Abstract
Fluorescent proteins are widely used to study molecular and cellular events, yet this traditionally relies on delivery of excitation light, which can trigger autofluorescence, photoxicity, and photobleaching, impairing their use in vivo. Accordingly, chemiluminescent light sources such as those generated by luciferases have emerged, as they do not require excitation light. However, current luciferase reporters lack the brightness needed to visualize events in deep tissues. We report the creation of chimeric eGFP-NanoLuc (GpNLuc) and LSSmOrange-NanoLuc (OgNLuc) fusion reporter proteins coined LumiFluors, which combine the benefits of eGFP or LSSmOrange fluorescent proteins with the bright, glow-type bioluminescent light generated by an enhanced small luciferase subunit (NanoLuc) of the deep sea shrimp Oplophorus gracilirostris. The intramolecular bioluminescence resonance energy transfer (BRET) that occurs between NanoLuc and the fused fluorophore generates the brightest bioluminescent signal known to date, including improved intensity, sensitivity and durable spectral properties, thereby dramatically reducing image acquisition times and permitting highly sensitive in vivo imaging. Notably, the self-illuminating and bi-functional nature of these LumiFluor reporters enables greatly improved spatio-temporal monitoring of very small numbers of tumor cells via in vivo optical imaging and also allows the isolation and analyses of single cells by flow cytometry. Thus, LumiFluor reporters are inexpensive, robust, non-invasive tools that allow for markedly improved in vivo optical imaging of tumorigenic processes.
Introduction
A number of in vivo imaging technologies, for example magnetic resonance imaging (MRI), positron emission tomography (PET), PET-MRI, PET-computed tomography (PET-CT) and ultrasound have been developed and used in the clinic (1,2). The prohibitive costs and laborious nature of MRI and PET has limited their use for pre-clinical investigations of developmental and pathological processes, and for monitoring the response of disease to therapeutic agents. To address this issue, a variety of bioluminescent imaging (BLI) and fluorescence imaging reporter systems have been developed for preclinical studies, yet these reporters lack the in vivo penetration (sensitivity) or duration and strength (intensity) of signal that are needed to provide quantitative, real-time and inexpensive in vivo imaging (3–5). For example, the routinely used ATP-dependent firefly and click beetle luciferases, as well as the ATP-independent Renilla and Gaussia luciferases, are limited by light absorption and by their reported physical instability to conditions manifest in vivo, including changes in temperature, pH and urea concentration (6). As a consequence, the utility of these luciferases reportedly benefit from imaging with long acquisition times, often in excess of 5 minutes, and use within nude (nu/nu) or shaved mice since less fur or lighter fur allows more signal to reach the detector. Collectively, these features limit their utility, particularly in more high-throughput, pre-clinical drug screening efforts (7–9).
Some multi-modal imaging reporters have been developed that permit the analysis or isolation of single cells by methods such as flow cytometry and fluorescence-activated cell sorting (FACS), respectively. However, the signal intensities of these reporters have limited sensitivity in vivo, and the cassettes encoding these reporters are large, thus restricting their application when using viral delivery methods that require space to encode transgenes or shRNAs (10–14). Accordingly, a compact multi-modal reporter having enhanced signal intensity is needed for preclinical cancer studies. To meet this need, a variety of reporters and knock-in mouse models have been developed that allow one to monitor, albeit at low resolution, the development and progression of neoplastic disease, and its response to therapeutics (2,9,15–18).
The ideal in vivo reporter should combine the benefits of high fluorescent signal intensity with the low background associated with bioluminescent molecules, which would permit single cell analysis as well as spatial and temporal monitoring in live animals. However, the utility of fluorescent molecules is hindered in vivo by the requirement for externally provided excitation light that generates auto-fluorescence and has limited penetration due to absorption by tissues. Conversely, bioluminescent enzymes are limited by wide variations in signal intensity and duration. To resolve these problems, bioluminescence resonance energy transfer-based (BRET) reporters employing direct fusion of a donor luciferase moiety and a fluorescent acceptor moiety have emerged as promising tools for monitoring complex biological processes, including tumor development and progression (19,20). The majority of BRET reporters are designed with Renilla luciferase (RLuc) and variants thereof, which serve as the donor molecule to a yellow fluorescent acceptor molecule, although firefly luciferase (FLuc) BRET fusions have also been made. While several BRET reporter fusions have been described, these reporters suffer from sub-optimal acceptor activation, due to the poor overall levels and kinetics of light production generated by most luciferases, which is in part due to auto-inactivation by enzymatic by-products (4,21–28). To overcome these challenges, we utilized the enhanced small luciferase subunit (NanoLuc) of the deep-sea shrimp Oplophorus gracilirostris, which displays extremely bright, stable, glow-type luminescent properties and physical stability, with >150-fold brighter luminescence compared to firefly and renilla luciferases and >2 hours signal half-life (6,29).
Here we report the creation and markedly improved imaging properties of novel BRET reporters we coin LumiFluors, which are fusions of enhanced Green Fluorescent Protein (eGFP, QY=0.6) or long stokes shift mOrange (LSSmOrange, QY=0.45; a red-shifted GFP variant, (30)) to NanoLuc (GpNLuc and OgNLuc, respectively). Specifically, we document that these LumiFluors are highly sensitive optical reporters for monitoring tumorigenesis, and mechanistically show that the much brighter in vivo signals of these BRET reporters is due to intramolecular energy transfer from the intense luminescent signal of NanoLuc to the fused fluorophore. This creates an optical reporter that is activated without the need for UV excitation, has little auto-fluorescence, and that can be used to FACS sort cells that stably or inducibly express these reporters. Further, the small size of LumiFluor reporter cassettes allows their incorporation into several viral vector delivery systems. Finally, assessments of the GpNLuc and OgNLuc LumiFluor reporters in both solid and soft tumor models demonstrated exquisitely sensitive monitoring of tumor development at both shallow and deep tissue levels and facile analyses of tumor cells ex vivo by flow cytometry. Thus, LumiFluor reporters are broadly applicable and highly sensitive optical reporter tools that can be used for real-time, non-invasive in vivo spatio-temporal monitoring of molecular and cellular events.
Materials and Methods
Cell lines and luciferase assays
HEK293T cells (ATCC; CRL-11268) were maintained in DMEM medium (Invitrogen) supplemented with 10% FBS (Premium Select; Atlanta Biologicals), GlutaMAX (Invitrogen), and PSG (penicillin, streptomycin, and L-glutamine, Invitrogen). Human Raji Burkitt lymphoma cells (ATCC; CCL-86) were transduced with concentrated RIEP retroviral particles (plasmid was kindly provided by C. Miething, Uniklinikum Freiburg, Freiburg in Breisgau, Germany) in the presence of Ecotropic Receptor Booster (Clontech). Cells were then selected with puromycin (1μg/ml) and maintained in RPMI 1640 media supplemented with 10% FBS and PSG. All cell lines procured from ATCC were characterized by Short Tandem Repeat (STR) profiling. NSCLC A549-pBABE and A549-LKB1 cells were previously characterized and kindly provided by Dr. Frederic J. Kaye (31) and maintained in RPMI 1640 media supplemented with 10% FBS and PSG antibiotics. Mouse lymphoma cell lines were generated by crossing the Eu-Myc transgenic mouse with the Rosa(26)rtTA transgenic mouse (JAX#006965). At the age of 8 weeks, offspring carrying Eu-Myc and Rosa(26)rtTA alleles were closely monitored for tumor development. Lymphomas were then harvested and homogenized in PBS with 10% FBS and the erythrocytes were lysed. The cells were then filtered through a 40 μm nylon filter and plated in 45% IMDM (with 25 mM HEPES) (GIBCO), 45% DMEM (high glucose, GIBCO), 10% FBS with 1% penicillin/streptomycin, 4 mM L-glutamine, 25 μM β-mercaptoethanol, 1× sodium pyruvate and 10 ng/ml mouse IL7 (R&D Systems). The cells were passaged several times to create a stably growing cell line. All cells were cultured in standard, humidified conditions (37°C, 5% CO2).
Transfection of HEK293T cells for luciferase assays was carried out using Lipofectamine 2000 (Life Technologies). Luminescence was measured 24 hr post-transfection on an Envision plate reader (Perkin Elmer) following addition of Nano-Glo Luciferase Assay Substrate (Promega).
Construction and expression of the GpNLuc and OgNLuc LumiFluor reporters
The enhanced Green Fluorescent Protein (eGFP) or long stokes shift mOrange (LSSmOrange) cDNAs lacking a stop codon were PCR amplified and cloned with 5′ BspMI and 3′ EcoRV restriction enzyme sites into the pRetroX-Tight-Puro vector (Clontech) in place of the PuromycinR cassette. Orientation of the eGFP and LSSmOrange inserts and the lack of a stop codon were confirmed by sequencing. In-frame fusion of an DISGG peptide linker and NanoLuc to eGFP or LSSmOrange was achieved by a restriction enzyme-free, two step PCR cloning protocol. Briefly, two separate sets of PCR primers were designed with overlapping regions of homology to the new pRetroX-eGFP or pRetroX-LSSmOrange vectors and NanoLuc. These were then used to amplify each respective region, and then transformed into competent E. coli. Recombination of the DISGG peptide linker-NanoLuc fragment into the pRetroX-eGFP and pRetroX-LSSmOrange vectors was confirmed by sequencing.
In vitro of characterization of recombinant luciferases
For in vitro characterization of recombinant luciferases, luminescence intensity was measured in white opaque 384-well microplates (OptiPlate-384 HS, PerkinElmer Inc.) using 2104 EnVision Multilabel Plate Reader (PerkinElmer Inc.). The assay reagent contained 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 50 μM Nano-Glo substrate (furimazine, FZ; Promega Corporation) or 100 μM coelenterazine; CLZ (Biosynth International, Inc.). Luminescence intensity was recorded 3.0 min after adding the assay reagent to the respective luciferase dilutions (5×10−2 – 1.56×10−3 μM). Emission spectral scans of all recombinant luciferases (50 nM each) were performed in white opaque 96-well microplates (OptiPlate-96, PerkinElmer Inc.) using SpectraMax M5 fluorescence microplate reader (Molecular Devices, LLC). Emission spectra were recorded from 390 nm to 600 nm using the integration time of 1000 ms with 5-nm step increments. The assay reagent for the emission spectral scans contained 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 50 μM FZ) (Promega Corporation) or 100 μM CLZ (Biosynth International, Inc.).
Subcutaneous tumor xenografts
A549-pBABE-GpNLuc and A549-LKB1-GpNLuc cells were cultured, dissociated via trypsin digestion and suspended in a 1:1 mixture of PBS and Matrigel (BD Biosciences). Cell suspensions of either A549-pBABE or A549-LKB1 (200μL) containing 5×102, 5×103, 1×105 or 5×105 cells were subcutaneously implanted as shown (Fig. 3) in 4–6 week old NOD/SCID mice. In vivo luminescence of transplanted cells was measured on days 1 and 2 post implantation and once every seven days using an in vivo imager (IVIS Spectrum; Xenogen).
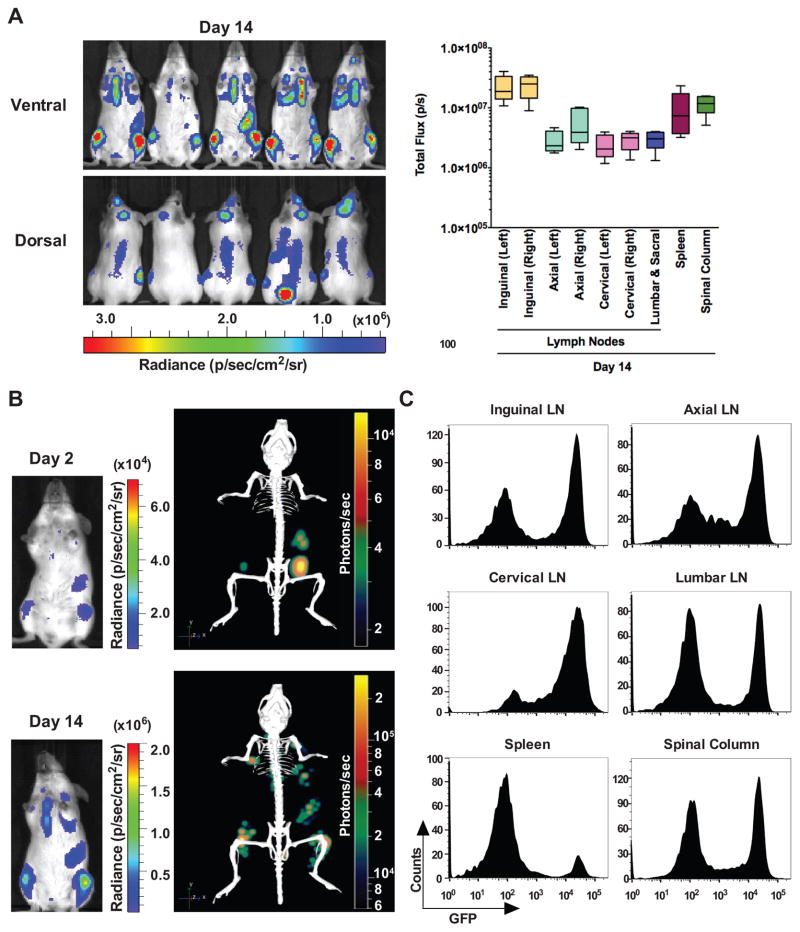
Figure 3.
Longitudinal bioluminescence imaging quantification and flow cytometry analyses of GpNLuc-expressing Eμ-Myc lymphoma transplants. A, allografts of Eμ-Myc mouse lymphoma cells stably expressing GpNLuc (1 × 106) that were injected i.v. into syngeneic Albino C57Bl/6 recipient mice (n = 10). Left, furimazine was injected intravenously (i.v.) and ventral and dorsal bioluminescence images were captured from day 1 to day 14 post-transplantation (lens aperture = f/1; image exposure time = 6 seconds; binning = 8; field of view = 22.6 cm; and emission set to open filter). Right, quantification of bioluminescent signal intensities in vivo from indicated lymph nodes and tissues colonized by B cell lymphoma. B, direct comparison of tumor burden on day 2 versus 14 post-transplantation by 2D (left) or 3D (right) bioluminescence imaging. Representative images are shown. C, ex vivo confirmation of tumor burden by flow cytometry analyses of surgically resected lymph nodes and tissues identified by BLI. Graphs are representative of mice analyzed on day 14 post-transplantation (n = 3).
Xenograft tumor volume measurements
To determine tumor volume by external caliper, the greatest longitudinal diameter (length) and the greatest transverse diameter (width) were determined. Tumor volume based on caliper measurements were calculated using the modified ellipsoidal formula, Tumor volume = 1/2(length × width2), as described (32).
Orthotopic NSCLC xenografts
A549-pBABE-GpNLuc and A549-LKB1-GpNLuc cells were dissociated via trypsin digestion, suspended in PBS and 5×105 cells were injected into the tail-vein of NOD/SCID mice. The luminescence signal from implanted tumors cells were measured once every seven days using an in vivo imager (IVIS Spectrum; Xenogen).
Orthotopic Eμ-Myc lymphoma allografts or Burkitt lymphoma xenografts
Eμ-Myc or Burkitt lymphoma cells stably expressing GpNLuc were resuspended in PBS and 1×106 sorted GFP+ cells were injected via tail vein into syngeneic Albino C57Bl/6 or NOD/SCID recipients, respectively. The luminescence signal from implanted tumors cells were measured once every seven days unless otherwise specified using an in vivo imager (IVIS Spectrum or Bruker Xtreme Optical and X-ray small animal imaging system). Tissues infiltrated with tumor cells that were identified by bioluminescence imaging (BLI) were collected for flow cytometry analysis.
Bioluminescence imaging
In vitro BLI was performed using an IVIS Spectrum one min after addition of Nano-Glo Luciferase Assay Substrate (furimazine; 2-furanylmethyl-deoxy-coelenterazine) following the manufacturer’s specifications (Promega). In vivo bioluminescent imaging was performed on isoflurane-anesthetized animals 5 min after injection of the indicated doses of furimazine either i.p. or i.v. tail-vein. Images were captured with open filter and acquisition times of 60 seconds or less at the indicated settings. Data were analyzed using Living Image software.
Results
Development and analyses of bifunctional LumiFluor BRET reporters
Brighter reporter proteins that exhibit durable signal emission are needed for spatial and temporal imaging of molecular and cellular processes in vivo. The advent of intramolecular BRET fusion reporters has made significant strides in this effort, where the ability to auto-illuminate fused fluorescent probes generates higher quantum yields than that offered by luciferase molecules alone (33). However, previous attempts at creating chimeric fluorescent-bioluminescent fusions have primarily employed the flash-type light-emitting properties of Renilla luciferase (RLuc), and variants thereof, but these fail to generate the intense, stable and durable signals required for truly sensitive in vivo imaging applications (Table 1).
Table 1.
BRET reporter fusion pairs
| Name | Fluorophore (exmax/emmax) | Peptide Linker | Molecular Modeling |
Luciferase (emmax) | Substrate | Spectral Separation |
a BRET Ratio | b BRET Effciency | Reference |
|---|---|---|---|---|---|---|---|---|---|
| LumiFluor - GpNLuc | EGFP (488/509) | 5c aa - DISGG | Yes | NanoLuc (460) | d FZ | 49 nm | 2.60 ± 0.02 | 3.02 ± 0.10 | |
| LumiFluor - OgNLuc | LSSmOrange (437/572) | 5 aa - DISGG | No | NanoLuc (460) | FZ | 112 nm | e ND | ND | |
| Nano-lantern | VenusΔC10 (515/528) | 2 aa - GT | No | Renilla_Rluc8ΔN3_S257G (480) | f CLZ-h | 48 nm | g2.26 ± 0.03 | g2.82 ± 0.00 | 22 |
| NR | EYFP (513/527) | 12 aa - SGLRSRAQALAT | No | Renilla_Rluc (480) | i CLZ | 47 nm | NR | NR | 26 |
| BAF-Y | EYFP (513/527) | 12 aa - SGLRSAAQALAT | No | Renilla_Rluc (480) | CLZ | 47 nm | NR | NR | 26 |
| eBAF-Y | EYFP (513/527) | 12 aa - SGLRSAAQALAT | No | Renilla_Rluc8 (485) | CLZ | 42 nm | NR | NR | 26 |
| BRET1 | EYFP (513/527) | 11 aa - RARDPRVPVAT | No | Renilla_Rluc (480) | CLZ | 47 nm | NR | NR | 27 |
| BRET2 | GFP2 (400/511) | NR | No | Renilla_Rluc (400) | j CLZ-400a | 111 nm | NR | NR | 28 |
| NR | GFP2 (400/511) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc-C (480 or 400) | CLZ or CLZ-400a | 31 or 111 nm | NR | NR | 25 |
| NR | GFP2 (400/511) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc-M (485 or 400) | CLZ or CLZ-400a | 26 or 111 nm | NR | NR | 25 |
| NR | GFP2 (400/511) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8 (485 or 400) | CLZ or CLZ-400a | 26 or 111 nm | NR | NR | 25 |
| BRET3 | mOrange (548/564) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8 (480) | CLZ | 84 nm | 0.79 ± 0.01 | NR | 24 |
| BRET3.1 | mOrange (548/564) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8 (515) | k CLZ-v | 49 nm | 0.74 ± 0.02 | NR | 23, 24 |
| BRET4.1 | TagRFP (555/584) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8 (515) | CLZ-v | 69 nm | NR | NR | 24 |
| BRET5 | TagRFP (555/584) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8.6 (535) | CLZ | 49 nmn | NR | NR | 24 |
| BRET6 | TurboFP (588/635) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8.6 (535) | CLZ | 100 nm | 0.58 ± 0.02 | NR | 24 |
| BRET6.1 | TurboFP (588/635) | 18 aa - SGSSLTGTRSDIGPSRAT | No | Renilla_Rluc8.6 (570) | CLZ-v | 65 nm | 0.78 ± 0.04 | NR | 24 |
| TurboLuc | TurboFP635 | 14 aa - QSTVPRARDPPVAT | No | Firefly_Fluc2 (560) | d-Luciferin | 75 nm | NR | NR | 21 |
BRET Ratio = [Fusion Reporter Bioluminescent emission (long wavelength)/Bioluminescent emission (short wavelength)] - [Donor Only Bioluminescent emission (long wavelength)/Donor Only Bioluminescent emission (short wavelength)]ref 23
BRET Efficiency = (Fusion Reporter Acceptor Peak emission/Fusion Reporter Donor Peak emission)
aa = Amino Acid
FZ = Furimizine; 2- furanylmethyl-deoxy-coelenterazine
ND = Not Determined
CLZ-h = Coelenterazine-h; 2-Deoxycoelenterazine
Calculated in this report
NR = Not Reported
CLZ = Coelenterazine
CLZ-400a = Coelenterazine-400a; 1-bisdeoxycoelenterazine (DeepBlueCTM)
CLZ-v = Coelenterazine-v
Brightness of luciferase proteins is a function of quantum yield, catalytic rate and sensitivity to product inhibition (33). Thus, we reasoned that one could engineer a more robust, auto-regulatory BRET fusion reporter using the enhanced luciferase variant of Oluc-19 (NanoLuc; NLuc), previously generated by directed evolution. Notably, NanoLuc produces three orders of magnitude more luminescence than RLuc when provided with an optimized substrate (furimazine), due to combined improvements in all three parameters governing brightness (6). We predicted that the intense and stable glow-type light emitted by NanoLuc, which ranges from 440–480 nm, could be successfully employed for BRET to excite fluorescent proteins having a high quantum yield such eGFP in cis, by substrate-dependent chemical energy transfer (Fig. 1A). The crystal structure of eGFP (34) and molecular modeling of the structure of NanoLuc were used to guide the design of the chimeric reporter (Fig. 1B). Using this model, a short flexible (5-residue) amino acid linker between the N-terminal eGFP and C-terminal NanoLuc (GpNLuc LumiFluor) moieties was optimized to allow independent folding of the two proteins and to maintain the close physical proximity (range of 30 – 70 Å) required for efficient intramolecular energy transfer (Fig. 1B and Table 1).
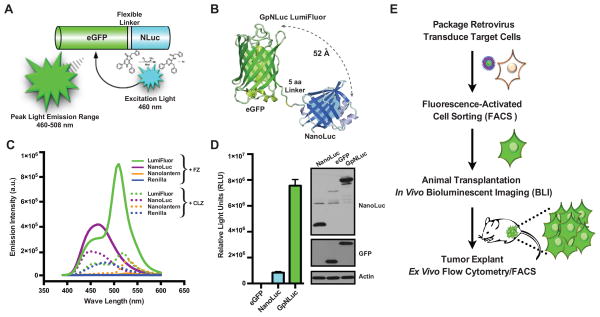
Figure 1.
Development and validation of an eGFP-NanoLuc (GpNLuc) bifunctional LumiFluor reporter. A, schematic of the GpNLuc reporter. The N-terminus of GpNLuc is derived from eGFP, which is followed by a flexible 5-residue linker (DISGG), and the C-terminus is derived from NanoLuc. Following hydrolysis of its substrate furimazine, the light emitted by the NanoLuc moiety activates the eGFP moiety via bioluminescence resonance energy transfer (BRET) in cis. B, structural model and functional evaluation of the GpNLuc LumiFluor. A model of GpNLuc was generated by combining the structure of eGFP (pdb4EUL) with a model of NanoLuc based on sequence homology to fatty acid binding protein (pdb1B56). The in-frame 5-residue DISGG linker was added between the C-terminus of eGFP and the N-terminus of NanoLuc. The distance between the NanoLuc active site and the eGFP fluorophore ranges between 30 to 70 Å based on this model, with a mean of 52 Å. C, normalized spectral emission scans of native proteins. Equimolar amounts of expressed and purified recombinant NanoLuc, Renilla, GpNLuc, and Nano-lantern BRET fusion proteins were aliquoted and emission intensities measured in triplicate in the presence of either furimazine (FZ; 50 μM) or coelenterazine (CLZ, 100 μM). D, expression and functional comparison of the GpNLuc fusion reporter to eGFP and NanoLuc alone. Left, HEK293T cells were transfected with equal concentrations of each respective retroviral construct and luciferase assays were performed 24 hr post-transfection (n = 4; mean ± s.e.m.). Right, western blot analyses of whole cell lysates from HEK293T cells transfected with NanoLuc (lane 1), eGFP (lane 2), or GpNLuc (lane 3). E, approach used to validate the functional utility of the bifunctional GpNLuc reporter for in vivo bioluminescent imaging and ex vivo flow cytometry analyses.
To rigorously evaluate the optical properties of the optimized GpNLuc BRET reporter fusion, we compared GpNLuc directly to another recently described BRET fusion reporter based on enhanced RLuc (RLuc8.6) and a YFP variant (Venus), Nano-lantern-YNL, whose spatial arrangement was optimized by circular permutations of Venus rather than molecular modeling (Table 1) (22). As expected given their optimized donor/acceptor configurations, analysis of purified GpNLuc compared to Nano-lantern protein revealed similar BRET ratio and efficiency profiles. However, the GpNLuc BRET ratio was 3–4.5-fold greater compared to that reported for BRET3 and BRET6 fusion reporters (Fig. 1C and Table 1). Further, while Nano-lantern exhibits improved brightness over other reported BRET fusions (22), the GpNLuc LumiFluor displays 70-fold higher peak emission intensity, with 85-fold more luminescence than Nano-lantern when provided furimazine, or 8-fold higher peak emission intensity, with 45-fold more luminescence than Nano-lantern when provided with each luciferases preferred substrate, despite similar energy transfer characteristics (Fig. 1C and Supplementary Fig. S1A).
Given its compact size, the GpNLuc fusion was cloned into a retrovirus (pRetroX-Tight-MCS-GpNLuc) and used to generate cell lines stably expressing the reporter (Supplementary Fig. S1B). To verify the spectral characteristics and proper expression of the GpNLuc fusion protein (46-kDa), HEK293T cells were transiently transfected with increasing concentrations of this retroviral construct constitutively expressing GpNLuc or with equivalent concentrations of eGFP alone or NanoLuc alone (Fig. 1D and Supplementary Fig. S1C). GpNLuc has a 10-fold increase in total light output over NanoLuc alone, which was slightly higher but similar to that observed by analysis of purified protein, and, not surprisingly, several orders of magnitude more intense than eGFP in the absence of excitation light. Importantly, fluorescence microscopy confirmed functional eGFP activity of GpNLuc, and luciferase assays confirmed concentration-dependent luciferase activity of this chimeric reporter (Supplementary Fig. S1C). Finally, western blot analysis confirmed the presence of the predicted 46-kDa GpNLuc fusion protein (Fig. 1D). Thus, both the eGFP and NanoLuc moieties of the GpNLuc chimera are functional and their fusion creates a markedly improved BRET reporter that can be used to transduce, image and FACS sort target cells to allow, for example, the evaluation of tumor cell growth in vitro and in vivo (Fig. 1E).
Fluorescent-bioluminescent properties of GpNLuc in reporter tumor cells ex vivo
To characterize the in vitro properties of GpNLuc reporter, mouse Eμ-Myc lymphoma and human A549 non-small cell lung cancer (NSCLC) tumor cell lines were transduced with retroviruses expressing GpNLuc and stable clones selected by FACS sorting GFP+ cells. These GpNLuc-expressing tumor cells were then serially diluted and the total light emitted was imaged using a cooled CCD camera following treatment with furimazine (Supplementary Fig. 2A, B, top). Quantifying bioluminescence as a function of cell number revealed that the minimum number of detectable cells was about 8–16 cells/well for both cell lines, which is 40–60 fold better than the numbers of NSCLC cells that are required for detection using conventional firefly luciferase (35).
The imaging data were also evaluated using a photo multiplier tube-based (PMT) plate reader equipped with an enhanced luminometer capable of ultra-sensitive luminescence measures less than 5 amol/well. The minimum number of detectable cells by fluorescence was approximately 31,000 lymphoma and 125,000 NSCLC cells/well. Strikingly, however, this number was as low as 4–8 cells/well for both cell lines when assessing luminescence intensity (Supplementary Fig. 2A, B, bottom). The highly sensitive detection of GpNLuc versus eGFP fluorescence alone corresponds to >3 orders of magnitude more light signal and to detecting 8,000–30,000 fold fewer cells, respectively. Finally, stable GpNLuc expression and single cell analysis of serially passaged cells showed that GpNLuc-expressing lymphoma and NSCLC cell lines were ≥95% GFP positive after two weeks of culture, validating their use for non-invasive in vivo imaging (Supplementary Fig. 2C, D).
GpNLuc signal intensity and sensitivity in vivo
Blue-shifted light emissions are scattered by tissues and are absorbed by hemoglobin in vivo. Thus, the narrow, blue-shifted emission range of NanoLuc is not optimal for penetrating mammalian tissues and sensitive in vivo optical imaging (36). To initially assess the utility of the enhanced spectral profile and intense signal properties of the GpNLuc LumiFluor in vivo, subcutaneous xenografts were performed with varying numbers of (LKB1-null) A549-GpNLuc NSCLC cells (A549-GpNLuc), and these were compared to A549-GpNLuc cells that were also engineered to express the tumor suppressor LKB1 (A549-LKB1-GpNLuc) (Fig. 2A). Previous reports have claimed the ability to detect fewer than 10 cells in vivo using conventional luciferases (7,8). However, in these studies images were captured with an open filter and acquisition times of 5 minutes or more, and in some cases several days post-transplant. To test the in vivo sensitivity of the GpNLuc LumiFluor reporter, images were captured as indicated with an open filter and acquisition times of 60 seconds or less. Longitudinal monitoring with bioluminescent imaging (BLI) revealed that ≤500 GpNLuc-expressing cells are easily detected using brief image acquisition times on the first day post transplant and that GpNLuc effectively tracks the inhibitory affects of LKB1 on NSCLC tumor growth (Fig. 2A and Supplementary Fig. S3A–C). Quantitation of signal-to-noise ratios for 500 cells revealed that the GpNLuc signal is 2–3 orders of magnitude above background signal generated in control mice similarly injected with substrate; thus, far fewer cells can be successfully imaged. To define the optimal dose of furimazine substrate and the stability of the resulting GpNLuc signal, subcutaneous xenografts were established with 500,000 GpNLuc-expressing tumor cells, and tumors were allowed to develop to 1500-mm3. Recipient mice were then administered with specific doses of furimazine and followed by periodic BLI (Fig. 2B). Analysis of signal intensity and stability revealed that 250 μg/kg and 500 μg/kg furimazine produced 12–16 fold higher signal than a 50 μg/kg dose, although all three doses displayed a remarkably stable signal output, with a t½ of 40 minutes (Fig. 2B). These remarkable in vivo properties for the GpNLuc LumiFluor are in stark contrast with the apparent rapid in vivo signal decay rate of secreted NanoLuc, which has a t½ of 5–10 minutes (36).
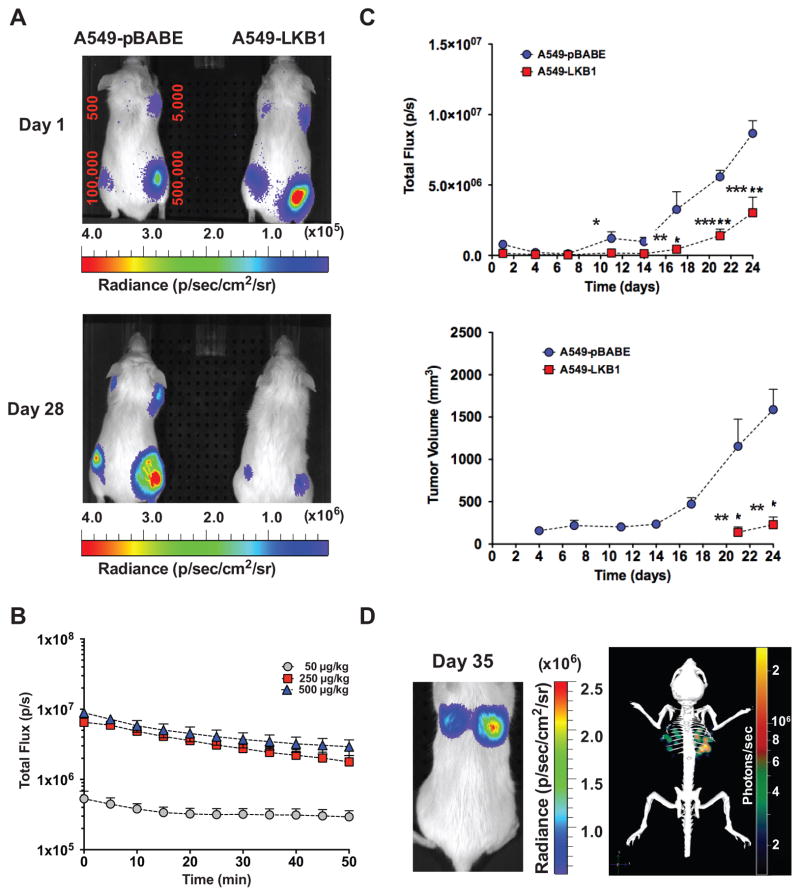
Figure 2.
GpNLuc signal strength and stability in A549 NSCLC xenograft and orthotopic transplants. A, A549-GpNLuc cells or A549 cells engineered to also express the LKB1 tumor suppressor (A549-LKB1-GpNLuc) were injected subcutaneously into the front and rear flanks of recipient NOD/SCID mice with the indicated numbers of tumor cells to gauge limits of signal detection (minimum number of detectable cells) 1 day following injection. Furimazine was injected intraperitoneally (i.p.) and bioluminescence images were captured for the two cohorts, which were monitored longitudinally from day 1 to day 28 post-transplantation (lens aperture = f/1; image exposure time = 60 seconds on Day 1 or 7 seconds on Day 28; binning = 8; field of view = 13.3 cm; and emission set to open filter). B, in vivo dose-response kinetics of GpNLuc signal strength. Mouse subcutaneous xenografts were established with A549-GpNLuc cells (5 × 105) and signal strength was monitored temporally in response to i.p. furimazine administration at the indicated doses when tumor volume reached 1500 mm3 (n = 3). C, direct comparison of subcutaneous tumor growth monitored temporally by bioluminescent imaging (BLI; top) and caliper measurements (bottom) for mouse xenografts (5 × 105 cells) from A549-GpNLuc or A549-LKB1-GpNLuc cohorts (n = 3). A significant difference was detectable between the A549-GpNLuc and A549-LKB1-GpNLuc cohorts on day 11 by BLI but not until day 21 by caliper measurements (*P < 0.05; **P < 0.01; ***P < 0.001). D, tissue penetrating ability of GpNLuc signal was evaluated by orthotopic transplantation of A549-GpNLuc cells (1 × 106) injected intravenously (via tail vein) into NOD/SCID mice. Furimazine was injected intravenously (i.v.) and 2D (left) and 3D (right) bioluminescence images were captured. Images are representative of mice monitored longitudinally from day 1 to day 49 post-transplantation (lens aperture = f/1; image exposure time = 60 seconds; binning = 8; field of view = 6.6 cm; and emission set to open filter).
The sensitivity of the GpNLuc reporter was also assessed by a direct comparison of BLI and external caliper measurements in subcutaneous xenografts (Fig. 2C). Temporal analysis identified a significant difference between the A549-GpNLuc and A549-LKB1-GpNLuc NSCLC cohorts as early as day 11 post-transplant using BLI, which was not evident until day 21 using caliper measurements. We next tested the ability of the GpNLuc signal to penetrate through deep tissue using an orthotopic lung tumor model. As few as 500,000 GpNLuc-expressing A549 NSCLC cells were injected via tail-vein into recipient mice, allowed to colonize the lungs, and recipients were followed by longitudinal BLI monitoring (Fig. 2D). Three-dimensional (3D) reconstruction performed using the Living Image DLIT algorithm confirmed that GpNLuc signal was easily detected from deep within the lungs and ex vivo BLI of these surgically resected lungs validated the DLIT reconstruction (Fig. 2D, Supplementary Fig. S4A–C and Supplementary Video S1). Notably, the intensity of the GpNLuc signal allowed the detection of micro-metastases at regional lymph nodes (Supplementary Fig. S4D).
GpNLuc monitoring of soft tumors by BLI and flow cytometry
In vivo monitoring of models of hematological malignancies is challenging, as experimental parameters often rely on end-point analysis such as overall survival, or periodic blood sampling, WBC cell counts and flow cytometry analyses. To test the utility of GpNLuc LumiFluor reporter in detecting such malignancies, we established orthotopic allografts following i.v. (tail vein) transplantation of two independently derived Eμ-Myc B cell lymphomas expressing the GpNLuc reporter (Fig. 3 and Supplementary Fig. S5). Longitudinal monitoring revealed a progressive increase in signal intensity, which increased by more than two orders of magnitude on day 14 post-transplant versus that manifest on days 1 and 2 (Fig. 3A and Supplementary Fig. S6, S8A). Temporal analysis confirmed that monitoring tumor development in deep tissues with this LumiFluor reporter is technically feasible given the detection of lymphoma cells in the lungs of recipient mice (Supplementary Fig. S5C, D). Homing and colonization of lymphoma cells into the spleen, inguinal lymph nodes and spinal bone marrow was easily and strongly detected as early as 2 days post-transplant, followed shortly thereafter by detection within the axial, cervical and lumbar/sacral lymph nodes (Fig. 3B and Supplementary Fig. S5E, S6). This represents a significantly reduced time frame for detection compared to the two weeks required for most leukemia models using conventional luciferase reporters having inferior light emission (15). Differences in observed signal intensity and half-life could be attributed to the route of substrate administration. While an i.p. administration route was used for monitoring the NSCLC tumor models (Fig. 2), substrate was administered via an i.v. route for the Eμ-Myc B cell lymphoma orthotopic allografts or human Burkitt lymphoma xenografts, and analysis of signal intensity and stability revealed that a 250 μg/kg dose administered i.v. also displays stable signal output (Supplementary Fig. 7A). Both DLIT 3D-reconstruction with the IVIS Spectrum and optical imaging coupled with X-ray performed with a Bruker In Vivo Xtreme optical/X-ray imager confirmed the anatomic origins of observed signals (Fig. 3B, Supplementary Fig. S6B, S7B and Supplementary Video S2). Finally, ex vivo BLI of surgically resected tissues or flow cytometry analyses of surgically resected tissues on day 14 following lymphoma transplant validated these findings (Fig. 3C and Supplementary Fig. S5D, E, and Fig. S8).
Enhanced output of LumiFluor reporters requires intramolecular energy transfer
To test if increased light output and broader optical profile generated via the intramolecular BRET within GpNLuc truly enabled more sensitive in vivo imaging, Y67A and Y67C GpNLuc substitution mutants within the chromophore of the eGFP (37) moiety were generated (Fig. 4A). As a control, mutation of the adjacent threonine residue not predicted to disrupt the eGFP chromophore (T66G) was also generated in GpNLuc. Finally, a second LumiFluor reporter was generated that has an even broader optical profile, by fusing long stokes shift mOrange (LSSmOrange, a red-shifted GFP variant) to NanoLuc (OgNLuc). HEK293T cells were transfected with equal concentrations of retroviral constructs constitutively expressing NanoLuc, the GpNLuc or OgNLuc LumiFluors, or the GpNLuc point mutants (Fig. 4A). As predicted, like GpNLuc, there were 10-fold increases in total light output of OgNLuc or GpNLuc-T66G over that of NanoLuc alone, and there was a marked attenuation in light output by the GpNLuc-Y67A and GpNLuc-Y67C mutants. Further, FACS analyses of retrovirus transduced Eμ-Myc B cell lymphomas confirmed that OgNLuc displays a red-shifted fluorescent signal and that the GpNLuc-Y67C mutant cannot generate a GFP signal (Fig. 4B and Supplementary Fig. S9).
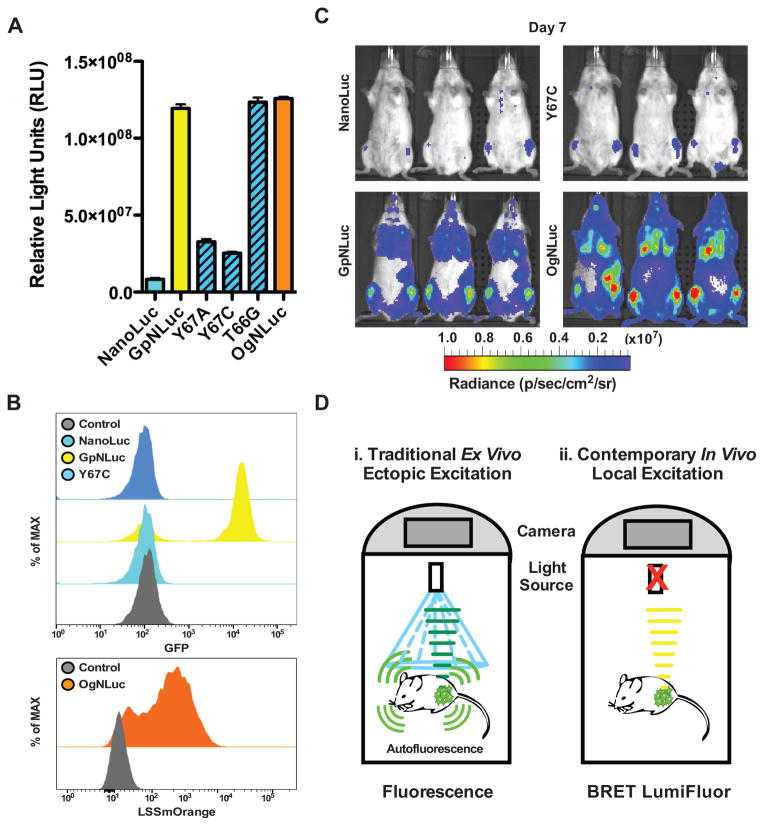
Figure 4.
Intramolecular BRET in LumiFluors drives fluorophore excitation/emission and is essential for sensitive in vivo imaging. A, comparison of catalytically inactive GpNLuc-Y67A and GpNLuc-Y67C mutants, as well as the red-shifted LSSmOrange-NLuc (OgNLuc) fusion, to either GpNLuc or NanoLuc alone. HEK293T cells were transfected with equal concentrations of each respective retroviral construct and luciferase assays were performed 24 hr post-transfection (n = 3; mean ± s.e.m.). B, flow cytometric analysis of Eμ-Myc mouse lymphoma cells, along with serially passaged Eμ-Myc lymphoma cells engineered to express NanoLuc, GpNLuc, GpNLuc-Y67A, GpNLuc-Y67C, GpNLuc-T66G or OgNLuc, confirmed effects of mutagenesis on the fluorescence excitation capacity of NanoLuc on eGFP and on LSSmOrange. C, allografts of Eμ-Myc mouse lymphoma cells expressing either NanoLuc, GpNLuc, GpNLuc-Y67C, or OgNLuc (1 × 106) were injected i.v. into syngeneic Albino C57Bl/6 recipient mice (n = 3). Furimazine was injected i.v. and ventral bioluminescence images (BLI) were captured on day 7 post-transplantation (lens aperture = f/1; image exposure time: NanoLuc = 25 seconds, GpNLuc = 6 seconds, GpNLucY67C = 40 seconds, or OgNLuc = 3 seconds; binning = 8; field of view = 22.6 cm; and emission set to open filter). D, model comparing and contrasting conventional in vivo fluorescent imaging to new methods offered by GpNLuc and OgNLuc LumiFluor reporters. Ectopic excitation of fluorescent reporters in vivo results in significant autofluorescence whereas local excitation of fluorophores by intramolecular energy transfer from a fused NanoLuc partner prevents global autofluorescence and augments overall signal output and detection.
To compare their in vivo activity, BLI analyses of orthotopic Eμ-Myc lymphomas allografts expressing these reporters were performed. These analyses confirmed that the BLI potential of the GpNLuc-Y67C mutant was comparable to NanoLuc alone, with much inferior in vivo optical properties requiring 4–6 fold longer exposure times (25 and 40 seconds, respectively) versus the GpNLuc LumiFluor (6 seconds) (Fig. 4C). Notably, despite having an apparent equivalent total light output in vitro and half the exposure time in vivo (3 seconds), OgNLuc displayed an even greater (2–4 fold) increase for in vivo signal output relative to GpNLuc, likely owing to its red-shifted emission that is capable of enhanced tissue penetration. Based on these image acquisitions, signal-to-noise ratios were quantified and revealed that GpNLuc and OgNLuc have a signal working range between 3–4 orders of magnitude above background signal generated in control mice similarly injected with substrate. Thus, compared to conventional in vivo fluorescent or bioluminescent imaging, the GpNLuc and OgNLuc LumiFluor BRET reporters generate a robust, high-intensity signal that is perfectly suited for sensitive, non-invasive in vivo optical imaging (Fig. 4D).
Discussion
The development of luciferase molecules having enhanced light-emitting properties such as NanoLuc is as active arena of study (4,5,38–41). However, due to absorption and scattering of the blue-shifted, short wavelength light emitted by NanoLuc, this reporter cannot penetrate tissues and the signal generated by NanoLuc has a short half-life in vivo (6,36). Here we describe the generation and characterization of a novel class of in vivo BRET imaging reporters coined LumiFluors that overcome these deficiencies. Specifically, LumiFluors have the desired fluorescent-bioluminescent spectral and optical properties that allow sensitive imaging both ex vivo and in vivo, and they also allow one to isolate and fully characterize target cells using flow cytometry. Indeed, the enhanced strength, stability and duration of signal, and the deep tissue penetration capabilities of the GpNLuc and OgNLuc LumiFluors dramatically reduce image acquisition times making them more desirable than conventional reporters for in vivo imaging. The increased sensitivity and imaging speed offered by LumiFluors provides improved monitoring of tumor development and the detection of small metastatic lesions (42).
The development and characterization of the GpNLuc and OgNLuc BRET reporters was achieved by a molecular modeling-guided approach that resulted in a short, flexible peptide linker that enables highly efficient donor energy transfer to the paired acceptor to generate intense bioluminescent signals. Oplophorus gracilirostris luciferase (OLuc) naturally has high quantum yields (QY) (29), but even brighter signals were achieved by pairing the enhanced version of OLuc (NanoLuc) to high QY fluorophores (eGFP QY=0.6 and LSSmOrange QY=0.45). Compared to NanoLuc alone, LumiFluor BRET reporters are more than 10-fold brighter and thus display increased tissue penetration thereby overcoming current limitations associated with reporters used for in vivo imaging. A key advantage of LumiFluor reporters is that they provide the user with the ability to non-invasively monitor specific cell populations in vivo and to then isolate these cells by FACS. This is, for example, particularly useful for characterization of sub-populations and heterogeneity in primary and metastatic tumors, and in circulating tumor cells, in orthotopic or even genetically engineered mouse models (GEMM). Moreover, their strength of signal suggests that LumiFluors may allow one to locate, isolate and characterize rare cancer stem/initiating cells and dormant/resistant tumor cells.
A potential limitation to the use of LumiFluor reporters is their dependence on the substrate furimazine. While furimazine was previously shown to be stable in media in the presence of serum (6), it is a coelenterazine analog and coelenterazine is known to be a substrate for multidrug resistance (MDR1) P-glycoprotein (PGP), which can lead to its rapid export from cells that express MDR1, thereby impacting signal intensities (43). Future experiments will evaluate the transport properties of furmazine by MDR1 as well as its ability to cross the blood-brain barrier.
Many uses of LumiFluors are feasible, for example as reporters in GEMM, as fusions with proteins to monitor real-time biological processes in cells, and as biosensor tags for antibodies or small molecule probes that home to select target cells, which could be used to visualize responses in vivo and to determine the margins of select tissues and/or tumors, to aid in surgical procedures (17,44,45). Finally, LumiFluors can also be used in traditional BRET assays to study protein:protein or ligand:protein interactions by developing a split LumiFluor reporter for complementation assays (46,47).
The fluorescent component of LumiFluor reporters also permits multiplexing. For example, the distinct spectral characteristics of GpNLuc and OgNLuc allow one to simultaneously monitor signals coupled to multiple molecular and cellular events in either in vitro or in vivo formats. Importantly, the sustained intense signals produced by these two LumiFluors also increases the confidence for monitoring rare coincident events in preclinical models, for example the interplay of immune cells with tumors, tumor-stromal interactions and how these, and the fate of primary tumors and micrometastases, are affected by treatment with therapeutics. Finally, engineering the NanoLuc moiety of LumiFluors, so that it emits light at different wavelengths, should allow for the intramolecular activation of a broad spectrum of fluorescent proteins that will expand the repertoire and imaging capabilities of these novel reporters.
Supplementary Material
Acknowledgments
The authors thank Promega Corporation for their generous gifts of plasmids for NanoLuc, the NanoLuc substrate furimazine and the polyclonal antibody to NanoLuc and Dr. Frederic J. Kaye for the NSCLC A549-pBABE and A549-LKB1 cell lines. We also thank Sollepura D. Yogesha, Krishna Chinthalapudi, Rangarajan Erumbi and Pengfei Fang for helpful technical assistance and Drs. Shawn Hingten, Chad Pecot, and Klaus Hahn and members of the Amelio Lab for helpful comments, suggestions and scientific review of this manuscript.
Grant Support
This work was supported by a Howard Temin Pathway to Independence Award in Cancer Research from the National Cancer Institute R00-CA157954 and UNC University Cancer Research Funds (A.L.A.), NIH/NCI R01 grant CA167093 (J.L.C.), by a Swiss National Science Foundation Postdoctoral Fellowship (F.X.S.), by a NIH/NIGMS T32 training grant GM007092 (A.M.M.), and by monies from the State of North Carolina to UNC-Chapel Hill. This work was also supported in part by NCI Comprehensive Cancer Center Grants P30-CA076292 awarded to the H. Lee Moffitt Cancer Center & Research Institute and P30-CA016806 awarded to the Lineberger Comprehensive Cancer Center.
Footnotes
Disclosure of Potential Conflicts of Interest: F.X. Schaub, J.L. Cleveland, and A.L. Amelio declare a patent application related to this work. The remaining authors declare that they have no conflict of interest.
Authors’ Contributions
Conception and design: A.L. Amelio
Development of Methodology: A.L. Amelio
Acquisition of data: F.X. Schaub, M.S. Reza, C.A. Flaveny, W. Li, S. Hoxha, A.L. Amelio
Analysis and interpretation of data: F.X. Schaub, M.S. Reza, C.A. Flaveny, A.M. Musicant, M. Guo, J.L. Cleveland, A.L. Amelio
Writing, review and/or revision of the manuscript: M. Guo, J.L. Cleveland, A.L. Amelio
Administrative, technical, or material support: A.M. Musicant, M. Guo
Study supervision: J.L. Cleveland, A.L. Amelio
References
- 1.Youn H, Hong K-J. In vivo Noninvasive Small Animal Molecular Imaging. Osong Public Health and Research Perspectives. 2012;3(1):48–59. doi: 10.1016/j.phrp.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puaux A-L, Ong LC, Jin Y, Teh I, Hong M, Chow PKH, et al. A Comparison of Imaging Techniques to Monitor Tumor Growth and Cancer Progression in Living Animals. International Journal of Molecular Imaging. 2011;2011(1):1–12. doi: 10.1155/2011/321538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nature Reviews Cancer. 2005;5(10):796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harbor protocols. 2012;2012(8) doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- 5.Kocher B, Piwnica-Worms D. Illuminating cancer systems with genetically engineered mouse models and coupled luciferase reporters in vivo. Cancer discovery. 2013;3(6):616–29. doi: 10.1158/2159-8290.CD-12-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS chemical biology. 2012;7(11):1848–57. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14342–46. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J-B, Urban K, Cochran E, Lee S, Ang A, Rice B, et al. Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PLOS ONE. 2010;5(2):e9364. doi: 10.1371/journal.pone.0009364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edinger M, Sweeney TJ, Tucker AA, Olomu AB, Negrin RS, Contag CH. Noninvasive assessment of tumor cell proliferation in animal models. Neoplasia (New York, NY) 1999;1(4):303–10. doi: 10.1038/sj.neo.7900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin RA, Felsen CN, Yang J, Lin JY, Whitney MA, Nguyen QT, et al. An optimized triple modality reporter for quantitative in vivo tumor imaging and therapy evaluation. PLOS ONE. 2014;9(5):e97415. doi: 10.1371/journal.pone.0097415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. European journal of nuclear medicine and molecular imaging. 2004;31(5):740–51. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 12.Ray P, De A, Min J-J, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64(4):1323–30. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67(7):3085–93. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 14.Yan X, Ray P, Paulmurugan R, Tong R, Gong Y, Sathirachinda A, et al. A transgenic tri-modality reporter mouse. PLOS ONE. 2013;8(8):e73580. doi: 10.1371/journal.pone.0073580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christoph S, Schlegel J, Alvarez-Calderon F, Kim Y-M, Brandao LN, Deryckere D, et al. Bioluminescence imaging of leukemia cell lines in vitro and in mouse xenografts: effects of monoclonal and polyclonal cell populations on intensity and kinetics of photon emission. Journal of Hematology & Oncology. 2013;6(1):1–1. doi: 10.1186/1756-8722-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contag PR. Whole-animal cellular and molecular imaging to accelerate drug development. Drug discovery today. 2002;7(10):555–62. doi: 10.1016/s1359-6446(02)02268-7. [DOI] [PubMed] [Google Scholar]
- 17.Kocher B, Piwnica-Worms D. Illuminating cancer systems with genetically engineered mouse models and coupled luciferase reporters in vivo. Cancer discovery. 2013;3(6):616–29. doi: 10.1158/2159-8290.CD-12-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152(1–2):340–51. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De A, Jasani A, Arora R, Gambhir SS. Evolution of BRET Biosensors from Live Cell to Tissue-Scale In vivo Imaging. Frontiers in endocrinology. 2013;4:131. doi: 10.3389/fendo.2013.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacres H, Michie M, Wang J, Pfleger KDG, Trowell SC. Effect of enhanced Renilla luciferase and fluorescent protein variants on the Förster distance of Bioluminescence resonance energy transfer (BRET) Biochem Biophys Res Commun. 2012;425(3):625–29. doi: 10.1016/j.bbrc.2012.07.133. [DOI] [PubMed] [Google Scholar]
- 21.Mezzanotte L, Blankevoort V, Löwik CWGM, Kaijzel EL. A novel luciferase fusion protein for highly sensitive optical imaging: from single-cell analysis to in vivo whole-body bioluminescence imaging. Analytical and Bioanalytical Chemistry. 2014;406(23):5727–34. doi: 10.1007/s00216-014-7917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K, Chang Y-F, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nature communications. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):12060–65. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De A, Ray P, Loening AM, Gambhir SS. BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23(8):2702–09. doi: 10.1096/fj.08-118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De A, Loening AM, Gambhir SS. An improved bioluminescence resonance energy transfer strategy for imaging intracellular events in single cells and living subjects. Cancer Res. 2007;67(15):7175–83. doi: 10.1158/0008-5472.CAN-06-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino H, Nakajima Y, Ohmiya Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nature methods. 2007;4(8):637–39. doi: 10.1038/nmeth1069. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(1):151–56. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dionne P, Mireille C, Labonte A, Carter-Allen K, Houle B, Joly E, et al. BRET2: efficient energy transfer from Renilla luciferase to GFP2 to measure protein-protein interactions and intracellular signaling events in live cells. Luminescence Biotechnology. 2002:539–55. [Google Scholar]
- 29.Shimomura O, Masugi T, Johnson FH, Haneda Y. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry. 1978;17(6):994–98. doi: 10.1021/bi00599a008. [DOI] [PubMed] [Google Scholar]
- 30.Shcherbakova DM, Hink MA, Joosen L, Gadella TWJ, Verkhusha VV. An Orange Fluorescent Protein with a Large Stokes Shift for Single-Excitation Multicolor FCCS and FRET Imaging. J Am Chem …. 2012;134(18):7913–23. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao C, Gao R, Zhang M, Amelio AL, Fallahi M, Chen Z, et al. Role of LKB1-CRTC1 on Glycosylated COX-2 and Response to COX-2 Inhibition in Lung Cancer. Journal of the National Cancer Institute. 2015;107(1):dju358–dju58. doi: 10.1093/jnci/dju358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer chemotherapy and pharmacology. 1989;24(3):148–54. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 33.Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harbor protocols. 2012 doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- 34.Arpino JAJ, Rizkallah PJ, Jones DD. Crystal structure of enhanced green fluorescent protein to 1. 35 Å resolution reveals alternative conformations for Glu222. PLOS ONE. 2012;7(10):e47132. doi: 10.1371/journal.pone.0047132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins DE, Oei Y, Hornig YS, Yu S-F, Dusich J, Purchio T, et al. Bioluminescent imaging (BLI) to improve and refine traditional murine models of tumor growth and metastasis. Clinical & experimental metastasis. 2003;20(8):733–44. doi: 10.1023/b:clin.0000006815.49932.98. [DOI] [PubMed] [Google Scholar]
- 36.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, et al. NanoLuc reporter for dual luciferase imaging in living animals. Molecular imaging. 2013;12(7):1–13. [PMC free article] [PubMed] [Google Scholar]
- 37.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. Understanding GFP posttranslational chemistry: structures of designed variants that achieve backbone fragmentation, hydrolysis, and decarboxylation. Journal of the American Chemical Society. 2006;128(14):4685–93. doi: 10.1021/ja056635l. [DOI] [PubMed] [Google Scholar]
- 38.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein engineering, design & selection : PEDS. 2006;19(9):391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 39.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nature methods. 2007;4(8):641–43. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 40.Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Löwik C. Sensitive Dual Color In Vivo Bioluminescence Imaging Using a New Red Codon Optimized Firefly Luciferase and a Green Click Beetle Luciferase. PLOS ONE. 2011;6(4):e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mezzanotte L, Aswendt M, Tennstaedt A, Hoeben R, Hoehn M, Löwik C. Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast media & molecular imaging. 2013;8(6):505–13. doi: 10.1002/cmmi.1549. [DOI] [PubMed] [Google Scholar]
- 42.Kaijzel EL, van der Pluijm G, Löwik CWGM. Whole-body optical imaging in animal models to assess cancer development and progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(12):3490–97. doi: 10.1158/1078-0432.CCR-07-0402. [DOI] [PubMed] [Google Scholar]
- 43.Pichler A, Prior JL, Piwnica-Worms D. Imaging reversal of multidrug resistance in living mice with bioluminescence: MDR1 P-glycoprotein transports coelenterazine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(6):1702–07. doi: 10.1073/pnas.0304326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lima-Fernandes E, Misticone S, Boularan C, Paradis JS, Enslen H, Roux PP, et al. A biosensor to monitor dynamic regulation and function of tumour suppressor PTEN in living cells. Nature communications. 2014;5:4431. doi: 10.1038/ncomms5431. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen QT, Tsien RY. Fluorescence-guided surgery with live molecular navigation--a new cutting edge. Nature Reviews Cancer. 2013;13(9):653–62. doi: 10.1038/nrc3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfleger KDG, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nature methods. 2006;3(3):165–74. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 47.Gammon ST, Villalobos VM, Roshal M, Samrakandi M, Piwnica-Worms D. Rational design of novel red-shifted BRET pairs: Platforms for real-time single-chain protease biosensors. Biotechnology progress. 2009;25(2):559–69. doi: 10.1002/btpr.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.