Abstract
On the male X and female active X chromosome (Xa), the macrosatellite repeat (MSR) DXZ4 is packaged into constitutive heterochromatin characterized by CpG methylation and histone H3 tri-methylated at lysine-9 (H3K9me3). In contrast, DXZ4 on the female inactive X chromosome (Xi), is packaged into euchromatin, is bound by the architectural protein CCCTC-binding factor, and mediates Xi-specific long-range cis contact with similarly packaged tandem repeats on the Xi. In cancer, male DXZ4 can inappropriately revert to a Xi-like state and other MSRs have been reported to adopt alternate chromatin configurations in response to disease. Given this plasticity, we sought to identify factors that might control heterochromatin at DXZ4. In human embryonic stem cells, we found low levels of 5-hydroxymethylcytosine at DXZ4, and that this mark is lost upon differentiation as H3K9me3 is acquired. We identified two previously undescribed DXZ4 associated non-coding transcripts (DANT1 and DANT2) that are transcribed towards DXZ4 from promoters flanking the array. Each generates transcript isoforms that traverse the MSR. However, upon differentiation, Enhancer of Zeste-2 silences DANT1, and DANT2 transcription terminates prior to entering DXZ4. These data support a model wherein DANT1 and/or DANT2 may function to regulate constitutive heterochromatin formation at this MSR.
Keywords: Macrosatellite, DXZ4, X chromosome inactivation, Human embryonic stem cells, Euchromatin and heterochromatin, Long noncoding RNA
Introduction
Macrosatellite repeat (MSR) DNAs are among the largest tandem-repeats in our genome. Each MSR is unique to one or two chromosomal locations, such as the ZAV array at chromosome 9q32 (Tremblay et al. 2010), or the RS447 array at chromosome 4p16.1 and 8p23 (Okada et al. 2002), and consist of a variable number of large (1–12 kilobase (kb)) repeat units that share greater than 90% inter-repeat unit sequence identity (Schaap et al. 2013; Tremblay et al. 2010; Warburton et al. 2008). The function of MSRs is unclear, although at least one has been shown to compensate for centromere loss and function as a neocentromere (Hasson et al. 2011). Some MSRs contain an open reading frame (ORF) in each repeat unit and their transcription is normally restricted to the testis, but this regulation is frequently lost in cancer resulting in their reactivation, earning them the name of cancer-testis loci (Cheng et al. 2011). Presumably, their multi-copy arrangement reflects a need for a large amount of gene product in the testis. Of the MSRs with coding potential, the best characterized is D4Z4 due to its central role in the onset of the common neuromuscular disorder facioscapulohumeral muscular dystrophy (FSHD) (Wijmenga et al. 1992). D4Z4 is found at 4q35 (van Deutekom et al. 1993) and 10q26 (Deidda et al. 1995), and in approximately 95% of FSHD cases, a 4q35 allele with fewer than 10 repeat units on a specific genetic background (Lemmers et al. 2002) results in the inappropriate expression and translation of the ORF in skeletal muscle (Lemmers et al. 2010), activating various germline and innate immunity genes that inhibit muscle differentiation (Geng et al. 2012).
Other MSRs show no obvious coding potential, yet are universally expressed (Chadwick 2008; Tremblay et al. 2010). Retention of these repeats in our genome suggests that they likely fulfill some purpose outside of simply coding for proteins. Among this group, the X-linked MSR DXZ4 is particularly unusual due to its location on a sex chromosome. DXZ4 is unique to Xq23 and consists of a GC-rich tandem repeat of between 12–120 uninterrupted 3 kb repeat units (Fig. 1a) (Giacalone et al. 1992; Schaap et al. 2013; Tremblay et al. 2011). Males are hemizygous for DXZ4 whereas in females, DXZ4 is exposed to the effects of X chromosome inactivation (XCI). XCI is a female specific phenomena whereby early in development, one of the two X chromosomes is largely rendered transcriptionally inert in order to balance X-linked gene dosage with males (Lyon 2002). A central player in this process is the long non-coding RNA (lncRNA) X-inactive specific transcript (XIST), which is expressed exclusively from the chosen inactive X chromosome (Xi) and associates with the chromosome territory in cis (Brockdorff et al. 1992; Brown et al. 1992). XIST triggers a cascade of events that ultimately result in the repackaging of the Xi DNA into facultative heterochromatin, an arrangement that is faithfully maintained throughout all subsequent somatic cell divisions (Chadwick and Willard 2003a). In males and on the female active X chromosome (Xa), DXZ4 is packaged into constitutive heterochromatin characterized by CpG methylation, histone H3 tri-methylated at lysine 9 (H3K9me3) and association of heterochromatin protein 1 (Chadwick 2008; Giacalone et al. 1992; Zeng et al. 2009). In contrast, DXZ4 on the Xi is organized into a euchromatic configuration characterized by CpG hypomethylation, histone acetylation and histone H3 di-methylated at lysine 4 (H3K4me2) (Chadwick 2008; Giacalone et al. 1992). Additionally, the architectural protein CCCTC-binding factor (CTCF) (Lobanenkov et al. 1990) specifically associates with DXZ4 on the Xi (Chadwick 2008; Chadwick and Willard 2003b), an interaction that is conserved in primates (McLaughlin and Chadwick 2011) and mouse (Horakova et al. 2012a; Yang et al. 2015). Intriguingly, in contrast to the surrounding heterochromatic chromosome, several other large tandem repeat DNA on the Xi are packaged into euchromatin, are bound by CTCF, and make frequent Xi-specific long-range contact with DXZ4 (Horakova et al. 2012b); as such, these X-linked tandem repeats may contribute to the alternate 3-dimensional configuration of the Xi relative to that of the Xa (Teller et al. 2011). Further evidence to support an important role for these MSRs on the Xi comes from functional analysis of the mouse homologue of one of the MSRs that makes Xi-specific contact with DXZ4 in humans (Horakova et al. 2012b; Rao et al. 2014). Yang and colleagues demonstrated that the lncRNA transcribed from the Firre locus (Hacisuleyman et al. 2014) is required for maintenance of histone H3 tri-methylated at lysine 27 (H3K27me3) and sub-nuclear localization of the Xi (Yang et al. 2015).
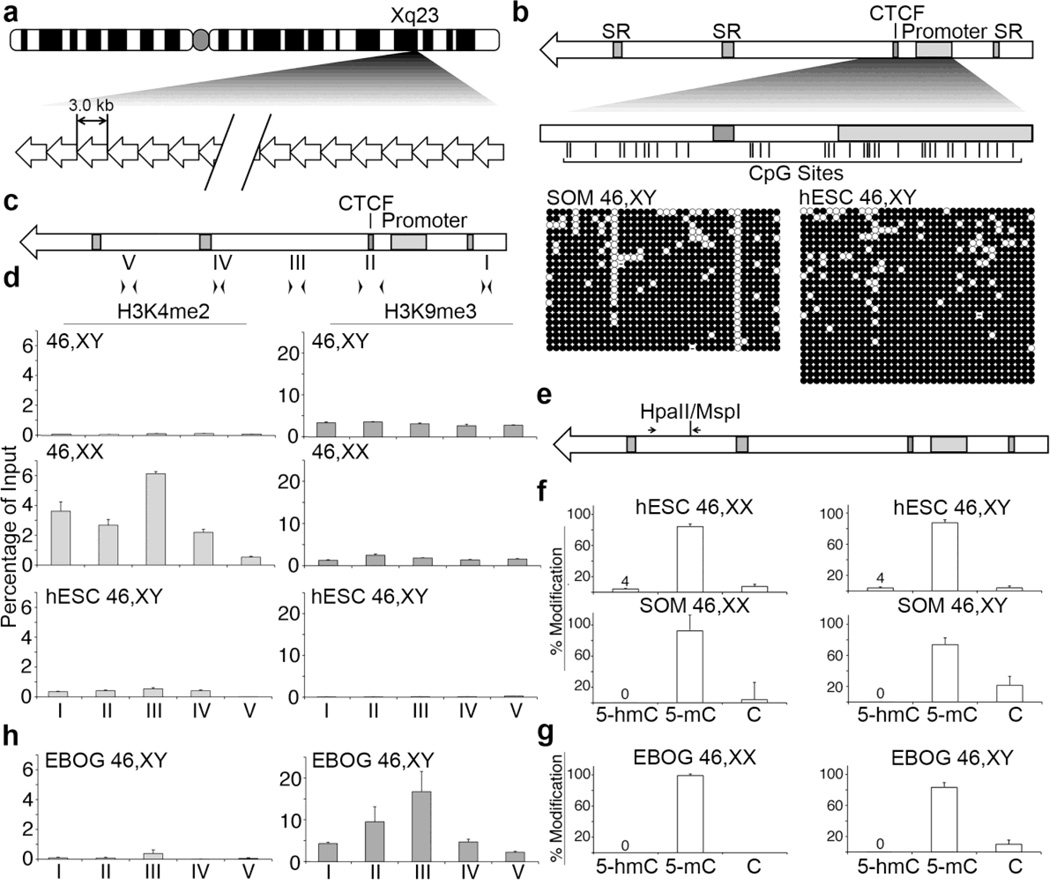
Fig. 1. Chromatin characterization in pluripotent and differentiated hESC.
(a) Ideogram of the human X chromosome indicating Xq23 and the location of DXZ4. A schematic representation of DXZ4 is shown beneath the ideogram, composed of 12–120 copies of the 3 kb repeat unit. (b) Schematic representation of a single DXZ4 monomer (top). Annotated features (shaded boxes) include simple repeats (SR), the CTCF binding site and the internal promoter. The region assessed by Bisulfite Sequencing (BiS) analysis is expanded immediately below, and vertical lines indicate the locations of all 36 CpG residues. The BiS profiles for male somatic fibroblasts (1139; SOM 46,XY), and male hESCs (H1; hESC 46,XY) are shown immediately below. The black (methylated) and white (unmethylated) circles indicate methylation status, while each horizontal row is the BiS profile from a single clone. A sequence that has diverged and is no longer a CpG is indicated by a dash. (c) Schematic DXZ4 monomer showing the location of primer sets used for qChIP: I (F6.R20), II (F23.R14), III (F17.R8), IV (F11.R22) and V (F4.R19). (d) qChIP data for H3K4me2 (left) and H3K9me3 (right) at DXZ4 for male (46,XY) and female (46,XX) somatic cells, and male hESCs (hESC 46,XY). Each data set is graphed as a percent of input. Error bars indicate standard deviation from the mean of triplicate qPCR reactions from two or more replicate ChIP experiments. (e) DXZ4 schematic indicating the location of the CpG that is part of a HpaII/MspI recognition site and the location of primers used for qRT-PCR. (f) Graphs showing the quantitation of percent 5-hmC, 5-mC, and C at the HpaII/MspI site. Female samples (46,XX) are shown on the left and include H9 (hESC) and IMR90 (SOM), whereas male samples (46,XY) are indicated on the right and include H1 (hESC) and 1140 (SOM). Data shown is from three independent biological replicates. (g) Quantitation of percent 5-hmC, 5-mC, and C at the HpaII/MspI site in female H9 (left) and male H1 (right) EBOG. (h) Graphs showing qChIP data as in part-(D) above, but for EBOG derived from male hESC (EBOG 46,XY).
This unusual epigenetic regulation of MSR chromatin states is not unique to DXZ4, as a shift toward a more euchromatic organization and binding by CTCF is also observed at D4Z4 in the context of FSHD (Ottaviani et al. 2009; van Overveld et al. 2003; Zeng et al. 2009). Furthermore, in some male cancers, DXZ4 can also revert to a Xi-like chromatin configuration (Moseley et al. 2012). These data suggest that in somatic cells, MSR are normally arranged into a constitutive heterochromatin state, but that a triggering event such as cellular transformation or repeat copy number contraction can compromise this programming, resulting in the potential for an inappropriate gain of function.
Interestingly, despite displaying features of constitutive heterochromatin, male DXZ4 and the female Xa allele are actively transcribed into lncRNAs (Chadwick 2008; Tremblay et al. 2011). What the purpose of the lncRNA is and if it has a role in maintaining the chromatin state of the MSR is unknown, although novel small RNA species originating from DXZ4 (Chadwick 2008) may mediate Argonaute-dependent DNA methylation at the tandem repeat (Pohlers et al. 2014). Nevertheless, at D4Z4, a lncRNA has been implicated in facilitating the switch toward a more euchromatic organization for the MSR in FSHD (Cabianca et al. 2012). Therefore, consistent with their role in developmentally regulating chromatin states (Gendrel and Heard 2014), lncRNAs are suitable candidates for directing MSR chromatin packaging.
To date, the chromatin state of DXZ4 has only been examined in somatic and transformed cells. Therefore, we sought to determine the chromatin arrangement of DXZ4 in human embryonic stem cells (hESCs). Given the innate issues relating to XCI status in female hESCs and the compromised organization of the Xi (Lessing et al. 2013; Minkovsky et al. 2012), we focused primarily on the male X to explore the constitutive heterochromatin arrangement at this allele that closely resembles the organization of MSRs on the autosomes. Here we report that DXZ4 resides in an alternate chromatin state in hESCs, that adoption of a constitutive heterochromatin organization for the MSR is developmentally regulated, and that the chromatin alteration is coupled to changes in expression of two DXZ4 associated novel lncRNAs.
Materials and Methods
Novel transcripts
Sequences associated with this study have been deposited with GenBank under the accession numbers [GenBank: KM192212, GenBank: KM192213, GenBank: KM192214, GenBank: KM192215, GenBank: KM192216, GenBank: KM192217]. The HUGO Gene Nomenclature Committee (HGNC) approved gene names and symbols DANT1 and DANT2.
Cells
Human Telomerase immortalized cell lines hTERT-RPE1 (X4000-1 46,XX retinal pigment epithelia) and hTERT-BJ1 (C4001-1 46,XY foreskin fibroblast) were originally obtained from Clontech, but are now available from the American Type Culture Collection (ATCC). Human embryonic Stem Cell lines H9 (WA09, 46,XX) and H1 (WA01, 46,XY) were obtained from WiCell Research Institute. The following human cells lines were obtained from ATCC: primary skin fibroblast cells CCD-1139Sk (CRL-2708, 46,XY) and CCD-1140Sk (CRL-2714, 46,XY); male colorectal adenocarcinoma cell lines DLD-1 [CCL-221] and HCT116 (CCL-247); fetal lung fibroblasts IMR-90 (CCL186, 46,XX) and WI-38 (CCL-75, 46,XX). Fetal human dermal fibroblasts were obtained from ScienCell Research Laboratories (Catalog Number 2300). All cells were maintained according to the supplier recommendations.
Immunofluorescence
Immunofluorescence was performed on cells grown directly on slides as previously described (Chadwick and Willard 2002). Except, the maximum slide seed area was delimited using a Super PAP Pen (IM3580, Beckman Coulter®) and coated with 700µl of Matrigel™ or 0.1% Gelatin prior to seeding hESCs or EBOGs, respectively. Additionally, hESCs were allowed to adhere for 1hr in complete StemPro® media supplemented with ROCK Inhibitor Y-27632 (ROCKi, SCM075, Millipore™), the media was then replaced with fresh complete StemPro® media (-ROCKi), and allowed to recover overnight. Antibodies used for indirect Immunofluorescence included rabbit anti-H3K4me2 (07–030, EMD Millipore), mouse anti-H3K27me3 (ab6002, abcam), rabbit anti-NANOG (3580S, Cell Signaling Technology), mouse anti-SSEA4 (4755S, Cell Signaling Technology), goat anti-SOX2 (AF2018, R&D Systems), and rabbit anti-OCT4 (2750S, Cell Signaling Technology). Conjugate secondary antibodies (Alexa-Fluor®) were obtained from Life Technologies Corporation. DNA was counterstained using the VECTASHIELD® mounting medium with DAPI from VECTOR Laboratories. Imaging was performed on an Olympus IX71 operated by the DeltaVision pDV, deconvolved with softWoRx 5.5.1 (DeltaVision), and compiled using Adobe Photoshop CS6 (Adobe Systems).
Epimark assay and bisulfite/oxidative bisulfite sequencing
Genomic DNA was isolated using the Qiagen Tissue Midiprep Kit (Qiagen, Valencia, CA, USA). Oxidation of 5-hydroxymethycytosine was performed as described by Booth et al (2012), with exceptions. Briefly, both the control and oxidized samples were treated in parallel by denaturing with 5mM NaOH, incubation on ice during oxidation step, and purified using the Qiagen QIAquick PCR Purification kit including four-600µl PE washes. Bisulfite modification was subsequently performed on both the control and oxidized samples using the Qiagen EpiTect Bisulfite Kit according to the manufacturer’s instructions. PCR amplification of the DXZ4 target region was performed using the oligos listed in Supplementary Table 1. The PCR products were cloned and sequenced as previously described (Moseley et al. 2012). Additionally, 5-hydoxymethylcytosine frequency at a single CpG was assayed using the NEB Epimark® 5-hmC and 5-mC Analysis kit (E3317S). The products were analyzed by qPCR, on a Bio-Rad CFX96 Real-Time System with a C100™ Thermal Cycler using EvaGreen qPCR Mastermix (Mastermix-S, Applied Biological Materials), using the DXZ4 oligos described in Supplementary Table 1. Quantitation of the 5-hydroxymethylcytosine qPCR data was generally performed according to the New England BioLabs Epimark 5-hmC and 5-mC Analysis Kit manual, except that the SQ values for each sample were used. Briefly, both the Epimark assay and the qPCRs were performed in triplicate. Average SQ values for each sample were normalized against the reference gene (MYT-1). The percent 5-hmC, 5-mC, and C of the total DNA was calculated as follows: %5-hmC = ((Unglucosylated MspI digest x (glucosylated undigested DNA / unglucosylated undigested DNA) – glucosylated MspI digest)/ glucosylated undigested DNA) x 100; % 5-mC = (glucosylated HpaII digest - Unglucosylated MspI digest x (glucosylated undigested DNA/unglucosylated undigested DNA)/glucosylated undigested DNA) x 100; %C = (glucosylated undigested DNA-glucosylated HpaII digest)/glucosylated undigested DNA) – glucosylated MspI digest)/glucosylated undigested DNA) x 100.
Promoter luciferase assay
DNA fragments upstream of DANT1 and DANT2 exon-1 were generated by PCR using the Qiagen Hotstar Taq Plus and cloned into pDrive (Qiagen). Primers used can be found in Supplementary Table 1. The inserts were sequence verified, subcloned into pGL4.10, transfected, and assayed for luciferase activity as previously described (Horakova et al. 2012b). A pGL4.10 promoterless construct was used to normalize the expression between 293T and H9 while pGL3 was used as a positive control.
RNA FISH
RNA FISH was performed on cells grown directly on slides as described above. Spectrum Orange and Spectrum Green direct-labeled probes were prepared by nick translation according to manufacturer’s instructions (Abbott Molecular). Labeled probes were ethanol precipitated with 25 µg of human Cot-1 DNA (Invitrogen) and resuspended in 100 µl of Hybrisol VII (MP Biomedicals). The probes included two BAC clones 2272M5 (DXZ4 repeat) and RP11–761E20 (DANT2) that were obtained from Invitrogen and an XIST plasmid pX1644 (Chadwick and Willard 2002). A DANT1 FISH probe was generated using the genomic sequence spanning the region defined by the oligos CD173052-F & CD173052-R. The corresponding fragment was PCR amplified and TA cloned into pDrive (Qiagen) before sequence verification. Cells were fixed and extracted for 10 minutes at room temperature in 1x phosphate buffered saline (PBS) containing 3.7% formaldehyde and 0.1% Triton X-100. Slides were then washed for 2 minutes each in 1x PBS before dehydrating for 2 minutes each in 70%, 80% and 100% ethanol. Slides were then air-dried. Probes were denatured for 10 minutes at 72°C and transferred to 37°C for 1 hour to block repeats before applying to the slide. Probe mixtures were sealed under coverglass using rubber cement. Hybridization was performed in a humidified chamber at 37°C for 16 hours. RNA FISH was washed twice at room temperature for two minutes each in 50% formamide with 2x Saline-Sodium Citrate (SSC) followed by one wash of three minutes in 37°C 50% formamide 2xSSC and finally three minutes in 37°C 2xSSC. DNA was counterstained as described above. Imaging was performed, deconvolved, and compiled as described above. Controls were included to show the RNA-specific nature of the RNA FISH and to demonstrate that the treatment was specific to RNA. Cells on three additional slides were fixed and dehydrated as above. However, before application of the FISH probe, two slides were incubated at 37°C for 1 hour with 100 µl of PBS supplemented with 7 units of RNaseA (Qiagen) before washing three times for two minutes each with PBS before dehydration. For one RNaseA treated slide, RNA FISH was performed as above. For the remaining RNaseA treated and non-treated slide, DNA FISH was performed by denaturing the samples at 83°C for 10 minutes in 70% formamide, 2xSSC before dehydrating for 2 minutes each in 70% and 100% ethanol. Denatured probes were applied to dried slides and incubated overnight at 37°C in a humidified chamber. RNA FISH was washed as above, while DNA FISH was washed by first incubating slides for 8 minutes each at 42°C in 50% formamide 2xSSC, followed by 8 minutes in 42°C 2xSSC.
Standard and quantitative reverse transcription PCR
Total RNA was isolated and reverse transcribed as previously described (Horakova et al. 2012b). Standard reverse transcription PCR (RT-PCR) was performed as previously described (Horakova et al. 2012b) using the oligos listed in Supplementary Table 1. Human tissue cDNA samples were prepared as described previously (Tremblay et al. 2010). Quantitative PCR was performed on a Bio-Rad CFX96 Real-Time System with a C100™ Thermal Cycler using EvaGreen qPCR Mastermix (Mastermix-S, Applied Biological Materials), using the oligos in Supplementary Table 1, and normalized to GAPDH expression. For qrtPCR the samples were analyzed generally using the ΔΔCT method (Livak and Schmittgen 2001), except that the Cq values instead of CT values were used. Briefly, the expression of reference gene for each sample was normalized against the reference gene by subtracting the Cq value of the from the Cq value of the reference gene to generate the ΔCq. ΔCq for each sample was then normalized to the ΔCq values of the reference sample (H9) to give the ΔΔCq. The fold difference in expression was then determined by taking the log10 of each sample. There average expression ratios and standard deviations were then determined.
hESC Differentiation into Embryoid Body Out-Growths (EBOGs)
ESCs were cultured on BD Matrigel™ hESC-qualified Matrix (354277, BD Biosciences)-coated 60mm CELLSTAR® cell culture dish (628160, Greiner bio-one) using complete StemPro® hESC SFM media (StemPro®, A10007-01, Gibco®) supplemented with 2% Methylsulfoxide (MX1456, EMD) for 24hrs and grown to ~90% confluence. ESCs were disaggregated using StemPro® Accutase® Cell Dissociation Reagent (A11105-01, Gibco®) into 5–20 cell clumps and seeded to 3 wells of a CorningTM Ultra-low attachment 6-well dish (07–200–601, Fisher Scientific) using StemPro® media, lacking recombinant Human bFGF (PHG0264, Gibco®) & 2-mercaptoethanol (βME, 21985, Gibco®), supplemented with 10µM SB431542 (SB, S1067, Selleckchem) [10µM SB-StemPro® (-bFGF, -βME)]. EBOG generation was performed essentially as described (Mahmood et al. 2010) with modifications described by others (Awaya et al. 2012; Chetty et al. 2013). The cells were allowed to differentiate into embryoid bodies (EBs) with media changes on days 3, 6, & 8 using 10µm SB-StemPro® (-bFGF, -βME) media. On Day 10 the EBs were split 1:3 and seeded to 0.1% Gelatin (G-2500, Sigma)-coated NunclonTM 6-well cell culture dish (140675, Thermo Scientific) in chemically defined medium (CDM, (Mahmood et al. 2010)) supplemented with 1µM SB (1µM SB-CDM). These were allowed to further differentiate and expand as EB-OGs for over 11 passages, with fresh 1µM SB-CDM replacement every 2–3 days.
Quantitative Chromatin Immunoprecipitation (qChIP)
ChIP was performed essentially as previously described (Moseley et al. 2012), except cells were fixed for 8 min in 1xPBS containing 0.75% or 1% formaldehyde (hESC or somatic cells, respectively). Fixed cells were sheared at 4°C on High power with 30s ON and 30s OFF for 5 minutes per round using the Diagenode Bioruptor 300 Sonication System attached to a refrigeration circulator. Somatic Cells were sonicated a total of 4 rounds, while hESCs were sonicated for 6 rounds, with brief vortex and centrifugation at 4°C between each round to obtain an average shear size of 250–300bp. Antibodies used for ChIP were obtained from EMD Millipore and include mouse anti-EZH2 (17–662), rabbit anti-H3K27me3 (07–449), rabbit anti-H2K4me2 (07–030), rabbit anti-H3K9me3 (07–523), and rabbit anti-CTCF (07–729). Immunoprecipitated DNA was assessed by quantitative PCR (qPCR) on a Bio-Rad CFX96 Real-Time System with a C100™ Thermal Cycler using EvaGreen qPCR Mastermix (Mastermix-S, Applied Biological Materials). All oligos used, described in Supplementary Table 1, were obtained from Eurofins MWG Operon. Data was analyzed using Bio-Rad CFX Manager 3.1. Data is shown as percentage of input for each sample. This was determined by dividing the SQ values of the ChIP sample from the corresponding Input sample. Compensation for each samples was then performed by subtracting the percent input of the no-antibody control from the antibody-treated sample. The average and standard deviation from the percent of input was then determined for every sample.
Results
DNA methylation and chromatin of DXZ4 in human embryonic stem cells
Previously, we have shown that CpG residues in the vicinity of the bi-directional promoter and encompassing the CTCF binding site of individual DXZ4 monomers are mostly methylated in male somatic cells (Chadwick 2008). The status of CpG methylation at DXZ4 in hESCs is unknown. Therefore, we assessed the same interval in male hESCs using bisulfite sequencing (BiS) (Fig. 1b). We found that similar to male somatic cells (Fig. 1b, left panel), DXZ4 CpGs are methylated in the male hESCs (Fig. 1c, right panel).
These data suggest that DXZ4 is already arranged into constitutive heterochromatin in pluripotent cells. Therefore, in order to further explore this possibility, we performed chromatin immunoprecipitation (ChIP) with antibodies to the euchromatin marker H3K4me2 as well as the heterochromatin marker H3K9me3, and assessed DXZ4 for the presence of either chromatin modification by qPCR using primer pairs regularly spaced throughout a single DXZ4 repeat unit (Fig. 1c). As anticipated, H3K4me2 was a prominent feature of female somatic cells (corresponding to the Xi allele of DXZ4), whereas H3K9me3 was detected at DXZ4 in both male and female somatic cells (Fig. 1d, top panels). However, low levels of H3K4me2 and no H3K9me3 could be detected at DXZ4 in male hESCs (Fig. 1d, bottom panels).
BiS analysis does not distinguish between 5’-methylcytosine (5-mC) and a similar, but functionally distinct modification 5’-hydoxymethylcytosine (5-hmC)[Reviewed in (Cadet and Wagner 2014)]. Given that in primate and mouse brain, as well as in mouse and human ESCs, 5-hmC is preferentially associated with active chromatin marks at enhancers and promoters (Ficz et al. 2011; Stroud et al. 2011; Szulwach et al. 2011b)[Reviewed by (Branco et al. 2012; Chopra et al. 2014)] we sought to determine if the DNA methylation detected at DXZ4 in hESCs could reflect the presence of 5-hmC. We quantified the amount of 5-hmC at DXZ4 using two complementary approaches; the EpiMark assay from New England Biolabs, and oxidative Bisulfite Sequencing (oxBiS).
The EpiMark assay from New England Biolabs involves first treating the target DNA with T4 β-glucosyltransferase that adds a glucose moiety to 5-hmC, but not 5-mC. The DNA is then subjected to restriction endonuclease digestion with HpaII or its isoschizomer MspI before qPCR across the digest site in order to determine what fraction of the DNA is uncut. HpaII will not cut sites marked by 5-mC or 5-hmC, whereas MspI is insensitive to either modification and will cut the target site. However, MspI is incapable of digesting DNA in which 5-hmC is glucosylated; therefore, template DNA containing this modification will remain uncut and detectable by qPCR. Primers were designed to amplify across a single HpaII/MspI recognition sequence in DXZ4 that could be used to analyze 5-hmC at a single CpG (Fig. 1e). By this assay, we consistently detected 5-hmC at DXZ4 in both male and female hESCs, but not somatic cells (Fig. 1f).
The oxBiS assay uses potassium perruthenate to oxidize 5-hmC to 5-formylcytosine, which after bisulfite treatment is converted to uracil along with unmethylated cytosines. When compared to traditional BiS profiles derived from the same sample, that were not first oxidized, it is possible to determine the percent of 5-hmC by subtracting the percent methylation in the oxBiS profile from that detected by BiS. Consistent with the EpiMark assay, oxBiS analysis revealed that 1.5% (male H1 hESCs) and 2.7% (female H9 hESCs) of the BiS signal corresponded to 5-hmC at DXZ4, while none was detected in somatic cells (Supplementary Fig. 1a). Although the percent 5-hmC found at DXZ4 was low, but present in both assays, these levels are consistent with those previously reported at the promoters of various pluripotency markers in hESCs (Szulwach et al. 2011a).
To determine if 5-hmC at DXZ4 is lost upon differentiation, we repeated the EpiMark assay on DNA extracted from embryoid body outgrowths (EBOGs) derived from the male and female hESCs. Appropriate differentiation was validated by the loss of pluripotency markers and gain of anticipated EBOG-associated gene expression (Supplementary Fig. 2). Results showed that differentiation into EBOGs was coupled with a loss of 5-hmC (Fig. 1g). Next, we sought to determine if the differentiation associated loss of 5-hmC correlated with change to DXZ4 chromatin by repeating the H3K4me2 and H3K9me3 qChIP analysis. We found that differentiation of hESCs into EBOG resulted in gain of H3K9me3 at DXZ4 coupled with a reduction in the already low H3K4me2 levels (Fig. 1h and Supplementary Fig. 1b). Therefore, these data indicate that the formation of constitutive heterochromatin at DXZ4 is developmentally regulated.
lncRNA expression in the region surrounding DXZ4
Several lncRNAs, including XIST, have been shown to play central roles in developmentally linked chromatin change [Reviewed in (Jeon et al. 2012)]. Therefore, we explored the possibility that developmentally regulated lncRNAs may be associated with DXZ4. DXZ4 is itself transcribed primarily from the Xa and male X in somatic cells through a promoter contained within each repeat unit (Chadwick 2008; Tremblay et al. 2011). However, we began by further characterization of the genomic interval around DXZ4 and searched for evidence of any lncRNAs not originating directly from the MSR itself.
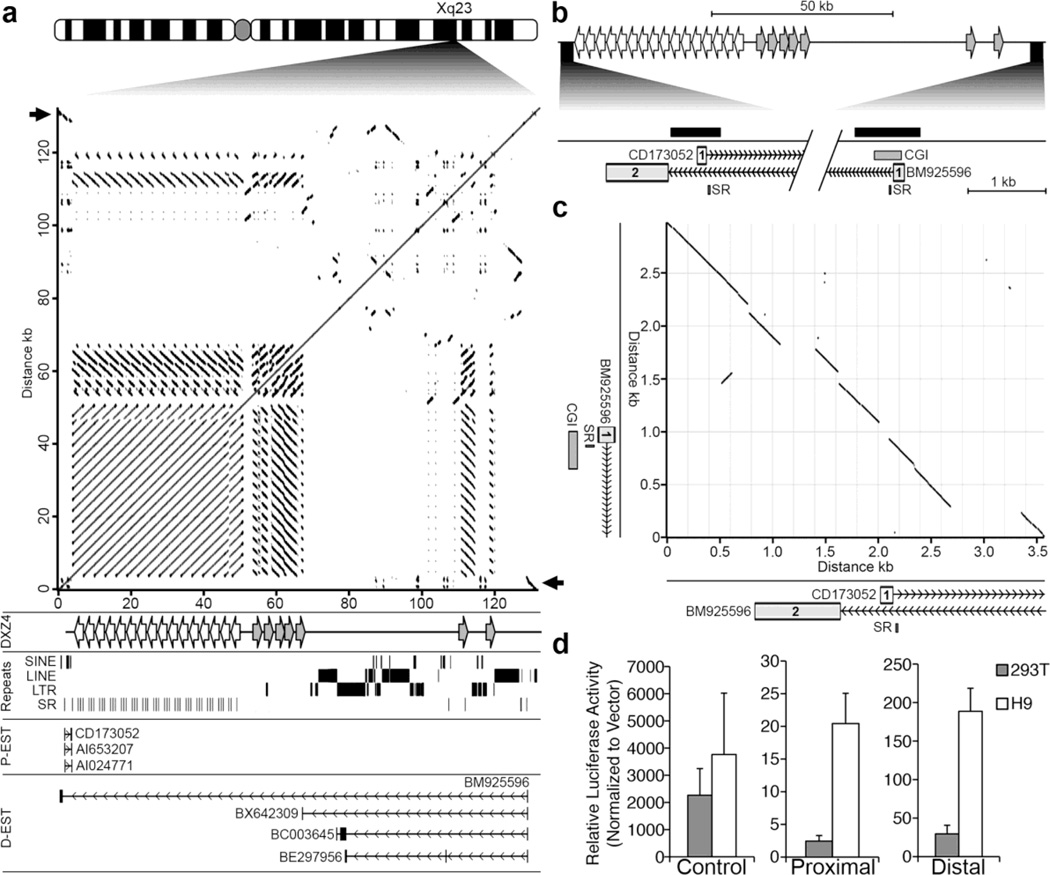
Pair-wise alignments clearly show the tandem arrangement and location of DXZ4, but also reveal the presence of numerous closely related DXZ4-like sequences located distal and inverted relative to the main DXZ4 array (Fig. 2a). Examination of annotated spliced expressed sequence tags (ESTs) and messenger RNA (mRNA) in the immediate vicinity surrounding DXZ4 revealed two clusters of spliced ESTs. One cluster originates immediately proximal to DXZ4 and these are transcribed toward the array before terminating within 0.6 kb of the first DXZ4 monomer at the proximal edge (Fig. 2a, P-EST). One member of this cluster is derived from hESCs (CD173052), whereas the other two are from a breast tumor (AI024771) and pooled germ cell tumors (AI653207). The second cluster originates approximately 80 kb distal to DXZ4 (Fig. 2a, D-EST). These ESTs come from male brain (BM925596), an unknown adult source (BX642309) and a rhabdomyosarcoma (BE298956 and BC003645). All four ESTs are transcribed toward DXZ4, but intriguingly, BM925596 traverses the entire DXZ4 array before splicing to an exon <400 bp upstream of the beginning of the proximal cluster (Fig. 2b), indicating that the two putative transcriptional units overlap and are anti-sense relative to one another. Pairwise alignment of the DNA sequence encompassing the beginning of the proximal and distal EST clusters reveal these sequences share extensive inverted sequence identity with one another (Fig. 2c) and likely originate from a common ancestral sequence. Interestingly, there is a noticeable break in the sequence identity of exon-1 in the proximal and distal ESTs, but a conserved pyrimidine-rich simple repeat (SR) sequence (TTTCC)n is located immediately downstream of each exon-1. Each member of the proximal cluster originate close to one another at their 5’ end, as do members of the distal cluster. Therefore, we reasoned that the sequence encompassing and extending upstream of the first exon of each cluster likely possesses promoter activity. The presence of a CpG island (CGI) at exon-1 of the distal cluster supports this notion (Fig. 2b and c).
Fig. 2. Related promoters flanking DXZ4 drive transcription toward the array.
(a) Ideogram of the human X chromosome with the location of DXZ4 at Xq23 expanded. Beneath the ideogram is a pair-wise alignment of the DXZ4 interval corresponding to nucleotides 114,955,568 bp to 115,088,136 bp (hg19) adapted from the output of the Basic Local Alignment Search Tool at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with the align two or more sequence option. The black arrows point to the region of inverted homology that is the focus of part-C (described below). Immediately beneath, is indicated (DXZ4) the location of the main DXZ4 array (left-facing white arrows) as well as the DXZ4-related inverted repeats (right-facing grey arrows), followed by the location of repeat classes (Repeats), annotated ESTs or mRNAs originating proximal to DXZ4 (P-EST) that are transcribed from left to right, and distal ESTs (D-EST) that are transcribed from right to left. Vertical lines represent exons, and horizontal lines represent introns with chevrons indicating direction of transcription. Each EST or mRNA was extracted from data contained in the Human mRNAs and Spliced ESTs annotations on the UCSC Genome Browser. (b) Schematic map of the DXZ4 interval, with the region corresponding to the beginning of the proximal and distal ESTs expanded immediately below. Exon-1 of proximal EST CD173052 is shown (“1” containing white-box), as are exons 1 and 2 of BM925596 (grey boxes containing “1” and “2”, respectively). The locations of a conserved simple repeat (SR) and a CpG island (CGI) are also shown while the solid black bars indicate the locations of the genomic fragments cloned and used to assess for promoter activity in part-D (described below). The diagonal break corresponds to the genomic DNA between the candidate promoters. (c) Pair-wise alignment of the DNA sequence around exon-1 of the proximal ESTs to exon-1 of the distal ESTs, corresponding to nucleotides 114,955,273–114,958,855 (proximal region – x-axis) and 115,084,395–115,087,382 (distal region – y-axis) of the human X chromosome (hg19). An expansion of the schematic map from B corresponding to the x and y-axis are beneath and left of the alignment. (d) Relative promoter activity for the Proximal and Distal promoter candidate sequences as assessed in hESCs (H9) and 293T cells. Firefly luciferase activity is normalized to that of a constitutively active co-transfected Renilla luciferase construct, and is graphed relative to activity detected from a promoter-less vector. Data shown represents the mean of three separate transfection experiments performed in triplicate and error bars indicate standard deviation. Data for the Control sample was derived from a construct containing a strong promoter and enhancer as a positive control.
In order to test the fragments for promoter activity, the candidate regions were PCR amplified (Fig. 2b), TA-cloned and sequence verified before subcloning upstream of a promoterless luciferase reporter gene. Both the proximal and the distal promoter-candidate constructs, as well as a control construct consisting of a powerful SV40 promoter and enhancer were introduced separately into 293T and H9 hESCs before assessing for luciferase activity. As expected, the control construct generated robust luciferase activity at comparable levels in both cell types (Fig. 2d, left graph). Interestingly, proximal promoter activity was only two-fold higher than background levels in 293T cells, but showed obvious and reproducible activity at 20-fold higher than background in hESCs (Fig. 2d, middle graph), whereas, the distal promoter candidate showed activity in both cell types but displayed higher activity in hESCs (Fig. 2d, right graph). These data confirm the location of promoters for proximal and distal EST clusters, and suggest that expression from the proximal promoter is unlikely to be universal.
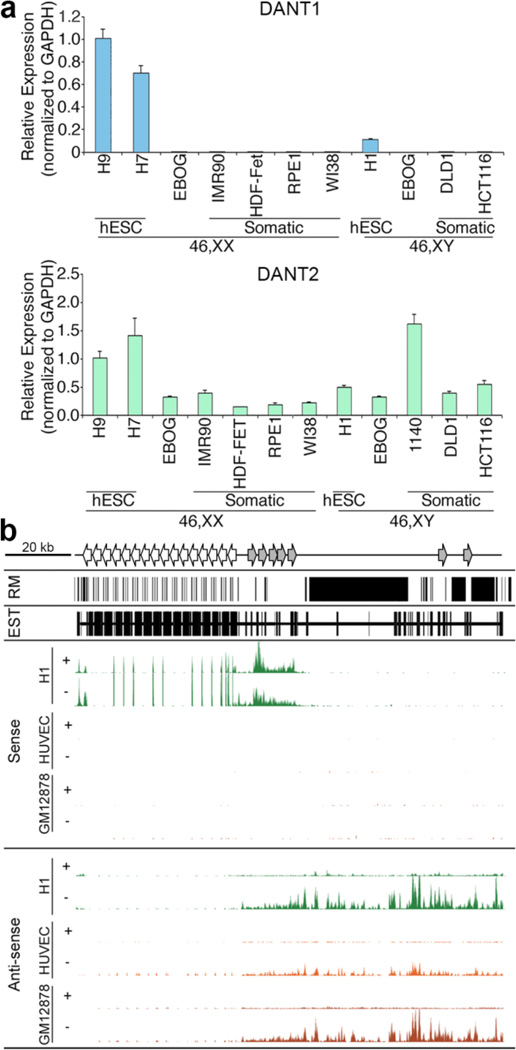
Next we sought to further characterize the transcripts associated with these promoters. For the distal promoter, the only exon present consistently in all ESTs is the first. Similarly, the proximal cluster show alternative splicing with the presence or absence of exon-2. Therefore, to assess total expression regardless of differential splice isoforms, we performed qRT-PCR with primers contained entirely within exon-1 of either the proximal EST cluster or distal EST cluster (Supplementary Table 1). Consistent with the promoter activity, the proximal transcript was only detected in hESCs, and was extremely low to undetectable in EBOG and all somatic samples tested (Fig. 3a, top panel). In contrast, the distal transcript could be detected in all cell types examined (Fig. 3a, bottom panel), and shows some variation in expression level between samples tested, although this may reflect differences in the cell type and passage of samples used.
Fig. 3. Characterization of the DANT1 and DANT2 lncRNAs.
(a) Graphs showing the expression of DANT1 (top) and DANT2 (bottom) lncRNA as determined by qRT-PCR in the various samples indicated. Primers used for qRT-PCR were contained within exon-1 of each gene amplifying a 57 bp or 100 bp amplicon, respectively (Supplementary Table 1). Data represents the mean of triplicate qPCR reactions and error bars indicate standard deviation. qRT-PCR data is normalized to GAPDH levels and graphed relative to expression in a female hESC sample (H9). (b) Genomic interval chrX:114,955,242–115,088,266 (hg19) showing the extent of repeat masked (RM) DNA, the location of all ESTs, and the RNA-seq data for the DXZ4 interval showing strand-specific sequencing for male hESC (H1), male umbilical vein endothelial cells (HUVEC) and female EBV transformed B-lymphocytes (GM12878) (Parkhomchuk et al. 2009). The location of the main DXZ4 array is represented at the top by the left-facing arrows, whereas the approximate location of inverted homologous DXZ4 monomers are represented by gray right-facing arrows. Sense-strand data is shown at the top, representing transcription from left-to-right, and anti-sense data is on the bottom, representing transcription from right-to-left. The “+” and “−” represent data from polyA+ and polyA- RNA sources, respectively. For each profile the y-axis is from 0–300 reads.
Using Open Reading Frame Finder, available within the National Center for Biotechnology Information suite (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi), the largest potential ORF for any of the proximal transcript splice isoforms was 204 nucleotides, whereas for distal transcript splice isoforms the longest ORF was 282 nucleotides. To determine if any of the short ORFs has any coding potential, the sequence of each isoform was entered into the Coding-Potential Assessment Tool, software that can reliably determine if a sequence has coding potential (Wang et al. 2013). None of the isoforms showed any coding probability using the standard default settings for human sequences. Therefore, we conclude that these represent novel lncRNAs. We refer to the proximal promoter driven transcript as DXZ4 Associated Non-coding Transcript, proximal (DANT1) and the distal promoter driven transcript as DXZ4 Associated Non-coding Transcript, distal (DANT2).
Exon-2 of DANT2 EST BM925596 is located upstream of DANT1 exon-1 (Fig. 2a and b) indicating that at least some DANT2 transcript completely traverses the DXZ4 tandem repeat and DANT1 transcriptional unit. To explore the DANT1 and DANT2 transcripts further, we examined publicly available next generation strand-specific RNA sequencing data sets (RNA-seq) (Parkhomchuk et al. 2009), using the long RNA-seq track on the University of California Santa Cruz genome browser (http://genome.ucsc.edu). The interval between DANT2 and DXZ4 is extensively repeat masked (Fig. 3b), characterized primarily by long terminal repeats (LTR) and long interspersed elements (LINE) (Fig. 2a), but a substantial proportion of the unique sequence is characterized by the presence of ESTs, indicating that the interval is extensively transcribed. Consistent with the qRT-PCR data (Fig. 3a) and promoter activity (Fig. 2d), sense strand DANT1 RNA-seq reads are only detected for the hESC sample, with comparable levels of transcript detected from both the polyadenylated (+) and nonpolyadenylated (−) samples (Fig. 3b, H1 top panel). As expected, DANT2 can be detected in all samples from the anti-sense strand, but in contrast to DANT1, it is almost exclusively from the nonpolyadenylated fraction (Fig. 3b, bottom panels). Furthermore, RNA-seq reads corresponding to exon-2 of EST BM925596 can be detected upstream of the DANT1 promoter (See small peaks at the far-left of the H1 Anti-sense profile in Fig. 3b), supporting the existence of this transcript isoform. The distinct lack of mapped RNA-seq reads throughout the DXZ4 MSR, despite dense representation of transcripts by the EST clones, could reflect reads that map to multiple locations being discarded from the RNA-seq profile, consistent with most MSR appearing as gaps in profiles of next generation sequence data tracks.
Validation of DANT1
The DANT1 RNA-seq profile suggests, that like DANT2, this lncRNA generates an array-traversing transcript (ATT) with the longest DANT1 transcript entirely contained within, but anti-sense to, the longest predicted DANT2 transcript (Fig. 3b, H1 top panels). Therefore, in order to validate these transcripts and the non-ATT annotated ESTs, we performed a series of RT-PCR analyses.
Initially, we performed RT-PCR with primers located in exon 1 and 3 of the short non-ATT EST CD173052 and confirmed the presence of this transcript in hESC (Fig. 4a, PCR-1). The EST entry for CD173052 terminates with a polyA sequence that is not present in the corresponding genomic DNA locus, suggesting that this transcript is polyadenylated. Using oligo-dT primed 3’ rapid amplification of cDNA ends (RACE), we confirmed that this transcript is indeed polyadenylated and therefore terminates before the DXZ4 array (data not shown).
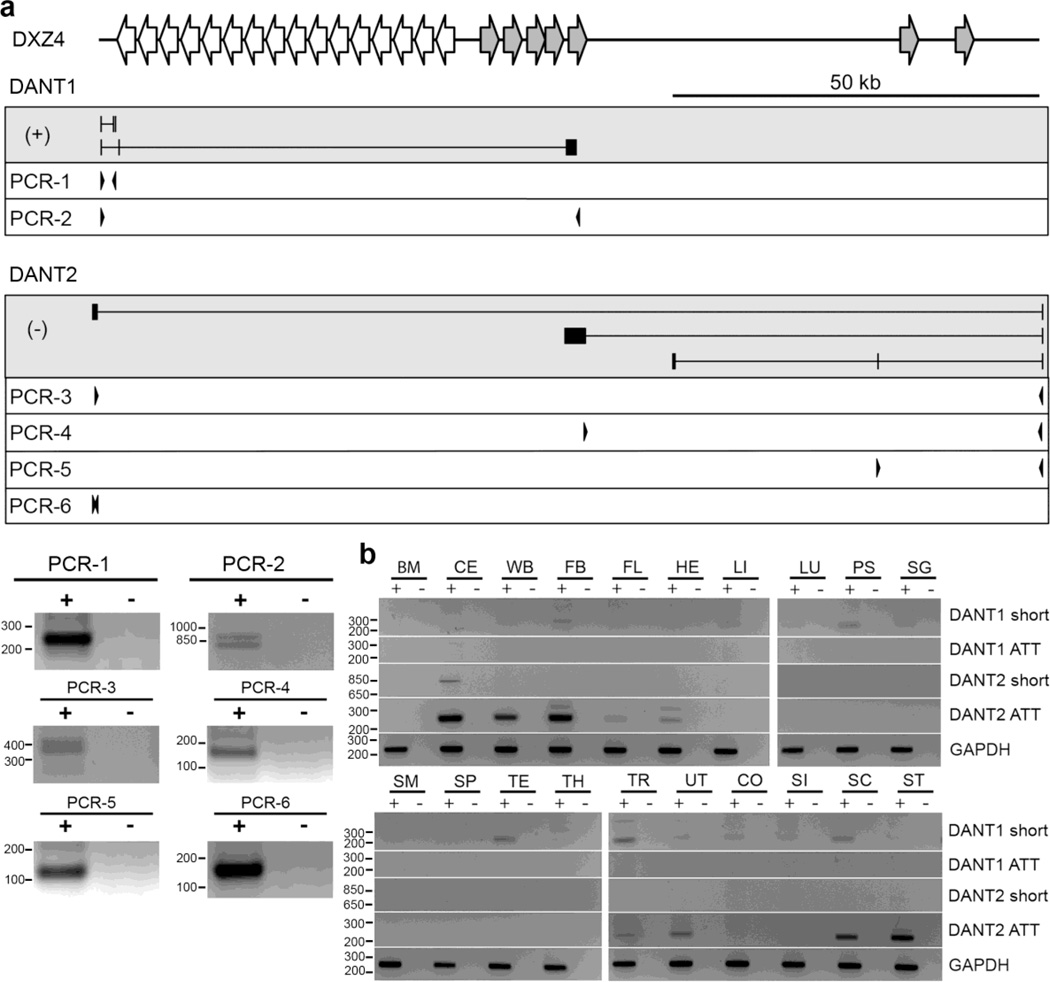
Fig. 4. Validation of the DANT1 and DANT2 short and array-traversing transcripts (ATT).
(a) Schematic map of the DXZ4 interval is shown at the top. Immediately below are maps of the transcripts in the grey-boxes with (+) or (−) indicating origins from the forward or reverse strand respectively. Primers used for RT-PCR’s 1–6 are shown beneath in the white boxes. Vertical lines represent exons and introns are horizontal lines. Representative RT-PCR results for PCR-1 through PCR-6 are shown as inverted ethidium bromide stained gel images. (b) Detection of DANT1 and DANT2 ATT and short transcripts in a variety of human tissues by RT-PCR, using the oligos described in Supplemental Table T1. The “+” and “−” for each sample indicate with and without reverse transcriptase, respectively. Sample key: bone marrow (BM), cerebellum (CE), whole brain (WB), fetal brain (FB), fetal liver (FL), heart (HE), liver (LI), lung (LU), prostate (PS), salivary gland (SG), skeletal muscle (SM), spleen (SP), testis (TE), thymus (TH), trachea (TR), uterus (UT), colon (CO), small intestine (SI), spinal cord (SC) and stomach (ST). Each is an inverted image of an ethidium bromide stained gel. Molecular weight marker sizes are indicated to the left of each gel image and are in bp.
In order to determine if in addition to this short transcript, DANT1 also transcribes an ATT isoform as the RNA-seq profile suggests, we looked at annotated ESTs within the DANT1 RNA-seq profile on the distal side of DXZ4. We postulated that if those ESTs that overlap the (+)-strand RNA-seq profile are genuinely part of DANT1, then it should be possible to physically link these ESTs to the DANT1 promoter by RT-PCR using a primer anchored in the ESTs and a primer in exon-1 of DANT1. We focused on a cluster of 16 EST entries within a unique 3kb region located 17kb distal to the edge of the main DXZ4 array. This region shares extensive but inverted DNA sequence identity to DXZ4, including the promoter and CTCF site, and is well represented in the DANT1 and DANT2 RNA-seq profiles in hESC (Supplementary Fig. 3a). We performed RT-PCR with a forward primer anchored in exon-1 of DANT1 and a reverse primer anchored in this EST cluster. This RT-PCR confirmed that DANT1 does indeed generate ATT, and identified two alternatively spliced isoforms that contain or do not contain the second annotated exon (Fig. 4a, PCR-2). Therefore, we conclude that DANT1 produces both non-ATT and ATT transcript isoforms.
Validation of DANT2
Next we sought to validate DANT2 and to determine if, like DANT1, it too produces both non-ATT and ATT isoforms. We started by attempting to validate the existence of DANT2-ATT by performing RT-PCR with a forward primer anchored in exon-1 and a reverse primer located in BM925596 exon-2, which is located upstream of the DANT1 promoter (Fig. 2b and 2c). This transcript was readily detected in hESCs, validating the existence of DANT2-ATT (Fig. 4a, PCR-3 and PCR-6). Interestingly, the RT-PCR resulted in several bands. These were TA-cloned and sequenced, revealing the existence of several novel spliced isoforms of DANT2-ATT, including one that spliced into the DXZ4 array itself (Supplementary Fig. 3b, transcript-e).
Notably, with the exception of exon-1, none of the DANT2-ATT isoforms included any of the exons contained in the other ESTs located in the distal interval between the DANT2 promoter and the DXZ4 array (See BX642309, BC003645 and BE297956 in D-EST of Fig. 2a). By RT-PCR, the existence of these spliced DANT2 transcripts were also validated (Fig. 4a, PCR-4 and 5).
Tissue distribution of DANT1 and DANT2 transcripts
So far, these data indicate that both DANT1 and DANT2 generate ATT isoforms, that DANT1 also produces short polyadenylated non-ATT isoforms and that both lncRNAs show highly variable exon inclusion. Furthermore, since males are hemizygous for the X chromosome and both the DANT1 and DANT2 ATTs could be detected in the same male hESC sample, transcription is occurring in both directions across DXZ4. Whether this is occurring simultaneously or mutually exclusively in different cells cannot be determined from these experiments. In addition, some of the spliced isoforms contain exons that are overlapping but anti-sense to one another, such as the example shown in Supplementary Fig. 3a.
Given the heterogeneity in DANT1 and DANT2 transcripts, we extended our analysis to assess their existence in a panel of twenty different human tissues. The short DANT1 transcript that is readily detected in hESCs was of particularly low abundance, and could only be confidently detected in prostate, testis, trachea and spinal cord (Fig. 4b, DANT1-short). In contrast, the ATT form of the DANT1 transcript could not be detected in any tissues (Fig. 4b, DANT1-ATT), and is therefore hESC-specific. The DANT2 hESC validated transcript (Fig. 2a, PCR-4), that includes an exon that is antisense to DANT1-ATT (Supplementary Fig. 3a), was only detected in cerebellum (Fig. 4b, DANT2-short). Finally, DANT2-ATT was particularly abundant in nervous tissue (cerebellum, whole brain, fetal brain and spinal cord), as well as a few other tissues. Therefore, even though qRT-PCR to exon-1 of DANT2 can be detected in all samples examined, the ATT form of DANT2 is not universal, indicating that DANT2 expression in the majority of tissues and cell types is non-ATT. Notably, transcription of DANT2 occurs from the same strand as the most abundant DXZ4 transcript (Chadwick 2008). Therefore, even though each DXZ4 monomer contains a promoter, it is feasible that some proportion of DXZ4 transcript corresponds to unspliced DANT2-ATT. Given that DANT2 and the major DXZ4 transcript are being transcribed in the same orientation, it is also possible that expression of DANT2 and DXZ4 may be associated in some way, with the expression of one influencing the expression of the other. To begin to test this, we determined the expression of DXZ4 and total DANT2 in the 20 different tissues by qRT-PCR. Interestingly, in most tissues, the relative levels of DANT2 correlate well with the relative levels of DXZ4 (Supplementary Fig. 4), a preliminary observation that warrants further investigation.
DANT1 and DANT2 lncRNA localization in hESCs and somatic cells
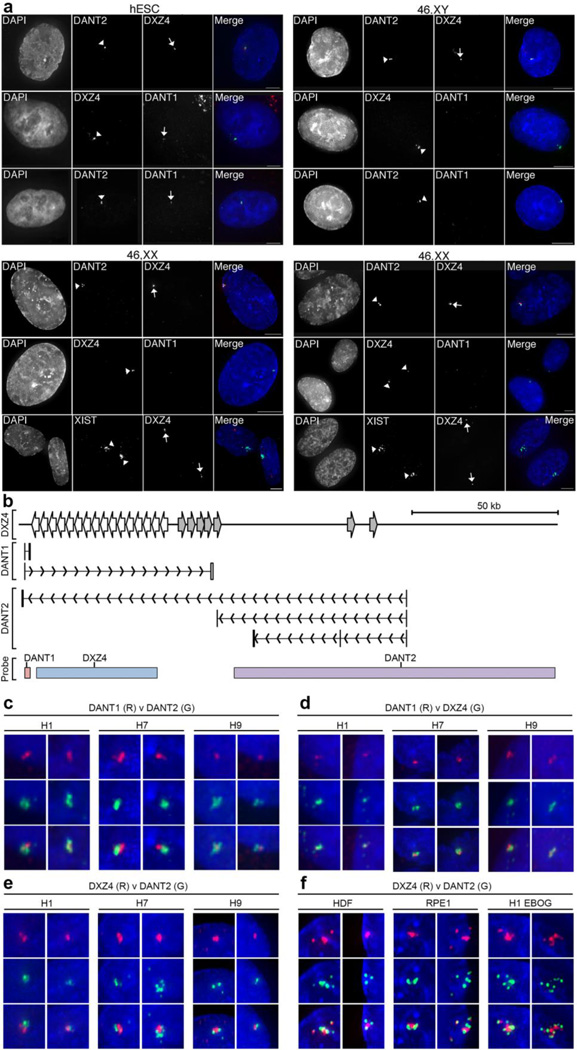
Previously, we have shown that DXZ4 is primarily expressed from the Xa (Chadwick 2008; Tremblay et al. 2011). To expand on these results in hESCs and evaluate the localization and allelic expression of the DANT1 and DANT2 lncRNAs, RNA FISH was performed in hESC and somatic cell lines. As anticipated based on the RT-PCR results, transcripts for DXZ4, DANT2 and DANT1 were readily detected in male (Fig. 5a) and female (Supplementary Fig. 5) hESC, but DANT1 could not be detected in male or female somatic cells (Fig. 5a). In contrast, DANT2 and DXZ4 were obvious in all male and female samples. However, in females, only a single DANT2 and DXZ4 signal was detected, that were always in close proximity, suggesting that they originate from the same chromosome. RNA FISH to DXZ4 and XIST showed the two signals were spatially distinct, consistent with predominant expression of DXZ4 and DANT2 from the Xa (Chadwick 2008; Tremblay et al. 2011). No RNA FISH signals could be detected if samples were first treated with RNaseA (Supplementary Fig. 6a), but the ability to detect DNA by FISH was unchanged (Supplementary Fig. 6b) indicating the specificity of the RNaseA treatment.
Fig. 5. RNA FISH Images showing allele-specific expression and spatial arrangement of DXZ4-associated lncRNAs.
(a) Detection of DANT1DANT2 and DXZ4 by direct-labeled RNA FISH probes in male H1 (hESC) and various male (1140: 46,XY) and female (46,XX: HDF-FET – left panel and RPE1 – right panel) somatic cells. For each sample column 1 shows nuclei counterstained with DAPI (white), column 2 shows labeled signals indicated by arrowheads. Column 3 shows signals highlighted by arrows. Column 4 consists of a merge of the DAPI-staining (blue) with direct-RNA FISH in columns 2 (green) and 3 (red). The white bars at the bottom right of the merged images indicate 5µm. In female somatic cell samples, the location of the Xi is defined by XIST RNA FISH. (b) Schematic map of the interval around DXZ4, indicating the location of short and ATT DANT1 and DANT2 transcriptional units. The location of probes used for RNA FISH is indicated at the bottom. The DANT1 probe is a cloned genomic fragment corresponding to DANT1-short exons 1–3, DXZ4 is BAC clone 2272M5 and DANT2 is BAC clone 761E20. Panels (c–f) show RNA hybridization signals for the direct-labeled FISH probes indicated (DANT1, DANT2 or DXZ4) labeled in red (R) or green (G) merged with DAPI staining of the nucleus (blue). Cell lines include male (H1) and female (H7 and H9) hESCs as well as H1 derived EBOG and female somatic fibroblasts (HDF) or epithelial (RPE1) cells. Overlapping signals appear yellow. Two representative examples are shown for each probe combination used on the various cell types. Each image is approximately 1µm across.
As described above, we validated the existence of ATT versions of DANT1 and DANT2 in hESC. Therefore, at least some nascent DANT1 and DANT2 transcripts overlap with DXZ4. Fig. 5b indicates the location of the three direct-labeled probes that were used in the RNA FISH experiments described above, relative to the different DANT1 transcript isoforms, indicating that it is possible for the probes to be detect more than their intended transcript. Therefore we examined close-up images of the RNA hybridization patterns for the different probe combinations. In three independent hESC cell lines, we consistently found that DANT1 and DANT2 signals were physically close to one another, but rarely overlapped (Fig. 5c). Likewise, DANT1 (Fig. 5d) and DANT2 signals (Fig. 5e) were physically close to DXZ4 transcripts, but with limited signal overlap. We extended this analysis to examine the spatial distribution of DANT2 and DXZ4 transcripts in somatic cells. DANT2-ATT is restricted to hESCs and a limited number of tissues (Figure 4) and cannot be detected in the cells examined (data not shown), thus we expect to only detect the expressed DANT2-short non-ATT isoform. Similar to what was observed in hESC, the DXZ4 and DANT2 signals were physically close with limited overlap (Fig. 5f). Therefore, even though the transcriptional units of the DANT1, DANT2 and DXZ4 loci overlap, their transcripts appear to be clustered and highly localized, but predominantly spatially separate.
Differentiation associated chromatin change at the DANT1 promoter
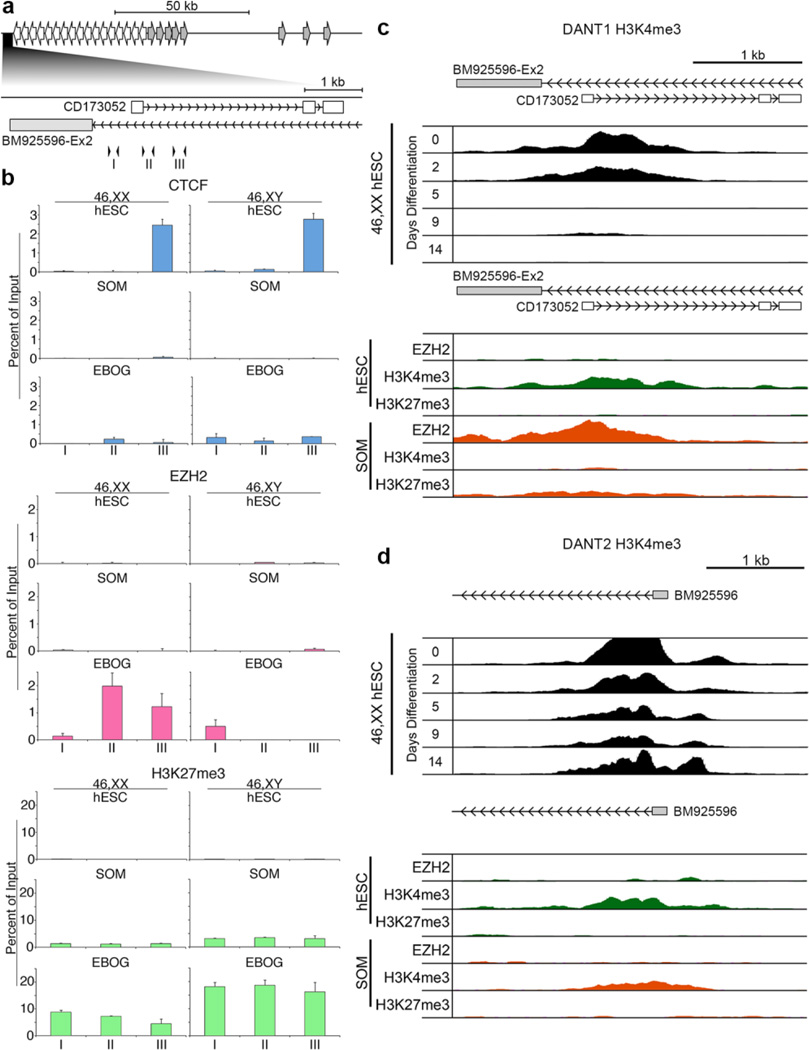
DANT1 expression is shut down as DXZ4 gains H3K9me3 and loses 5-hmC. Therefore, it is conceivable that the DANT1 lncRNA is linked to changes in DXZ4 chromatin. We sought to evaluate differences in chromatin at the DANT1 promoter in male and female hESC, their EBOG derivatives and somatic cells by qChIP using three sets of primers clustered around exon-1 (Fig. 6a). We found elevated levels of CTCF downstream of exon-1 in hESC that is largely absent in both EBOG and somatic cells (Fig. 6b). Given that DANT1 expression is lost upon differentiation, we reasoned this would be accompanied by acquisition of heterochromatin. Low levels of H3K9me3 can be observed at the DANT1 promoter in hESC that are similar to those seen in somatic cells, suggesting that silencing is unlikely to be mediated by H3K9me3, even though levels are slightly higher in EBOG (Supplementary Fig. 7). However, the Polycomb repressive complex 2 (PRC2) is a common form of differentiation associated gene silencing, characterized by a gain of the repression associated modification H3K27me3 catalyzed by the PRC2 complex subunit Enhancer of Zeste 2 (EZH2) [Reviewed in (Cao and Zhang 2004)]. We found that EZH2 is recruited to the DANT1 promoter in male and female EBOG and coincided with a gain in H3K27me3 (Fig. 6b). H3K27me3 was also a feature at the DANT1 promoter in male and female somatic cells, albeit at a lower level than that observed in EBOG (Fig. 6b). However, EZH2 was not obvious at DANT1 in somatic cells despite the presence of H3K27me3, the explanation for which is not immediately apparent. One potential interpretation of these data is that EZH2 association at the DANT1 promoter is transient in somatic cells, and therefore less readily detected.
Fig. 6. Chromatin organization and dynamics around the DANT1 and DANT2 promoters.
(a) Schematic map of the DXZ4 interval indicating the relative location of the DANT1 promoter. Immediately below this, the DANT1 region is expanded, corresponding to 114,956,035–114,959,533 bp of chromosome X (hg19) and the location of the short DANT1 EST (CD173052) and DANT2 ATT exon (BM925596-Ex2) are indicated. Inverted arrowheads correspond to the location of qChIP primer sets I (F1.R1), II (F2.R2) and III (F3.R3). (b) qChIP data for CTCF, EZH2, and H3K27me3. Each is graphed as a percent of input and is the mean of triplicate qPCR reactions. Error bars indicate standard deviation. Female (46,XX) data sets are on the left with male (46,XY) on the right. For each ChIP target, the top row shows data for female (H9) and male (H1) hESCs, middle row for female (RPE1) and male (BJ1) somatic cells and bottom row for EBOGs derived from the corresponding hESCs. (c) A map showing the location of DANT1 short EST (CD173052) and DANT2 ATT (BM925596-Ex2) corresponding to 114,956,077–114,959,451 bp of the X chromosome (hg19) is shown at the top. Immediately beneath this is publicly available H3K4me3 ChIP-seq data for H7 hESCs (46,XX hESC) prior to (day-0) and at various days post-differentiation (days 2–14) (Consortium et al. 2012). Below this is publicly available ChIP-seq data from the same interval for EZH2, H3K4me3 and H3K27me3 in male H1 (hESC) and male HUVEC somatic (SOM) cells (Consortium et al. 2012). (d) As in part-(c) above, but for DANT2 and interval 115,083,137–115,086,987 of the X chromosome (hg19).
Consistent with these observations, publicly available H7 hESC Chip-seq data for histone H3 tri-methylated at lysine 4 (H3K4me3), a marker for actively transcribed promoters (Santos-Rosa et al. 2002; Schneider et al. 2004), shows a broad peak around exon-1 of DANT1 (Consortium et al. 2012). This signal diminishes over a 2-week period as the cells are differentiated (Fig. 6c, top data set). Furthermore, this peak is specific to hESC, while EZH2 and H3K27me3 are restricted to differentiated cells (Fig. 6c, bottom data set). In contrast, and consistent with its continued expression, H3K4me3 at the DANT2 promoter is not lost upon hESC differentiation (Fig. 6d, top data set), although the signal does weaken, which may reflect the fact that the Xi allele is silenced upon differentiation (Fig. 5) reducing the signal by 50%. Furthermore, H3K4me3, but not EZH2 or H3K27me3, is a feature of the DANT2 promoter in hESC and somatic cells (Fig. 6d) consistent with the ubiquitous expression of this lncRNA.
Discussion
The organization of chromatin at MSRs in hESC is unknown. Here we describe our analysis of the X-linked MSR DXZ4, and report the identification of two novel lncRNAs. We found low levels of the euchromatic marker H3K4me2, and the absence of the repressive histone modification H3K9me3 characterizes DXZ4 in male hESC. Additionally, we observed low levels of 5-hmC intermingled with high levels of 5-mC. In mESC and hESC, 5-hmC is mostly associated with euchromatin and is primarily a feature of active promoters, enhancers, and gene bodies (Ficz et al. 2011; Stroud et al. 2011; Szulwach et al. 2011a). The levels of 5-hmC that we detected are consistent with those reported by single-base resolution analysis for transcription start sites (TSS) (Booth et al. 2012; Yu et al. 2012), as is its coexistence with regions of 5-mC enrichment (Booth et al. 2012; Yu et al. 2012). Given that the interval analyzed covers the internal DXZ4 promoter (Chadwick 2008), and that DXZ4 transcription can be detected in hESCs, the 5-hmC we observed may reflect proximity to the DXZ4 TSS. Alternatively, the low levels of 5-hmC and high levels of 5-mC are also in agreement with levels reported at intragenic sites (Yu et al. 2012), which is consistent with the fact that in hESC DXZ4 is completely contained within the transcriptional units of the DANT1 and DANT2 lncRNAs.
Differentiation of hESC is results in the loss of 5-hmC at DXZ4, accompanied by a decrease in H3K4me2 and a distinct gain of H3K9me3. These data indicate that acquisition of the constitutive heterochromatin state at DXZ4, seen at the MSR in males and at the female Xa allele in somatic cells (Chadwick 2008), is developmentally regulated. Intriguingly, change to DXZ4 chromatin is accompanied by alterations in the transcription of two novel flanking lncRNAs. The first lncRNA, DANT1, originates immediately proximal to DXZ4, and consists of alternatively spliced polyadenylated and non-polyadenylated transcripts that terminate either prior to entering the MSR (DANT1-short) or on the opposite side of the array (DANT1-ATT). While very low levels of DANT1-short can be detected in some tissues, high levels of this isoform and expression of DANT1-ATT are unique to hESC. Consistent with these data, the silencing of DANT1 is accompanied by recruitment of EZH2 to the promoter in differentiating hESC and deposition of the repressive histone modification H3K27me3 (Cao et al. 2002; Plath et al. 2003).
The second lncRNA, DANT2, originates 80 kb distal to DXZ4 and is primarily non-polyadenylated. Like DANT1, DANT2 is abundant in hESC, but the activity of its promoter is not developmentally regulated as DANT2 continues to be transcribed in a wide variety of somatic cell and tissue types. However, while differentiation does not appear to alter promoter activity, isoforms of the transcript are impacted. DANT2-ATTs splice to an exon immediately upstream of the DANT1 promoter and are readily detected in hESC, but not in EBOG (data not shown) or most tissues examined. The most notable exception is nervous tissue where the DANT2-ATT is readily detected. Given that constitutive heterochromatin is acquired at DXZ4 on differentiation, it is conceivable that this blocks DANT2 transcription across DXZ4 in those cell types that lack this transcript. Therefore, the detection of DANT2-ATT in some somatic cell types suggests that perhaps the chromatin state of DXZ4 differs in these tissues. Given that 5-hmC is more abundant in the central nervous system (Globisch et al. 2010), 5-hmC may be a characteristic of DXZ4 in nervous tissue. Therefore, the chromatin of the MSR may more closely resemble what was observed in hESC and reflect a role for DANT2 and DXZ4 in these tissues.
Interestingly, analysis of spliced isoforms of DANT1 and DANT2 revealed significant heterogeneity in exon content. With exception of exon-1 of both transcripts and exon-3 of the DANT1-short isoforms, few exons are common to the different DANT1 and DANT2 isoforms detected. Perhaps this reflects opportunistic splicing, whereby inclusion of an exon occurs if a suitable splice donor and acceptor are present in the primary transcript, while inconsistent inclusion would reflect poor matches with other sequence elements involved in splicing, reducing the overall frequency of retention in the various isoforms. The variable splicing may also suggest that the act of transcription at this locus is potentially more important than the RNA product itself.
Pair-wise alignment of the genomic interval around DXZ4 revealed that in addition to the main homogenous tandem repeat, many regions distal to the MSR share high sequence identity to DXZ4, but are inverted relative to the array. Despite differences in their transcription, the inverted promoters of DANT1 and DANT2 also share high sequence identity, suggesting that they are derived from a common ancestral sequence. Presumably, the ubiquitous expression of DANT2 and the hESC-specific transcription of DANT1 is encoded where the two promoters differ. The arrangement of promoters flanking DXZ4 differs substantially from what we have previously reported for the mouse homolog of the MSR (Horakova et al. 2012a). Mouse Dxz4 is also characterized by an uninterrupted tandem repeat, but the MSR is much smaller than its human homolog due to the inclusion of fewer repeat units. Although both Dxz4 and primate DXZ4 are expressed (Chadwick 2008; McLaughlin and Chadwick 2011; Tremblay et al. 2011), Dxz4 lacks an obvious promoter element within the repeat units. Instead, a single promoter is located distal to the array driving an ATT that terminates on the proximal side of the mouse MSR (Horakova et al. 2012a). Like DANT2, this transcript splices into an exon contained within a Dxz4 monomer and similar to both DANT1 and DANT2, various Dxz4 isoforms can be detected. Potentially, the massive expansion of the MSR in primates involved a duplication and inversion event after the diversion of the rodent and primate lineages, and as the DANT1/2 promoter sequences diverged, DANT1 acquired ES-specificity.
The transcriptional unit of the DANT1-ATT is entirely contained within the transcriptional unit of the DANT2-ATT, and DXZ4 is embedded within both. Due to the orientation of the promoters, transcription of DANT1 and DANT2 converge. As transcript for both can be detected at the same allele, the possibility of double stranded RNA (dsRNA) exists in hESCs. However, RNA FISH indicates that, while physically close, the transcripts are spatially distinct. Furthermore, while dsRNA is a conserved trigger of heterochromatin formation (Castel and Martienssen 2013), here constitutive heterochromatin formation at DXZ4 is coupled with the apparent loss of bi-directional transcription across the MSR, suggesting that dsRNA may not play a role.
In conclusion, similar to the FSHD-associated MSR (Cabianca et al. 2012), we have identified two novel lncRNAs associated with DXZ4, therefore lncRNAs may be common to MSRs in general. Given that the D4Z4 associated lncRNA impacts the chromatin state of the FSHD MSR, it is conceivable that DANT1 and/or DANT2 play some role in modulating the developmental regulation of chromatin at DXZ4. Silencing of DANT1 by PRC2 is differentiation dependent and coincides with a gain of constitutive heterochromatin at DXZ4. Therefore, DANT1 is a suitable candidate to determine if this lncRNA is mechanistically linked to the state of chromatin at the MSR. Since males are hemizygous for DANT1/DXZ4 and differentiation of male hESC results in gain of constitutive heterochromatin at the MSR, these are a suitable model cell type in which to investigate the underlying molecular mechanisms involved in assembly of H3K9me3 chromatin at large tandem repeat DNA, and the potential role of lncRNA in heterochromatin formation.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [GM073120 to B.P.C.]; and by a subaward from P01GM085354 to Dr. Stephen Dalton, University of Georgia.
Abbreviations
- 5-hmC
5’-hydoxymethylcytosine
- 5-mC
5’-methylcytosine
- ATCC
American type culture collection
- ATT
Array-traversing transcript
- BiS
Bisulfite sequencing
- CGI
CpG island
- ChIP
Chromatin immunoprecipitation
- CTCF
CCCTC-binding factor
- DANT1
DXZ4 Associated Non-coding Transcript, proximal
- DANT2
DXZ4 Associated Non-coding Transcript, distal
- dsRNA
Double stranded RNA
- EBOGs
Embryoid body out-growths
- EZH2
Enhancer of Zeste 2
- ESTs
Expressed sequence tags
- FSHD
Facioscapulohumeral muscular dystrophy
- H3K4me2
Histone H3 di-methylated at lysine 4
- H3K9me3
Histone H3 tri-methyated at lysine 9
- hESCs
Human embryonic stem cells
- HGNC
Human gene nomenclature committee
- kb
Kilobases
- LINE
Long interspersed elements
- lncRNA
Long non-coding RNA
- LTR
Long terminal repeats
- mRNA
Messenger RNA
- MSR
Macrosatellite repeat
- ORF
Open reading frame
- oxBiS
Oxidative bisulfite sequencing
- PBS
Phosphate buffered saline
- PRC2
Polycomb repressive complex 2
- qChIP
Quantitative chromatin immunoprecipitation
- qPCR
Quantitative PCR
- RACE
Rapid amplification of cDNA ends
- RT-PCR
Reverse transcription PCR
- SR
Simple repeat
- SSC
Saline-Sodium Citrate
- TSS
Transcription start sites
- Xa
Active X chromosome
- XCI
X chromosome inactivation
- Xi
Inactive X chromosome
- XIST
X inactive specific transcript
Footnotes
Ethical Standards
Experiments performed in this manuscript comply with the current laws of the USA.
Conflict of Interest
The author Debbie M. Figueroa declares they have no conflict of interest.
The author Emily M. Darrow declares they have no conflict of interest.
The author Brian P. Chadwick declares they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Awaya T, Kato T, Mizuno Y, et al. Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS One. 2012;7(12):e51638. doi: 10.1371/journal.pone.0051638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, et al. A Long ncRNA Links Copy Number Variation to a Polycomb/Trithorax Epigenetic Switch in FSHD Muscular Dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Wagner JR. TET enzymatic oxidation of 5-methylcytosine, 5-hydroxymethylcytosine and 5-formylcytosine. Mutation research Genetic toxicology and environmental mutagenesis. 2014;764–765:18–35. doi: 10.1016/j.mrgentox.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14(2):100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18(8):1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J Cell Biol. 2002;157(7):1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Barring gene expression after XIST: maintaining facultative heterochromatin on the inactive X. Semin Cell Dev Biol. 2003a;14:359–367. doi: 10.1016/j.semcdb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003b;12(17):2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- Cheng YH, Wong EW, Cheng CY. Cancer/testis (CT) antigens, carcinogenesis and spermatogenesis. Spermatogenesis. 2011;1(3):209–220. doi: 10.4161/spmg.1.3.17990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S, Pagliuca FW, Honore C, Kweudjeu A, Rezania A, Melton DA. A simple tool to improve pluripotent stem cell differentiation. Nat Methods. 2013;10(6):553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra P, Papale LA, White AT, et al. Array-based assay detects genome-wide 5-mC and 5-hmC in the brains of humans, non-human primates, and mice. BMC Genomics. 2014;15:131. doi: 10.1186/1471-2164-15-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deidda G, Cacurri S, Grisanti P, Vigneti E, Piazzo N, Felicetti L. Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genet. 1995;3(3):155–167. doi: 10.1159/000472291. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Noncoding RNAs and Epigenetic Mechanisms During X-Chromosome Inactivation. Annual review of cell and developmental biology. 2014 doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, et al. DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev Cell. 2012;22:1–14. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacalone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet. 1992;1(2):137–143. doi: 10.1038/ng0592-137. [DOI] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5(12):e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struc & Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Alonso A, Cheung F, et al. Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma. 2011;120(6):621–632. doi: 10.1007/s00412-011-0337-6. [DOI] [PubMed] [Google Scholar]
- Horakova AH, Calabrese JM, McLaughlin CR, Tremblay DC, Magnuson T, Chadwick BP. The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 2012a;13(8):R70. doi: 10.1186/gb-2012-13-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet. 2012b doi: 10.1093/hmg/dds270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev. 2012;22(2):62–71. doi: 10.1016/j.gde.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32(2):235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, et al. A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing D, Anguera MC, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitiatve PCR and the 2-ΔΔCT method. Methods. 2001 Dec;25(4):402–408. doi: 10.1006/meth.2001.1262. 2001, ISSN 1046-2023. [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5(12):1743–1753. [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation and human genetic disease. Acta Paediatr Suppl. 2002;91(439):107–112. doi: 10.1111/j.1651-2227.2002.tb03120.x. [DOI] [PubMed] [Google Scholar]
- Mahmood A, Harkness L, Schroder HD, Abdallah BM, Kassem M. Enhanced differentiation of human embryonic stem cells to mesenchymal progenitors by inhibition of TGF-beta/activin/nodal signaling using SB-431542. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25(6):1216–1233. doi: 10.1002/jbmr.34. [DOI] [PubMed] [Google Scholar]
- McLaughlin CR, Chadwick BP. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biology. 2011;12:R37. doi: 10.1186/gb-2011-12-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkovsky A, Patel S, Plath K. Concise review: Pluripotency and the transcriptional inactivation of the female Mammalian X chromosome. Stem Cells. 2012;30(1):48–54. doi: 10.1002/stem.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley SC, Rizkallah R, Tremblay DC, Anderson BR, Hurt MM, Chadwick BP. YY1 associates with the macrosatellite DXZ4 on the inactive X chromosome and binds with CTCF to a hypomethylated form in some male carcinomas. Nucleic Acids Res. 2012;40(4):1596–1608. doi: 10.1093/nar/gkr964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Gondo Y, Goto J, Kanazawa I, Hadano S, Ikeda JE. Unstable transmission of the RS447 human megasatellite tandem repetitive sequence that contains the USP17 deubiquitinating enzyme gene. Hum Genet. 2002;110(4):302–313. doi: 10.1007/s00439-002-0698-2. [DOI] [PubMed] [Google Scholar]
- Ottaviani A, Rival-Gervier S, Boussouar A, et al. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facioscapulohumeral dystrophy. PLoS Genet. 2009;5(2):e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37(18):e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pohlers M, Calabrese JM, Magnuson T. Small RNA Expression from the Human Macrosatellite DXZ4. 2014:G3. doi: 10.1534/g3.114.012260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1–16. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schaap M, Lemmers RJ, Maassen R, et al. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations: evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics. 2013;14:143. doi: 10.1186/1471-2164-14-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7(6):e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011b;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller K, Illner D, Thamm S, et al. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at >/= 20 Mb genomic length scales. Nucleus. 2011;2(5):465–477. doi: 10.4161/nucl.2.5.17862. [DOI] [PubMed] [Google Scholar]
- Tremblay DC, Alexander G, Jr, Moseley S, Chadwick BP. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics. 2010;11:632. doi: 10.1186/1471-2164-11-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay DC, Moseley S, Chadwick BP. Variation in Array Size, Monomer Composition and Expression of the Macrosatellite DXZ4. PLoS ONE. 2011;6(4):e18969. doi: 10.1371/journal.pone.0018969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2(12):2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, et al. Hypomethylation of D4Z4 in 4q–linked and non-4q–linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35(4):315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Hasson D, Guillem F, Lescale C, Jin X, Abrusan G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 2008;9:533. doi: 10.1186/1471-2164-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2(1):26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Yang F, Deng X, Berletch JB, et al. The lncRNA Firre anchors the inactive X chromsoome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Gen Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5(7):e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.