78. doi: 10.1111/febs.12955. [DOI] [PubMed] [Google Scholar]
- 45.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204(1):198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 46.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122(3):729–37. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dots indicate the distribution of cell spread areas, and grey bars indicate the number of cells found across the gradient region of a surface collagen density gradient substrate 15 hours post-seeding.
A. Rose plots of MSC migratory paths beginning at the origin on low, gradient, and high collagen density regions of PA-AA hydrogels. B. Tactic index, or the ratio of displacement to total distance traveled by cells on low, gradient, and high collagen density regions of PA-AA hydrogels. Each data point indicates one cell. (mean ± S.E.M.). C. Histograms depicting initial to final position displacement vector angles of MSCs on low, gradient, and high collagen density regions of PA-AA hydrogels. 0 angle indicates migration directly up the ligand gradient.
A. Rose plots of 3T3 migratory paths beginning at the origin on low, gradient, and high collagen density regions of PA-AA hydrogels. B. Tactic index, or the ratio of displacement to total distance traveled by cells on low, gradient, and high collagen density regions of PA-AA hydrogels. Each data point indicates one cell. (mean ± S.E.M.; ***p < 0.005). C. Histograms depicting initial to final position displacement vector angles of 3T3s on low, gradient, and high collagen density regions of PA-AA hydrogels. 0 angle indicates migration directly up the ligand gradient.
A. Rose plots of migratory paths beginning at the origin on the gradient region of PA-PEG-RGD hydrogels with RGD gradients of 3T3s treated with lysophosphatidic acid, bradykinin, nocodazole, and cytochalasin D. B. Rose plots of migratory paths beginning at the origin on the gradient region of PA-AA hydrogels with collagen gradients of 3T3s treated with lysophosphatidic acid.
FRET intensity ratio map of a blebbistatin treated cell (left) and an untreated cell (center), and measured FRET intensity ratio of undeformed protein underneath a blebbistatin treated cell and an untreated cell (right) seeded on a polyacryalmide hydrogels (right). (n = 8; ***p<0.005).