Abstract
The anticancer properties of cruciferous vegetables are well known and attributed to an abundance of isothiocyanates (ITCs) such as benzyl ITC (BITC) and phenethyl ITC (PEITC). While many potential targets of ITCs have been proposed, a full understanding of the mechanisms underlying their anticancer activity has remained elusive. Here we report that BITC and PEITC effectively inhibit deubiquitinating enzymes (DUBs), including the enzymes USP9x and UCH37, which are associated with tumorigenesis, at physiologically relevant concentrations and time scales. USP9x protects the anti-apoptotic protein Mcl-1 from degradation, and cells dependent on Mcl-1 were especially sensitive to BITC and PEITC. These ITCs increased Mcl-1 ubiquitination and either ITC treatment or RNAi-mediated silencing of USP9x decreased Mcl-1 levels, consistent with the notion that USP9x is a primary target of ITC activity. These ITCs also increased ubiquitination of the oncogenic fusion protein Bcr-Abl, resulting in degradation under low ITC concentrations and aggregation under high ITC concentrations. USP9x inhibition paralleled the decrease in Bcr-Abl levels induced by ITC treatment, and USP9x silencing was sufficient to decrease Bcr-Abl levels, further suggesting that Bcr-Abl is a USP9x substrate. Overall, our findings suggest that USP9x targeting is critical to the mechanism underpinning the well established anticancer activity of ITC. We propose that the ITC-induced inhibition of DUB may also explain how ITCs affect inflammatory and DNA repair processes, thus offering a unifying theme in understanding the function and useful application of ITCs to treat cancer as well as a variety of other pathological conditions.
Keywords: ITC, DUB, PEITC, BITC, cruciferous, ubiquitination, deubiquitinase, USP9x, UCH37, chemotherapeutic
Introduction
The dietary consumption of cabbage, broccoli and other cruciferous vegetables is associated with a decreased risk of cancer (1–3). The chemoprotective properties of these vegetables are attributed to isothiocyanates (ITCs) such as benzyl ITC (BITC), phenethyl ITC (PEITC) and sulforaphane (SFN) (Fig. 1A) (1,2). Plasma concentrations of ITCs can reach 0.25 μM from a single serving of broccoli, and intracellular concentrations can be 200-fold higher due to concentrative processes (3,4). Numerous studies demonstrate that these compounds have antiproliferative activity against tumors in both cell culture and animal models (2,3) and PEITC has entered clinical trials for lung and oral cancers(3). ITCs induce apoptosis in many cancer cell lines and exposure to BITC or PEITC for only 3 h inhibits cell growth with EC50 values of 1.8–17 μM (5). SFN also inhibits growth under these conditions, though the values of EC50 are typically much higher (50 μM). ITCs perturb many cellular processes, including DNA repair(3,6). autophagy (2), the inflammatory response (1) and the antioxidant response (1,2). ITCs also modulate the activity of several oncogenic proteins. For example, both PEITC and BITC reduce the levels of the anti-apoptotic protein Mcl-1 in leukemia cells (7–9) and PEITC induces the knockdown of Bcr-Abl kinase, the oncogenic fusion protein that causes chronic myeloid leukemia (10).
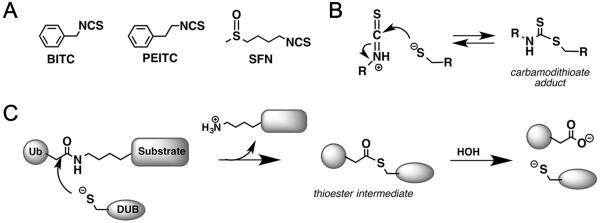
Figure 1. Proposed mechanism of DUB Inhibition.
A, Structures of naturally occurring ITCs. B, Proposed mechanism of ITC inhibition. C, Mechanism of DUB catalysis.
The molecular mechanisms underlying the anticancer properties of ITCs are under debate (1). ITCs are electrophiles that form reversible adducts with small molecule thiols such as glutathione (Fig. 1B) (3). Amines can form stable adducts with ITCs, although this reaction is not facile at neutral pH. Depletion of glutathione, and resulting generation of reactive oxygen species (ROS), is an appealing mechanism for the anticancer activities of ITCs (11). However, L-butathionine sulfoximine depletes glutathione and induces ROS to greater extents than PEITC, yet does not induce apoptosis (12). This finding discredits glutathione depletion/ROS production as the mechanism of anticancer activity. ITCs can also modify proteins at thiol and amine residues. At least thirty proteins have been reported to be potential ITC targets, including P450s, glutathione reductase, thioredoxin reductase, mutant p53, migration inhibitory factor, protein phosphatases and tubulin (1), but the functional consequences of ITC modification are usually unknown and the correlation with cellular phenotypes uncertain. Moreover, the reversible nature of ITC-thiol adducts suggests that Cys-modified proteins were unlikely to be identified in previous experiments. Therefore the wide array of potential ITC targets does not satisfactorily explain the pleiotropic cellular effects of ITCs.
Catalytic cysteine residues are generally very nucleophillic and react readily with electrophiles, so enzymes with catalytic cysteine residues are good candidates for ITC targets. Cysteine proteases are particularly attractive candidates for ITC inhibition because the thiol adduct resembles the thioester intermediate of peptide hydrolysis (Fig. 1B and Fig. 1C). The C=S bond is longer and more electronegative than C=O, and thus may resemble the transition state for peptide hydrolysis, potentially providing additional binding energy. ITCs are weak reversible inhibitors of papain (13), the prototypical cysteine protease from papaya, but the effects of ITCs on other cysteine proteases have not been investigated. Papain is distantly related to deubiquitinating enzymes (DUBs), the hydrolases that remove ubiquitin from target proteins and disassemble ubiquitin chains. DUBs regulate many important physiological processes, including protein degradation, DNA repair, autophagy and protein trafficking (6,14) and are potential targets for the treatment of many diseases, including cancer(15), neurodegeneration, inflammation and infection (14). We recognized that many of the phenotypes associated with dietary ITCs are also observed when cells are treated with DUB inhibitors. Therefore we hypothesized that DUBs might be targets of ITCs.
Here we report that both BITC and PEITC inhibit USP9X and UCH37 and other DUBs at physiologically relevant concentrations and time scales. DUB inhibition provides a molecular mechanism for the anticancer properties of dietary ITCs.
Materials and Methods
Detailed Methods are included in the Supporting Material.
Materials
All chemicals and reagents were from Sigma Aldrich unless otherwise stated. Solvents (except DMSO) were from Fisher (Pittsburg, PA). Other reagents used in this study: G5 isopeptidase inhibitor 1 (50-230-7928, Calbiochem); PEITC (Acros Organics); Bortezomib (Millennium Pharmaceuticals); Mini-Complete and PhosSTOP inhibitory cocktails (Roche Applied Science); bortezomib (LC laboratories); Alamar Blue® (Invitrogen); USP9x, USP7(catalytic domain), UCH-L3, Ubiquitin-AMC, Suc-Leu-Leu-Val-Tyr-AMC, RAP80 UIM Domains Agarose AM-120, and 20S human proteasome (Boston Biochem); normal goat IgG SC-2028 (Santa Cruz); TAMRA-ubiquitin propargylamide and Cy5-ubiquitin vinyl methyl ester (UbiQ); HA-ubiquitin vinylsulfone and HA-ubiquitin vinyl methyl ester were synthesized using standard methods previously described(16). The plasmid encoding the HA-1–75Ub-Intein-chitin binding domain fusion protein was a gift from Prof H. Ploegh (Whitehead Institute). BaF3 and BaF3/p210 cells were provided by Nathaniel Gray (Harvard University)
Antibodies
The following antibodies were used: anti-K48-linked ubiquitin, clone APU2 and anti-K63-linked ubiquitin, clone APU3 (Millipore); anti-PARP 9542, anti-cAbl 2862, anti-α-tubulin 2156, anti-Mcl-1 D35A5, anti-Flag 2368, (Cell Signaling Technologies); anti-actin clone AC-40 A3853, anti-GAPDH clone G9295, and anti-HA, Clone 3F10 (Roche); anti-ubiquitin, clone 6C1.17 (BD Pharmingen); anti-HSP70, anti-USP7, anti-UCH37, anti-USP24 and anti-USP9x (all rabbit monoclonal) (Abcam); HRP conjugated secondary antibodies (Abcam)
Tissue culture assays and preparation of cell lysates
B16/F10 and MCF7 cells were purchased from ATCC. BaF3 and BaF3/p210 cells were provided by Dr. Nathaniel Gray (Harvard Medical School, Boston, MA; obtained 2013), K562 cells were provided by Jeffrey Strovel (Avalon Pharmaceuticals; obtained in 2011), HeLa cells were provided by Benjamin F. Cravatt (The Scripps Research Institute; obtained in 2010), NIH/3T3 cells were provided by Dr. Rubio Ren (Brandeis University; obtained in 2012) and COS1 were provided by the Dr. Daniel Oprian (Brandeis University; obtained 2010). Cell lines were authenticated (9-Marker STR May 2015). The genetic profiles of K562, MCF-7, HeLa, BaF3, BaF3/p210 and NIH/3T3 cells were identical to reported genetic profiles. COS1 cells were confirmed to be of African green monkey in origin and free of all interspecies contamination.
Cells were cultured in DMEM (HeLa, COS1, NIH/3T3, B16-F10, and MCF-7) or RPMI (BaF3, BaF3/p210 and K562) supplemented with 10% heat inactivated FBS (DBS was used for NIH/3T3 cells), 1X glutaMAX, and 1% penicillin/streptomycin at 37 °C in a 5% CO2 humidified atmosphere. BaF3 cells were also supplemented with 1 ng/mL recombinant mouse interleukin-3 (rmIL-3, R&D Systems).
For lysate preparation, non-adherent cells were harvested by centrifugation, resuspended in 10 mM HEPES (pH 7.9), 5 mM MgCl2, 140 mM KCl, 1% NP40, protease and Phosphatase Inhibitor Cocktail II, lysed using 3 X freeze thaw cycles and clarified by centrifugation. Whole cell lysates were prepared by adding 0.1% SDS to the cells together with supernatant followed by sonication. Protein was analyzed by Western blot (protein, antibody dilution): K48-linked ubiquitin (6 μg, 1:9,000 antibody dilution; 30-40 μg, 1:20,000), PARP (30-40 μg, 1:1500), K63-linked ubiquitin (30-40 μg, 1:1,500), Mcl-1 (30-40 μg, 1:1,000), FLAG (30-40 μg, 1:6,000), cAbl (30-40 μg, 1:1,000), ubiquitin (10–20 μg, 1:14,000). Signals were normalized to actin (1:10,000 for 6 μg lysate; 1:30,000 for 30-40 μg lysate), α-tubulin (1:8,000) or GAPDH (1:35,000).
UbiquitinG76V-GFP assay
COS1 cells were transfected with an expression plasmid for ubiquitinG76V-GFP (plasmid 11941 from Addgene, from the laboratory of Nico Dantuma) using Mirus 2020 (Madison, WI). FACS was carried out on a Beckman FACS-Calibur. All data were analyzed using FlowJo V10, from TreeStar (Ashland, OR).
20S Proteasome assay
Proteasome activity was measured by monitoring the hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC in 50 mM potassium phosphate pH 7.6, 50 mM NaCl, 1 mM DTT at 25 °C. The K0.5 value for human 20S proteasome was experimentally determined to be 12 ± 2 μM (Hill coefficient = 2), in good agreement with literature values (17). The apparent K0.5 of proteasome activity of rabbit reticulocyte lysate was 28 ± 3 μM (Hill coefficient = 1.3).
Activity profiling of reactive cysteines
HeLa cell lysates were treated with either DMSO or PEITC (20 μM), followed by 100 μM of IA-alkyne. Click chemistry and peptide analysis was performed as described previously (18,19).
DUB activity profiling
Cell lysate was treated with ITC or 1% DMSO control, then treated with HA-UbVS, HA-UbVME, Cy5-Ub-VME or TAMRA-Ub-PA. Aliquots were removed and immediately quenched in 2X dithiothreitol loading buffer and frozen until analyzed. BaF3/p210 cells were treated with ITC or 0.1% DMSO control, harvested and washed two times with ice cold PBS. Cell pellets were then lysed with glass beads in ice cold 75 mM K2HPO4, pH 7.5, 150 mM NaCl and 250 mM sucrose. The clarified supernatant was incubated with Cy5-UbVME (250 nM) for 5 min at 37 °C. Aliquots were quenched and treated as above. Western blot analysis was carried out using standard methods. In-gel fluorescent scans were obtained using a GE Typhoon scanner.
Recombinant DUB assays
The hydrolysis of Ub-AMC was measured by monitoring the production of AMC at 37 °C in a black 96 well plate. Assay buffer contained 50 mM HEPES, pH 7.6, 100 mM NaCl, 0.75 mM BME. The fluorescence intensities were quantified with the appropriate AMC standard curves.
Proliferation Assays
Measured using the Alamar Blue method as described in the Supplementary Methods.
Immunoprecipitations
For Mcl-1 immunoprecipitations, COS1 cells were transiently transfected with an expression plasmid for 3XFLAG-tagged mouse Mcl-1 (Addgene plasmid 32978, from the lab of Joseph Opferman) using Mirus 2020 (Madison, WI). 3×Flag–Mcl-1 was immunoprecipitated with anti-Flag M2 magnetic beads and eluted with 500 μg/mL 3×Flag peptide.
For the isolation of polyubiquitinated proteins, BaF3/p210 cells were incubated with 5 μM BITC or PEITC or with 0.1% DMSO for 1 h at 37 °C. K562 cells were incubated with 5 μM BITC or PEITC or with 0.1% DMSO for 2 h at 37 °C. Poly-K63-linked ubiquitinated proteins were enriched with RAP80-UIM agarose (25 μL re-suspended slurry). For Bcr-Abl immunoprecipitation, the PEITC, BITC (5 μM) or DMSO treated cell lysates were adjusted to the same protein concentration (1 mg/mL) and pre-cleared with protein G (1 h at 4 °C). The cleared lysate was incubated with anti-cAbl (1 μL/100 μL lysate) overnight at 4°C. cAbl was immunoprecipitated with protein G beads.
Cell transfection and RNA interference
BaF3/p210 and K562 cells were transfected with siRNAs using the Amaxa Nucleofector II (Amaxa, Gaithersburg, MD). NIH/3T3/p210 cells were transfected using Dharmafect 1 (GE Dharmacon, Lafayette, CO). Pre-designed ON-TARGET plus siRNA pools (non-targeting and targeting USP9x) were obtained from Dharmacon. The following siRNA pools (Ms or Hu, Target sequence) were used:
Ms si-USP9x no. 09, 5′-CAGCAAAACUGUUCGUCAA-3′;
Ms si-USP9x no. 10, 5′-GGGCUAACGAUCUCAUUUA-3′;
Ms si-USP9x no. 11, 5′-GCUAAUGUGUAAAUGGCAA-3′;
Ms si-USP9x no. 12, 5′-GAUGAGGCUUCAAGAUAUA-3′;
Hu si-USP9x no. 06, 5′-AGAAAUCGCUGGUAUAAAU-3′;
Hu si-USP9x no. 07, 5′-ACACGAUGCUUUAGAAUUU-3′;
Hu si-USP9x no. 08, 5′-GUACGACGAUGUAUUCUCA-3′;
Hu si-USP9x no. 09, 5′-GAAAUAACUUCCUACCGAA-3′;
si-non-targeting no. 1, 5'-UGGUUUACAUGUCGACUAA-3'
si-non-targeting no. 2, 5'-UGGUUUACAUGUUGUGUGA-3'
si-non-targeting no. 3, 5'-UGGUUUACAUGUUUUCUGA-3'
si-non-targeting no. 4, 5'-UGGUUUACAUGUUUUCCUA-3'
Retroviral transduction
MSCV-p210-IRES-GFP vector obtained from the Ren laboratory (Brandeis University, Waltham, MA (20)) was used to produce retroviral pseudo-virus. NIH/3T3 cells were were transduced with virus GFP expressing NIH/3T3 cells were collected on a FACS Aria Flow Cytometer.
Results
ITCs increase the high molecular weight ubiquitin pool
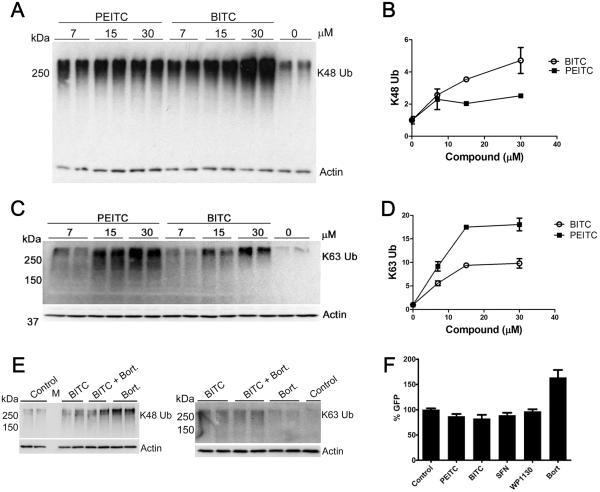
DUB inhibition can be revealed by the accumulation of high molecular weight ubiquitinated proteins (HMW-Ub). Inhibitors of the proteasome and P97 can also cause the accumulation of K48-linked ubiquitination, but as yet only DUB inhibitors are known to cause the accumulation of K63-linked ubiquitination. We examined the effects of BITC and PEITC on the HMW-Ub in the pro-B cell line BaF3/p210, which expresses Bcr-Abl kinase. Both BITC and PEITC caused the accumulation of K48-linked HMW-Ub (Fig. 2A and Fig. 2B), while SFN had no effect. Increases in K48-linked Ub could be observed within 4 h at 7 μM BITC or PEITC, and reached 2 to 4-fold at 15 μM. Similar results were obtained in BaF3 cells (Supplementary Fig. S1A and S1B). Both ITCs also caused a 9-18-fold increase in K63-linked Ub (Fig. 2C and 2D and Supplementary Fig. S1D). Treatment with the proteasome inhibitor bortezomib also increased the levels of K48-linked Ub but had no effect on K63-linked Ub (Fig. 2E). Thus the accumulation of both K48 and K63-linked HMW-Ub strongly suggests that BITC and PEITC inhibit DUBs.
Figure 2. Naturally occurring ITCs cause HMW-Ub accumulation in cells but do not inhibit the proteasome.
A, BaF3/p210 cells were treated with PEITC or BITC for 4 h. Whole cell lysates were analyzed by SDS-PAGE and probed for K48-linked Ub. Actin is shown as a loading control. Data are representative of two independent experiments and reproduced independently in the parent BaF3 cell line. B, Densitometry quantification of the blot shown in A. C, BaF3/p210 cells were incubated with BITC or PEITC for 4 h. Whole cell lysates were analyzed by SDS-PAGE and immunoblotted as indicated. Data are representative of two independent experiments and reproduced independently in the parent BaF3 cells. D, Quantification is shown in C. E, BaF3/p210 cells were treated with BITC (15 μM), BITC (15 μM) combined with bortezomib (20 nM) or bortezomib (20 nM) alone for 4 h. Whole cell lysates were analyzed by SDS-PAGE and western blot. `Control' denotes 0.1% DMSO vehicle only. Actin is shown as a loading control. Data are representative of two independent experiments. F, COS1 cells transiently expressing UbG76V-GFP were treated for 8 h with PEITC (12 μM), BITC (12 μM), SFN (12 μM), USP9x inhibitor WP1130 (5 μM), the proteasome inhibitor bortezomib (`Bort', 4 μM) or 0.1% DMSO (vehicle control). GFP was quantified by flow cytometry. Data presented are the mean ± s.d. of quadruplicate samples from three independent experiments. Significance was determined using the Student's t-test.
BITC and PEITC do not inhibit the flux through the ubiquitin proteasome system
The accumulation of K48-linked ubiquitin suggests that the anticancer effects of ITCs might arise from inhibition of flux through the ubiquitin-proteasome system, much like the anticancer effects of the proteasome inhibitor bortezomib. Therefore we used the UbG76V-GFP assay to monitor 26S proteasome activity and flux through the ubiquitin-proteasome system in live cells(21,22). This reporter protein consists of an ubiquitin linked to the N-terminus of GFP. The G76V mutation prevents the cleavage of ubiquitin from GFP, leading to its degradation by the 26S proteasome (22). When COS1 cells expressing UbG76V-GFP were treated with the proteasome inhibitor bortezomib, GFP fluorescence increased 1.5-fold, consistent with reports from other laboratories (e.g. (23); Fig. 2F). In contrast, BITC and PEITC did not increase GFP levels, indicating that proteasome activity was not inhibited. Similarly, the DUB inhibitor WP1130 also did not cause an increase in GFP levels (Fig. 2F). ITCs did not inhibit purified 20S proteasome or proteasome activity in rabbit reticulocyte lysate, contrary to a previous report(24) (Supplementary Fig. S2A and S2B and S2C). Collectively, these observations strongly suggest that the ITC-induced accumulation of HMW-Ub results from DUB inhibition.
PEITC does not perturb the global cysteine reactome
The methods used to identify ITC targets in previous reports were unlikely to detect modifications of cysteine residues in DUBs or other proteins. Therefore we used competitive cysteine reactivity profiling with an iodoacetamide-alkyne (IA-alkyne) probe (100 μM) to more thoroughly investigate the effects of ITCs on the global cysteine reactome of HELA cells (18). Over 1000 IA-alkyne-labeled cysteine residues were identified in HELA cells using mass-spectrometry analysis. Unfortunately, only two of these cysteine residues belonged to DUBs (UCHL1 and otubain 1), alluding to the poor affinity of IA-alkyne toward the active-site cysteines in DUBs, as well as the low abundance of DUBs relative to other cysteine-containing proteins in HELA lysates. Despite low coverage of DUBs, this cysteine profiling experiment provides a measure of the general promiscuity of PEITC across highly reactive cysteine residues in the proteome. Treatment of HELA lysates with PEITC (20 μM) significantly inhibited the labeling of only 14 out of 1400 profiled cysteines in at least one of two independent experiments (inhibition ≥67%; Supplementary Dataset 1), though in no case was labeling inhibited in both experiments. Seventy of the profiled cysteine residues are found in enzyme active sites or metal binding sites, including 6 dehydrogenases and 6 cysteine proteases (Supplementary Dataset 2). PEITC blocked the labeling of only one of these, mitochondrial phosphoenolpyruvate carboxykinase (Supplementary Fig. S3). These experiments demonstrate that PEITC is not a promiscuous cysteine modifying agent.
BITC and PEITC inhibit USP9x and UCH37
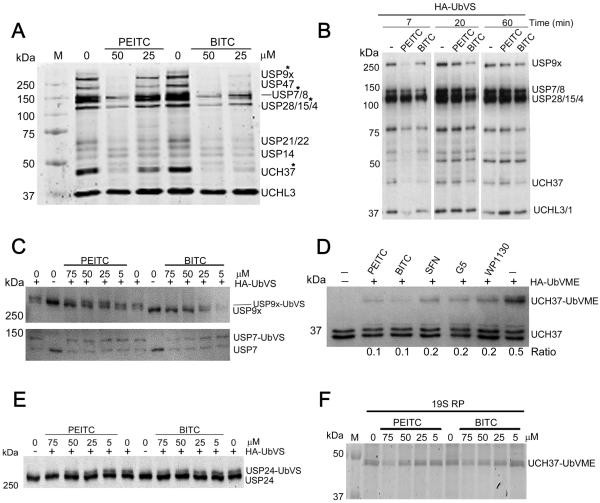
We turned to competitive activity profiling to identify which DUBs are inhibited by BITC and PEITC. In addition to the commonly used HA-tagged ubiquitin vinyl sulfone (HA-UbVS) and HA-tagged ubiquitin vinyl methyl ester (HA-UbVME) (25,26), we also used the more sensitive fluorogenic probes Cy5-UbVME (26) and TAMRA-ubiquitin propargylamide (TAMRA-UbPA) (27,28). These irreversible DUB inhibitors label 10–30 DUBs in cell lysates with varying repertoires depending on the probe and experimental conditions (25). The treatment of cell lysates with HA-UbVS produced the characteristic pattern of protein bands at 300, 150–100, 45, 36 kDa, generally ascribed to USP9x/USP24 (292 kDa), USP19 (146 kDa), USP7/8 (128 and 127 kDa, respectively), USP28/15 (122 and 112 kDa, respectively), UCH37 (38 kDa), UCH-L3 (26 kDa) and UCH-L1 (25 kDa) (Supplementary Fig. S4A) (25). Similar results, although with varying band intensities, were obtained with HA-UbVME, TAMRA-UbPA and Cy5-UbVME (Supplementary Fig. S4B, S4C and S4D). As expected, WP1130 and isopeptidase inhibitor G5 reduced the labeling of several DUBs (e.g., bands between 120 and 300 kDa), validating the experimental method (Supplementary Fig. S4A) (25,29).
BITC and PEITC inhibited the labeling of several DUBs in lysates prepared from cells pre-treated with ITCs (Fig 3A). When lysate was incubated with either BITC or PEITC and then diluted into a buffer containing HA-UbVS, the labeling increased with time, demonstrating that ITC inhibition was slowly reversible as expected (Fig. 3B). Labeled DUBs with molecular weights at 300 kDa, 135 kDa and 45 kDa were among the most susceptible to ITCs. These bands likely corresponded to three DUBs that are potential therapeutic targets for cancer: USP9x, USP7 and UCH37 (Fig. 3A, Supplementary Fig. S4A, S4B, S4C and S4D). The identities of USP9x and UCH37 were confirmed by observing the molecular weight shift upon labeling with immunoblotting (Fig. 3C and 3D and Supplementary Fig. S4E and S4F). USP24, a close relative of USP9x (16), was also a target of BITC and PEITC (Fig. 3E). However, immunoblotting revealed that the 128 kDa ITC target was not USP7 (Fig. 3C).
Figure 3. Naturally occurring ITCs inhibit USP9x and UCH37 in cell lysates and in living cells.
A, BaF3/p210 cells were incubated with PEITC or BITC for 4 h. The cells were harvested, washed, resuspended in lysis buffer and lysed with glass beads. After centrifugation, the clarified supernatant was normalized (0.6 mg/mL), then Cy5-UbVME (250 nM) was added and the mixture incubated at 37 °C for 5 min and analyzed by SDS-PAGE. Ingel fluorescent scan was obtained on Typhoon scanner. Data representative of three independent experiments. Asterisks denote DUB assignments that were verified by immunoblotting. In cases where the bands were not confirmed by immunoblotting, DUBs were identified based on accepted MW. B, A BaF3 cell lysate (12 mg/mL) was incubated with PEITC or BITC (250 μM), or with DMSO vehicle (1% final concentration) and then diluted 20-fold into buffer containing HA-UbVS (1.5 μM). Aliquots were removed at the indicated times resolved by SDS-PAGE and immunoblotted with anti-HA. `-' indicates no compound. C, A BaF3 cell lysate was incubated with PEITC or BITC, treated with HA-UbVS, resolved on a 6% polyacrylamide gel and immunoblotted with anti-USP9x and anti-USP7. `-' indicates vehicle control. D, A BaF3 cell lysate (1.5 mg/mL) was incubated with PEITC, BITC, or SFN (50 μM) or with G5 or WP1130 (10 μM) for 10 min prior to addition of HA-UbVME (1.5 μM). After 20 min aliquots were analyzed by immunoblotting with anti-UCH37 anti-body. `Ratio' represents UCH37-UbVME conjugate per total UCH37. Data representative of two independent experiments. `-' indicates no compound, vehicle only. E, A BaF3 cell lysate (1.5 mg/mL) was incubated with PEITC or BITC, treated with HA-UbVS (1.5 μM) and analyzed by immunoblotting with anti-USP24. Data are representative of two independent experiments. F, 19S regulatory particles (19S RP, 30 nM) were treated with PEITC or BITC for 30 min at which time Cy5-UbVME (300 nM) was added. After 20 min, aliquots were quenched in loading buffer and resolved by SDS-PAGE. Ingel scans were obtained on a GE Typhoon scanner.
The IC50 values for the inhibition of USP9x labeling were 27 ± 6 μM and 15 ± 3 μM for BITC and PEITC, respectively, in cell lysates (Supplementary Fig. S5A and S5B). The values of IC50 for the inhibition of UCH37 labeling were 22 ± 4 μM and 13 ± 1 μM for BITC and PEITC, respectively (Supplementary Fig. S5C). The values for USP24 with IC50 values of 56 ± 5 μM and 28 ± 5 μM for BITC and PEITC, respectively (Fig. 3E, Supplementary Fig S5D).
BITC and PEITC inhibit USP9x and UCH37 in vitro
The inhibition of USP9x was confirmed by assaying purified recombinant enzyme. PEITC inhibited labeling of recombinant USP9x with Cy5-UbVME with an IC50 value of 20 ± 2 μM, in good agreement with the lysate assays (Supplementary Fig. S6A and S6B). Both BITC and PEITC were slow binding competitive inhibitors of recombinant USP9x, with values of Ki = 25 ± 1 and 23 ± 2 μM, respectively (Supplementary Fig. S6C, S6D, S6E). The values of kon decreased with increasing substrate concentration, indicating competitive inhibition (Supplementary Fig. S6F).
UCH37 (also known as UCHL5) is a component of the 19S regulatory particle and the INO80 chromatin remodeling complex. The 19S regulatory particle activates UCH37, while UCH37 is inactive in the INO80 complex (30). Therefore we assayed the DUB activity of UCH37 in the 19S regulatory particle using activity profiling. Both PEITC and BITC inhibited UCH37 with values of EC50 of 36 ± 5 μM and 31 ± 6 μM, in reasonable agreement with the lysate assays (Fig. 3F and Supplementary Fig. S5E). No inhibition of recombinant UCHL3 or the catalytic domain of USP7 was observed, confirming the lysate assays (Supplementary Fig. S6A and S6B).
USP9x inhibition provides a molecular mechanism for the antileukemic effects of ITCs
Both USP9x and UCH37 are over-expressed in many cancers(30–33), so inhibition of these DUBs provides an attractive mechanism for the anticancer effects of ITCs (30,31,33). USP9x is known to protect the antiapoptotic protein Mcl-1 from ubiquitination and degradation(33). Intriguingly, ITCs decrease Mcl-1 levels in several cell lines (7–9), and similar decreases in Mcl-1 levels are observed when USP9x is either inhibited or silenced(32–34). These observations suggest that the USP9x may be a primary target for BITC and PEITC. In contrast, the role of UCH37 in cancer is not understood, and could involve either or both the 19S regulatory particle and the INO80 complex, or recently characterized complexes with SMAD proteins (30). Which of these complexes promote cancer, and whether the deubiquitinating activity of UCH37 is required, is not understood.
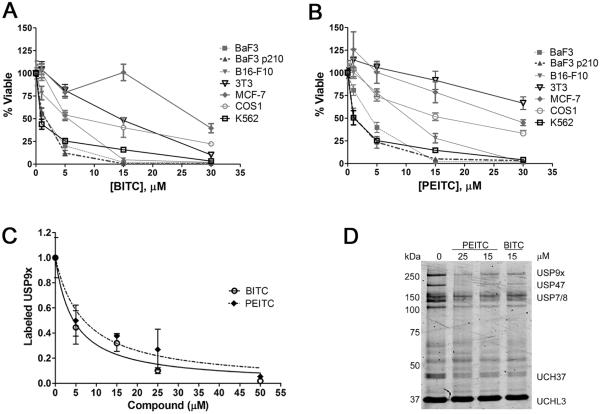
If the anticancer effects of BITC and PEITC arise from the inhibition of USP9x, then Mcl-1-dependent cells should be more sensitive to ITCs than other cell lines. Therefore we tested the ITC sensitivity of various cell lines (Fig. 4A and 4B). BaF3, BaF3/p210 and K562 cells are all dependent on Mcl-1 (35,36), and these cells are the most sensitive to BITC and PEITC, with EC50 values of 1–3 μM (Fig. 4A and 4B). In contrast, MCF7, NIH3T3 and COS1 cells are 8 to 40-fold less sensitive to BITC and PEITC, as expected given that these cells do not rely on Mcl-1. Importantly, MCF7 cells rely on the Mdm2, which is protected from ubiquitination and degradation by USP7(14). As shown above, BITC and PEITC do not inhibit USP7, which explains the resistance of these cells.
Figure 4. Mcl-1-dependent cells are more sensitive to BITC and PEITC.
A, Cells were incubated with BITC for 48 h. With the exception of K562 cells, viability was measured using the Alamar Blue method. Viability of K562 cells was measured with Promega CellTiterGlo assay. Data are presented as mean ± s.d. of at least triplicate samples. B, As in A, except cells were incubated with PEITC for 48 h. C, Lysates were prepared from BaF3/p210 cells treated with PEITC or BITC for 4 h. Samples were treated with Cy5-UbVME (250 nM) and resolved by SDS-PAGE. Data presented are the mean ± s.d. of at least two independent experiments. D, A representative in-gel scan for the experiment described in C.
USP9x inhibition occurs early in the ITC anticancer program
We further examined the effects of ITCs on BaF3/p210 cells to determine if the USP9x inhibition is an initiating event in their anticancer program. USP9x levels remained stable upon treatment with BITC (Supplementary Fig. S5F). USP9x inhibition was evident after treatment with either BITC or PEITC for 4 h (Fig. 4C and 4D). The values of IC50 for USP9x inhibition in cells were 4 ± 1 μM and 7 ± 2 μM for BITC and PEITC, respectively. These values are smaller than those obtained in lysates (see above). However, cells are known to concentrate ITCs by factors of hundreds (4), which can account for this higher potency. Importantly, the inhibition of USP9x in cells is similar to the inhibition of cell proliferation and viability.
As in other cell types, Mcl-1 exists as two major bands in BaF3 cells(37). The fast-mobility isoform (FM) is an N-terminally truncated product formed from the full-length, or slow-mobility (SM), isoform. The FM isoform has attenuated anti-apoptotic activity as well as a significantly longer half-life (37). A corresponding decrease in total Mcl-1 was also observed when BaF3/p210 cells were treated with BITC and PEITC (Fig. 5A, 5B and 5C). In contrast to this ITC induced decrease in Mcl-1, treatment with the proteasome inhibitors MG-132 and bortezomib increased Mcl-1 levels, as observed by others (38–40) (Fig. 5B, 5C and 5D). Bortezomib also induced the appearance of a lower molecular weight caspase cleavage Mcl-1 fragment that was not observed with either ITC (Fig. 5D) (41). MG-132 rescued Mcl-1 levels in ITC-treated cells, as expected if inhibition of USP9x increased ubiquitination and degradation (Fig. 5B and 5C). Importantly, BITC also decreased the levels of Mcl-1 in the presence of the translation inhibitor cycloheximide, further indicating that BITC promotes the degradation of Mcl-1 (Supplementary Fig. S7A, S7B and S7C). Similar results were obtained when BaF3 and K562 cells were treated with ITCs (Supplementary Fig. S7D, S7E and S7F).
Figure 5. ITCs promote the ubiquitination of Mcl-1.
A, BaF3/p210 cells were incubated with PEITC or BITC for 4 h. Whole cell lysates were analyzed by SDS-PAGE and western blot. `0 μM' indicates 0.1% DMSO vehicle only. Data are representative of four independent experiments. B, BaF3/p210 cells were treated with BITC (15 μM), BITC (15 μM) combined with MG-132 (20 μM) or MG-132 (20 μM) alone for 4 h. Whole cell lysates were analyzed by SDS-PAGE and immunoblotted as indicated. `Control' denotes 0.1% DMSO vehicle only. Actin is shown as a loading control. Data are representative of two independent experiments. C, Quantification of B, mean ± range of two experiments analyzed using the Student's t-test. D, BaF3/p210 cells were treated with BITC (15 μM), BITC (15 μM) combined with bortezomib (20 nM) or bortezomib (20 nM) alone for 4 h. Whole cell lysates were analyzed by SDS-PAGE and western blot. `Control' denotes 0.1% DMSO vehicle only. Actin is shown as a loading control. Data are representative of two independent experiments. E, COS1 cells transiently transfected with Flag-MCL1 were treated with PEITC (15 μM), PEITC (15 μM) together with bortezomib (300 nM), bortezomib alone (300 nM), BITC (15 μM), BITC (15 μM) together with bortezomib (300 nM), or with WP1130 (5 μM) for 2 h. Flag-MCL1 was immunopreciptated and blotted for K48-linked Ub and Flag. Control: DMSO vehicle only. `−' indicates IP of untransfected cells as an additional control. Data are representative of three independent experiments by two researchers.
The inhibition of USP9x should increase the ubiquitination of Mcl-1 (33). COS1 cells expressing Flag-Mcl-1 were treated with ITCs for 2 h and Flag-Mcl-1 was isolated by immunoprecipitation. Both BITC and PEITC, as well as WP1130, increased the levels of ubiquitinated Flag-Mcl-1 in a concentration dependent manner (Fig. 5E and Supplementary Fig. S8). Bortezomib alone also increased Flag-Mcl-1 ubiquitination, and the addition of bortezomib further increased ubiquitinated Flag-Mcl-1 in ITC-treated-cells. These data indicate that the ITC-induced decrease in Mcl-1 results from increased ubiquitination, as expected when USP9x is inhibited. These observations provide additional evidence that the mechanism of ITCs is distinct from proteasome inhibition or translation inhibition, and suggest that USP9x inhibition can account for the anticancer activity of ITCs in leukemia cells.
ITCs increase ubiquitination and degradation of Bcr-Abl
PEITC has been reported to cause the knockdown of Bcr-Abl kinase, the oncogenic fusion protein that causes chronic myelogenous leukemia (10). Both BaF3/p210 and K562 cells depend on Bcr-Abl, so the ITC sensitivity of these cells could derive from the depletion of Bcr-Abl as well as Mcl-1. We hypothesized that DUB inhibition could also be responsible for this ITC effect. Intriguingly, USP9x has been implicated in the regulation of Bcr-Abl (34,42). Therefore we examined the correlation of Bcr-Abl ubiquitination, PARP cleavage and USP9x inhibition.
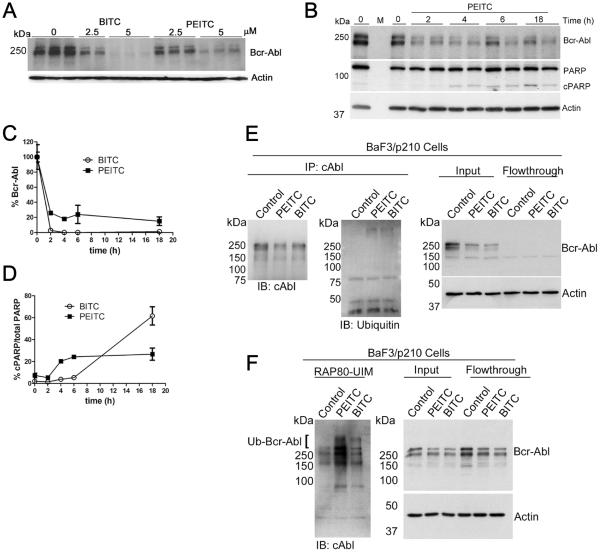
Both BITC and PEITC decreased the levels of total Bcr-Abl in BaF3/p210 cells (Fig. 6A, 6B and 6C and Supplementary Fig. S9A). Bcr-Abl knockdown was essentially complete after only 2 h treatment with BITC (5 μM), whereas PARP cleavage was not observed until 6 h (Fig. 6D). Likewise, Bcr-Abl was reduced to 20% of its initial level within 2 h after treatment with PEITC (5 μM), and PARP cleavage was observed at 4 h (Fig. 6D). No aggregation of Bcr-Abl kinase was observed at low ITC concentrations (5 μM), although aggregation was evident at high ITC concentrations (30 μM; Supplementary Fig. S9B, S9C and S9D).
Figure 6. DUB inhibition is an early event in ITC-induced knockdown of Bcr-Abl.
A, Whole cell lysates from BaF3/p210 cells treated with PEITC or BITC for 4 h were resolved by SDS-PAGE and immunoblotted as indicated. Data are representative of three independent experiments. B, Lysates from BaF3/p210 cells treated with PEITC (5 μM) were resolved by SDS-PAGE and immunoblotted as indicated. Data are representative of two independent experiments. C, Quantification of blots shown in B and in Supplementary Fig. S9A. Data are presented as mean ± range of two independent experiments. D, Quantification of blots shown in B and in Supplementary Fig. S9A. Data are presented as mean ± range of two independent experiments. E, BaF3/p210 cells were treated with 5 μM PEITC or BITC for 1 h. Bcr-Abl was immunoprecipitated from lysates (1 mg/mL) with anti-cAbl antibody. Actin is shown as a loading control. `Control”: DMSO, vehicle only. Data are representative of two independent experiments. F, BaF3/p210 cells were treated with 5 μM PEITC or BITC for 1 h. K63-linked polyubiquitinated proteins were isolated from lysates (1 mg/mL) using RAP80-UIM agarose. The presence of ubiquitinated Bcr-Abl is indicated with the bracket. Data are representative of two independent experiments and was repeated using K562 cells (see Supplementary Fig. S10C and S10D)
Two complimentary approaches were used to determine if ITC treatment increases ubiquitination of Bcr-Abl. First, Bcr-Abl was immunoprecipitated from lysates of BaF3/p210 cells. Significantly more ubiquitination was observed in Bcr-Abl isolated from ITC-treated cells than untreated cells (Fig. 6E and Supplementary Fig. S9E). Bcr-Abl ubiquitination predominantly involves K63-linkages, so we used RAP80-UIM-conjugated agarose resin to isolate K63-linked ubiquitinated proteins(34,43). More poly-ubiquitinated Bcr-Abl was recovered from ITC-treated BaF3/p210 cells than from untreated cells (Figure 6F and Supplementary Fig. S9F and S9G). A similar increase in Bcr-Abl ubiquitination was observed when K562 cells were treated with BITC and PEITC (Supplementary Fig. S10). Collectively, these observations demonstrate that ITC treatment increases the ubiquitination and degradation/aggregation of Bcr-Abl, strongly suggesting that the anti-leukemia program of ITCs results from DUB inhibition. Importantly, USP9x inhibition occurred on the same time scale as Bcr-Abl knockdown at low ITC concentrations (Fig. 6C and Supplementary Fig. S7E). Thus USP9x inhibition could be responsible for the increased ubiquitination of Bcr-Abl.
USP9x knockdown mimics the action of ITCs
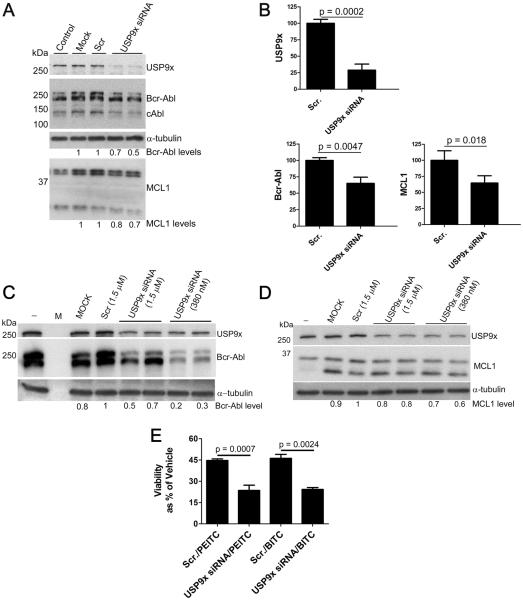
To further elucidate the role of USP9x inhibition in the knockdown of Mcl-1 and Bcr-Abl kinase, we used siRNA to decrease the level of USP9x in K562 cells. Optimal silencing occurred 24 h post transfection in these rapidly proliferating cells, resulting in a 70% decrease of USP9x (Fig. 7A and 7B and Supplementary Fig. S11A and S11B). The level of Mcl-1 decreased by approximately 30% in the USP9x silenced cells, as reported by others (Fig. 7B) (33,34). Bcr-Abl was also reduced by 30% in USP9x silenced cells. No insoluble Bcr-Abl aggregates were observed (Supplementary Fig. S11C). Mcl-1 and Bcr-Abl were also depleted when USP9x was silenced in BaF3/p210 cells (Fig. 7C and 7D and Supplementary Fig. 11D). Moreover, USP9x silencing resulted in significantly increased cell death in BaF3/p210 cells (Supplementary Fig. S11E), suggesting that the ITC induced inhibition of USP9x can, at least partially, account for the reduced viability and increased cell death observed upon ITC treatment. USP9x knockdown sensitizes cells to ITCs, as expected if USP9x were the primary target (Fig. 7E). However, electroporated cells appeared to be less sensitive to ITCs (Fig. 7E vs Supplementary Fig. S11F). We suggest two explanations for this observation: first, electroporation causes some cell death which increased background and decreased assay sensitivity; secondly, viability was measured after a short ITC treatment (6 h vs 24 or 48 h), to keep in the range of optimal siRNA induced Bcr-Abl knockdown. Knockdown of Bcr-Abl was also observed when USP9x expression was silenced in NIH/3T3 cells stably transfected with the Bcr-Abl gene (3T3/p210 cells) (Supplementary Fig. S11G). Thus experiments in three cell lines, with RNAi targeting USP9x from both human and mouse cells lines, confirm the role of USP9x in maintaining the levels of Bcr-Abl.
Figure 7. siRNA mediated silencing of USP9x decreases levels of Mcl-1 and Bcr-Abl.
A, K562 cells (1.5×106 cells) were transfected with non-targeting siRNA (Scr., 380 nM) or with USP9x siRNA (380 nM) using the Amaxa Nucleofector II (Program T-016). `Mock' indicates cells that were subject to electroporation with no siRNA. Cells were harvested 24 h post-transfection and immunoblotted as indicated. Data are representative of more then seven independent experiments. B, K562 cells (1.5×106 cells) were transfected with non-targeting siRNA (Scr., 380 nM) or with USP9x siRNA (380 nM) as in A, harvested 24 h post-transfection and analyzed by SDS-PAGE and immunoblot. Data presented are the mean ± s.d. of three independent experiments; two performed in duplicate and were analyzed using the student's t-test. Representative blot shown in A. (C–D) BaF3/p210 cells (2×106 cells) were transfected with non-targeting siRNA (Scr., 1.5 μM) or with USP9x siRNA (1.5 μM or 380 nM) using the Amaxa Nucleofector II (Program X-001). `Mock' indicates cells that were subject to electroporation with no siRNA. `−' indicates no treatment control. 48 h after transfection, cell lysates were analyzed by SDS-PAGE and immunoblotted as indicated. α-Tubulin is shown as a loading control. Mcl-1 protein level was determined by normalizing the top Mcl-1 band (long form with anti-apoptotic function) to the loading control. Data are representative of two experiments. E, BaF3/p210 cells (2×106 cells) were transfected with non-targeting siRNA (Scr) or with USP9x siRNA. 48 hours post-transfection, cells were treated with BITC or PEITC (100 μM) for 6 h then analyzed with Cell Titer Glow.
Discussion
The anticancer effects of BITC and PEITC are well established. Although ITCs were generally believed to deplete glutathione, inducing ROS, this mechanism has recently been discredited (11,12). We recognized that many of the effects attributed to ITCs are also properties of DUB inhibitors. Our data demonstrate that BITC and PEITC inhibit at least two DUBs that are potential anticancer targets, UCH37 and USP9x (30–32). Moreover, a strong correlation exists between elevated USP9x levels and poor prognosis (31). While the role of UCH37 in cancer is not well understood, USP9x protects the antiapoptotic/pro-survival protein Mcl-1 from ubiquitination and depletion (32,33). Thus the inhibition of USP9x can account for the ITC-induced decrease in Mcl-1. Leukemia cells are dependent on Mcl-1, and thus are especially sensitive to ITCs. However, Mcl-1 levels are elevated in many cancers, so the inhibition of USP9x provides a molecular mechanism for the broader anticarcinogenic activity of BITC and PEITC.
ITCs also increase the ubiquitination of Bcr-Abl, causing depletion via degradation at low concentrations and aggregation at high concentrations. A partial knockdown of USP9x also depletes Bcr-Abl. The simplest explanation for this observation is that Bcr-Abl is a substrate for USP9x, although we cannot rule out the involvement of other DUBs. The DUB inhibitor WP1130, also increases the ubiquitination and aggregation of Bcr-Abl, although this work was unable to demonstrate that USP9x inhibition accounted for these effects (34).
In addition to inducing growth arrest and apoptosis in cancer cells, ITCs perturb the inflammatory response (1) and DNA repair(3,6). DUBs also regulate these processes. At least eight DUBs are involved in inflammation (CYLD, A20, Cezanne, USP21, OTULIN, OTUD5, MCPIP1 and USP9x) (44),(45). Eight DUBs have also been implicated in DNA repair (OTUB1, USP1, USP3, USP11, USP16, USP47, BRCC36 and POH1). Given the multitude of physiological processes that are regulated by ubiquitination(6), DUB inhibition is likely to be the molecular mechanism underlying the pleiotropic effects of dietary ITCs. It is important to note that our study focused on just two structurally similar ITCs and two DUB targets. Other DUB targets remain to be identified. More structurally diverse ITCS such as sulphoraphane could inhibit different cysteine proteases or even other enzymes with cysteine nucleophiles.
DUBS, like other cysteine proteases, are challenging targets for drug discovery because potent inhibition usually requires the presence of electrophillic “warheads” that can react nonspecifically with other proteins(46). Many DUB inhibitors fall into this category. For example, G5(47) and b-AP15(48) are highly reactive dienones, which can cause cross-linking. WP1130(29) is an activated enone that has off target effects (49). Only a handful of selective DUB inhibitors have emerged, including the USP14-specific DUB inhibitor IU1, the USP7-specific inhibitors P5091 and HBX 19,818 and the USP1 inhibitor ML323(50). The reversible nature of ITC adducts and the relatively low intrinsic toxicity of this function offers a promising new avenue for the design of inhibitors for DUBs and other cysteine proteases. Since over 120 ITCs are available from both dietary and other natural sources, these compounds form a rich resource for lead compounds, drug discovery and functional food design.
Supplementary Material
Acknowledgements
We thank Arifa Ahsan for the TOC graphic, the Ploegh laboratory for the ubiquitin intein plasmid and Nelson Lau and Yuliya Sytnikova for assistance with the knockdown experiments.
Financial Support: This study was funded by the National Institutes of Health (grant GM100921 to LH) and a Sprout Grant from Brandeis University (to M.J.C.L). M.J.C.L. was supported by a Howard Hughes Medical Institute International Student Research Fellowship.
Footnotes
Disclosure of Potential Conflicts of Interest: F.E. declares competing financial interests as co-founder and shareholder of UbiQ.
Authors Contributions Conception and design: A.P. Lawson, M.J.C. Long, L. Hedstrom
Development of methodology: Acquisition of data (provided animals, acquired and managed patients,provided facilities, etc.): A.P. Lawson, M.J.C. Long, R.T. Coffey
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A.P. Lawson, M.J.C. Long, R.T. Coffey, Y. Qian, E. Weerapana, F. El Oualid, L. Hedstrom
Writing, review, and/or revision of the manuscript: A.P. Lawson, M.J.C. Long, F. El Oualid, E. Weerapana, L. Hedstrom
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A.P. Lawson, M.J.C. Long, R.T. Coffey, Y. Qian
Study supervision: E. Weerapana, L. Hedstrom
References
- 1.Mi L, Di Pasqua AJ, Chung FL. Proteins as binding targets of isothiocyanates in cancer prevention. Carcinogenesis. 2011;32(10):1405–13. doi: 10.1093/carcin/bgr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SV, Singh K. Cancer chemoprevention with dietary isothiocyanates mature for clinical translational research. Carcinogenesis. 2012;33(10):1833–42. doi: 10.1093/carcin/bgs216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta P, Kim B, Kim SH, Srivastava SK. Molecular targets of isothiocyanates in cancer: recent advances. Molecular nutrition & food research. 2014;58(8):1685–707. doi: 10.1002/mnfr.201300684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer JM, Teran-Garcia M, Jeffery EH. Enhancing sulforaphane absorption and excretion in healthy men through the combined consumption of fresh broccoli sprouts and a glucoraphanin-rich powder. The British journal of nutrition. 2012;107(9):1333–8. doi: 10.1017/S0007114511004429. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Tang L, Gonzalez V. Selected isothiocyanates rapidly induce growth inhibition of cancer cells. Molecular cancer therapeutics. 2003;2(10):1045–52. [PubMed] [Google Scholar]
- 6.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nature medicine. 2014;20(11):1242–53. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 7.Gao N, Budhraja A, Cheng S, Liu EH, Chen J, Yang Z, et al. Phenethyl isothiocyanate exhibits antileukemic activity in vitro and in vivo by inactivation of Akt and activation of JNK pathways. Cell death & disease. 2011;2:e140. doi: 10.1038/cddis.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou T, Li G, Cao B, Liu L, Cheng Q, Kong H, et al. Downregulation of Mcl-1 through inhibition of translation contributes to benzyl isothiocyanate-induced cell cycle arrest and apoptosis in human leukemia cells. Cell death & disease. 2013;4:e515. doi: 10.1038/cddis.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trachootham D, Zhang H, Zhang W, Feng L, Du M, Zhou Y, et al. Effective elimination of fludarabine-resistant CLL cells by PEITC through a redox-mediated mechanism. Blood. 2008;112(5):1912–22. doi: 10.1182/blood-2008-04-149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Trachootham D, Lu W, Carew J, Giles FJ, Keating MJ, et al. Effective killing of Gleevec-resistant CML cells with T315I mutation by a natural compound PEITC through redox-mediated mechanism. Leukemia. 2008;22(6):1191–9. doi: 10.1038/leu.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Lu W, Chen G, Zhang H, Jia Y, Wei Y, et al. Overcoming resistance to histone deacetylase inhibitors in human leukemia with the redox modulating compound beta-phenylethyl isothiocyanate. Blood. 2010;116(15):2732–41. doi: 10.1182/blood-2009-11-256354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu C, Hu W, Wu H, Hu X. No evident dose-response relationship between cellular ROS level and its cytotoxicity - a paradoxical issue in ROS-based cancer therapy. Scientific reports. 2014;4:5029. doi: 10.1038/srep05029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang CS, Tang WJ. Inhibition of papain by isothiocyanates. Biochimica et biophysica acta. 1976;452(2):510–20. doi: 10.1016/0005-2744(76)90202-3. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson B, Kumar S, Agarwal S, Eddins M, Marblestone JG, Wu J, et al. Discovery of Therapeutic Deubiquitylase Effector Molecules: Current Perspectives. Journal of biomolecular screening. 2014 doi: 10.1177/1087057114527312. [DOI] [PubMed] [Google Scholar]
- 15.Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, et al. An atlas of altered expression of deubiquitinating enzymes in human cancer. PloS one. 2011;6(1):e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chemistry & biology. 2002;9(10):1149–59. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 17.Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Molecular cell. 1999;4(3):395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- 18.Qian Y, Martell J, Pace NJ, Ballard TE, Johnson DS, Weerapana E. An isotopically tagged azobenzene-based cleavable linker for quantitative proteomics. Chembiochem : a European journal of chemical biology. 2013;14(12):1410–4. doi: 10.1002/cbic.201300396. [DOI] [PubMed] [Google Scholar]
- 19.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nature protocols. 2007;2(6):1414–25. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92(10):3829–40. [PubMed] [Google Scholar]
- 21.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nature biotechnology. 2000;18(5):538–43. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. The EMBO journal. 2011;30(15):2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Um JW, Im E, Lee HJ, Min B, Yoo L, Yoo J, et al. Parkin directly modulates 26S proteasome activity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(35):11805–14. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mi L, Gan N, Chung FL. Isothiocyanates inhibit proteasome activity and proliferation of multiple myeloma cells. Carcinogenesis. 2011;32(2):216–23. doi: 10.1093/carcin/bgq242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer HB, Nicholson B, Kessler BM, Altun M. Detection of ubiquitin-proteasome enzymatic activities in cells: application of activity-based probes to inhibitor development. Biochimica et biophysica acta. 2012;1823(11):2029–37. doi: 10.1016/j.bbamcr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong A, Merkx R, Berlin I, Rodenko B, Wijdeven RH, El Atmioui D, et al. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. Chembiochem : a European journal of chemical biology. 2012;13(15):2251–8. doi: 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekkebus R, van Kasteren SI, Kulathu Y, Scholten A, Berlin I, Geurink PP, et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. Journal of the American Chemical Society. 2013;135(8):2867–70. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer S, Weikart ND, Linne U, Mootz HD. Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorganic & medicinal chemistry. 2013;21(9):2511–7. doi: 10.1016/j.bmc.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 29.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer research. 2010;70(22):9265–76. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 30.Chen YJ, Ma YS, Fang Y, Wang Y, Fu D, Shen XZ. Power and promise of ubiquitin carboxyl-terminal hydrolase 37 as a target of cancer therapy. Asian Pacific journal of cancer prevention : APJCP. 2013;14(4):2173–9. doi: 10.7314/apjcp.2013.14.4.2173. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Hu Q, Liu W, He X, Cui L, Chen X, et al. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagnostic pathology. 2013;8(1):177. doi: 10.1186/1746-1596-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peddaboina C, Jupiter D, Fletcher S, Yap JL, Rai A, Tobin RP, et al. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC cancer. 2012;12:541. doi: 10.1186/1471-2407-12-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 34.Sun H, Kapuria V, Peterson LF, Fang D, Bornmann WG, Bartholomeusz G, et al. Bcr-Abl ubiquitination and Usp9x inhibition block kinase signaling and promote CML cell apoptosis. Blood. 2011;117(11):3151–62. doi: 10.1182/blood-2010-03-276477. [DOI] [PubMed] [Google Scholar]
- 35.Michels J, Johnson PW, Packham G. Mcl-1. The international journal of biochemistry & cell biology. 2005;37(2):267–71. doi: 10.1016/j.biocel.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Bose P, Grant S. Mcl-1 as a Therapeutic Target in Acute Myelogenous Leukemia (AML) Leukemia research reports. 2013;2(1):12–14. doi: 10.1016/j.lrr.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CR, Yang-Yen HF. The fast-mobility isoform of mouse Mcl-1 is a mitochondrial matrix-localized protein with attenuated anti-apoptotic activity. FEBS letters. 2010;584(15):3323–30. doi: 10.1016/j.febslet.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, et al. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer research. 2005;65(14):6282–93. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Galan P, Roue G, Villamor N, Montserrat E, Campo E, Colomer D. The proteasome inhibitor bortezomib induces apoptosis in mantle-cell lymphoma through generation of ROS and Noxa activation independent of p53 status. Blood. 2006;107(1):257–64. doi: 10.1182/blood-2005-05-2091. [DOI] [PubMed] [Google Scholar]
- 40.Yuan BZ, Chapman JA, Reynolds SH. Proteasome Inhibitor MG132 Induces Apoptosis and Inhibits Invasion of Human Malignant Pleural Mesothelioma Cells. Translational oncology. 2008;1(3):129–40. doi: 10.1593/tlo.08133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podar K, Gouill SL, Zhang J, Opferman JT, Zorn E, Tai YT, et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27(6):721–31. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 42.Brehme M, Hantschel O, Colinge J, Kaupe I, Planyavsky M, Kocher T, et al. Charting the molecular network of the drug target Bcr-Abl. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(18):7414–9. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Molecular cell. 2009;33(6):775–83. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. The Biochemical journal. 2015;467(1):191. doi: 10.1042/bj4670191. [DOI] [PubMed] [Google Scholar]
- 45.Park Y, Jin HS, Liu YC. Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(23):9433–8. doi: 10.1073/pnas.1221925110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colland F. The therapeutic potential of deubiquitinating enzyme inhibitors. Biochemical Society transactions. 2010;38(Pt 1):137–43. doi: 10.1042/BST0380137. [DOI] [PubMed] [Google Scholar]
- 47.Aleo E, Henderson CJ, Fontanini A, Solazzo B, Brancolini C. Identification of new compounds that trigger apoptosome-independent caspase activation and apoptosis. Cancer research. 2006;66(18):9235–44. doi: 10.1158/0008-5472.CAN-06-0702. [DOI] [PubMed] [Google Scholar]
- 48.D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature medicine. 2011;17(12):1636–40. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 49.Perry JW, Ahmed M, Chang KO, Donato NJ, Showalter HD, Wobus CE. Antiviral activity of a small molecule deubiquitinase inhibitor occurs via induction of the unfolded protein response. PLoS pathogens. 2012;8(7):e1002783. doi: 10.1371/journal.ppat.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Sidhu SS. Development of inhibitors in the ubiquitination cascade. FEBS letters. 2014;588(2):356–67. doi: 10.1016/j.febslet.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.