Abstract
Cyanobacterial diversity in soil crusts has been extensively studied in arid lands of temperate regions, particularly semi-arid steppes and warm deserts. Nevertheless, Arctic soil crusts have received far less attention than their temperate counterparts. Here, we describe the cyanobacterial communities from various types of soil crusts from Svalbard, High Arctic. Four soil crusts at different development stages (ranging from poorly-developed to well-developed soil crusts) were analysed using 454 pyrosequencing of the V3-V4 variable region of the cyanobacterial 16S rRNA gene. Analyses of 95 660 cyanobacterial sequences revealed a dominance of OTUs belonging to the orders Synechococcales, Oscillatoriales and Nostocales. The most dominant OTUs in the four studied sites were related to the filamentous cyanobacteria Leptolyngbya sp. Phylotype richness estimates increased from poorly- to mid-developed soil crusts and decreased in the well-developed lichenized soil crust. Moreover, pH, ammonium and organic carbon concentrations appeared significantly correlated with the cyanobacterial community structure.
Keywords: soil crust, Arctic, cyanobacteria, 454 pyrosequencing, 16S rRNA gene, soil chemistry
The study describes cyanobacterial community composition in soil crusts from Svalbard.
Graphical Abstract Figure.
The study describes cyanobacterial community composition in soil crusts from Svalbard.
INTRODUCTION
Soil crusts are considered to be characteristic of arid environment and are known to harbour diverse microbial communities (Elster et al. 1999). Cyanobacteria are the main primary producers in these formations (Belnap and Lange 2001), and many biotic and abiotic parameters are known to influence their development, such as water availability, soil geological properties, climate, presence of vascular plants and animal and human intervention (Elster 2002; Hu, Gao and Whitton 2012).
Soil cyanobacteria are well adapted to severe conditions, having developed a wide range of adaptive strategies that allow them to avoid the effect of extreme polar conditions such as long periods of desiccation and large temperature fluctuations (Moquin et al. 2012). For example, Microcoleus spp. are known to produce thick polysaccharidic sheaths which protect them against fluctuations in water availability and prevent desiccation (Belnap 2008). Mobility is another example of a survival strategy. Some soil cyanobacteria are motile and able to migrate to a more favourable environment, avoiding stressful conditions. However, the abundance and diversity of some cyanobacteria, mainly unicellular cyanobacteria, can be reduced under extreme conditions such as unstable and coarse soils (Rossi et al. 2012).
The community composition and nutrient availability of soil crusts can influence crust development (Li et al. 2010). Colourless soil crusts, which are usually poorly developed, are associated with a low biomass of major soil crust organisms, as well as low nutrient concentration (Housman et al. 2006). On the other hand, dark-coloured well-developed soil crusts have higher biomass and diversity of soil crust organisms as well as higher amounts of mineral nutrients (such as phosphorus and nitrogen) and organic carbon (Belnap 2008).
In high-latitude environments, soil crusts are common in the High Arctic, and have been reported in territories of the Russian, Canadian, and European Arctic as well as Greenland. Arctic soil crusts are known to be highly diverse, hosting complex microbial communities (Elster 2002). Next-generation sequencing technologies have greatly improved our knowledge about the microbial diversity of Arctic soil crusts, enabling a more comprehensive understanding (Chu et al. 2010; Schütte et al. 2010; Knelman et al. 2012). However, the identification of soil crust cyanobacteria has rarely been performed (Patova and Beljakova 2006; Strunecký, Elster and Komárek 2010; Pushkareva and Elster 2013). In general, these studies have shown that Arctic soil crusts are composed by cyanobacteria from the orders Chroococcales, Synechococcales, Oscillatoriales and Nostocales. However, much more information is needed to accomplish a full description of the cyanobacterial diversity in Arctic soils, and to better understand the impact of environmental factors on cyanobacterial diversity. Thus, here we aimed to describe the cyanobacterial communities in soil crusts from Petunia Bay (Svalbard) at different stages of development, using a comprehensive sampling scheme, and assess how nutrient availability influence their abundance and distribution.
MATERIALS AND METHODS
Study site and sampling
Petunia Bay is located in the northwestern branch of Billefjorden (Dickson Land, Svalbard). Average annual air temperature is approximately –6.5°C. Air temperatures above 0°C are generally recorded from June until the end of August or middle of September, with usual temperatures of 5°C –7°C (Rachlewicz, Szczucinsky and Ewertowski 2007). Annual precipitation in Petunia Bay is about 200 mm (Láska, Witoszová and Prošek 2012).
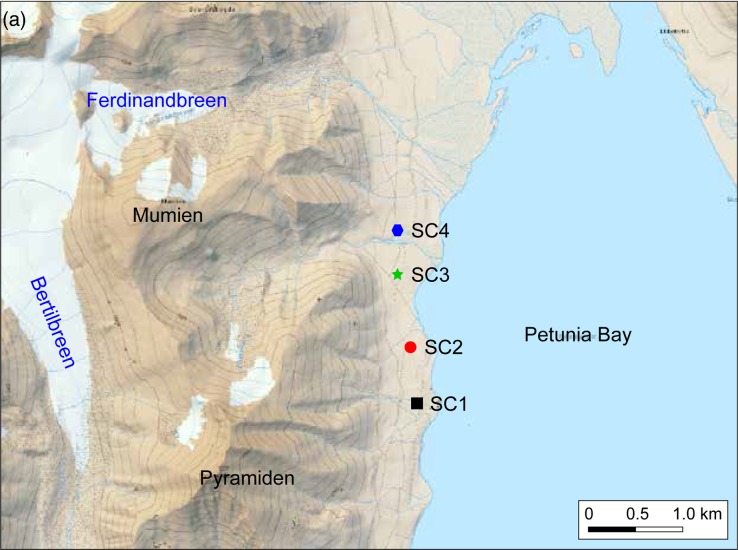
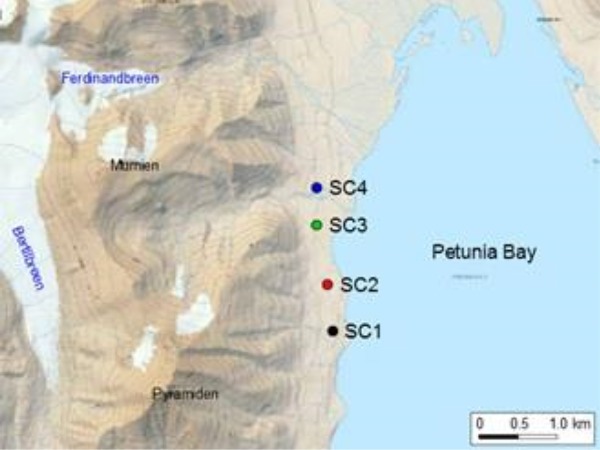
Soil crust samples were collected in August 2013 from four sites in the vicinity of Petunia Bay (78°40.961′—78°41.895′N, 16°26.661′—16°26.361′ E; Fig. 1a). Bedrocks are characterized as fluvial and glaciofluvial deposits from the Pleistocene and Holocene. Soil crusts covered around 80% of the area, but sparse vegetation was also present, including vascular plants such as Polygonum viviparum, Salix polaris, Carex rupestris and Dryas octopetala.
Figure 1.
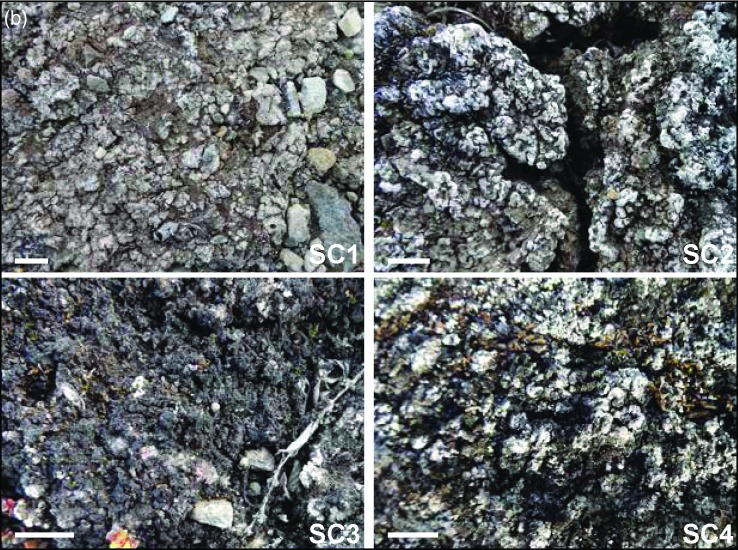
(a) Map of Petunia Bay, Billefjorden, central Svalbard showing the location of the sampling sites and (b) pictures of the studied soil crusts. Scale bar on picture (b) represents one centimeter.
Soil crusts differed macroscopically between the four sites and presented different levels of developmental growth. The sampling sites were located along 2 km walk in a homogenous area and appeared to be subjected to similar environmental conditions. (Fig. 1b). Soil crust in site SC1 was poorly developed and colonized by cyanobacteria alone. Sites SC2 and SC3 had soil crusts in mid development stage with a mixture of lichens and cyanobacteria. Finally, soil crust in site SC4 was well developed and had a rich lichen community dominated by the genera Cladonia and Collema. For pyrosequencing, nine soil crust samples of 2–3 cm depth were taken in each site within a 25 m2 area. These multiple samples were intended to take into account any possible patchiness of the communities. For chemical analysis, three soil crust samples at the same depth were collected in each site. Samples were kept at –20°C and transported in dry ice to the laboratory in Třeboň, Czech Republic.
Figure 1.
(Continued).
Chemical analysis
The three samples collected from each site were mixed and passed through a sieve (2 mm mesh). Analysed parameters included pH (in KCl buffer), conductivity, water content, total organic carbon, N-NH4, N-NO3, P-PO4, Ca2+, Mg2+, Na+ and K+. All analyses were performed according to the methodology described in Czech and European Union standards (ISO 10390, ISO 10523, ČSN EN 27888, ISO 11465, ČSN EN ISO 11732, ČSN EN ISO 13395 and ČSN EN ISO 15681-1). Significant differences in chemical properties between sites were determined using one-way ANOVA followed by Tukey's pairwise posthoc tests using the PAST 3 software (Hammer, Harper and Ryan 2001).
DNA extraction, PCR and pyrosequencing
Pyrosequencing analyses were carried out following the protocol of Pessi et al. (in preparation). DNA was extracted from the 36 soil crust samples using the PowerSoil DNA Isolation Kit (MOBIO, Carlsbad, CA, USA) according to manufacturer instructions. The V3-V4 region of the 16S rRNA gene was amplified using the cyanobacteria-specific primers 359F and 781Ra/781Rb (Nübel, Garcia-Pichel and Muyzer 1997) in separate reactions for each reverse primer (Boutte et al. 2006). A barcode sequences were added to both forward and reverse primers and were specific to each sample. PCR reactions consisted of 1× PCR buffer with 1.5 mM MgCl2, 1 mg ml−1 BSA, 200 μM of each dNTP, 0.2 μM of each primer, 1 U SUPER TAQ ‘plus’ DNA polymerase (HT Biotechnology, Cambridge, UK) and 4 ng μl−1 DNA in a final volume of 50 μl. The amplification comprised an initial denaturation step of 94°C for 2 min, followed by 35 cycles of 94°C for 45 s, 57°C for 45 s and 68°C for 45 s (for primer 781Rb) or 45 cycles of 94°C for 45 s, 60°C for 45 s and at 68°C for 45 s (for primer 781Ra), and a final extension of 68°C for 5 min. PCR reactions were performed in triplicates, which were pooled prior to purification. The 36 amplified PCR products were purified using the GeneJet PCR Purification Kit (Thermo Scientific, Waltham, MA, USA) and quantified using the Quant-iTPicoGreen dsDNA Assay Kit (Life Technologies, Carlsbad, CA, USA). Libraries were pooled in equimolar concentrations and sent to Beckman Coulter Genomics (Takeley, UK), where sequences were obtained using the 454 GS FLX+ Titanium platform (454 Life Sciences, Branford, CT, USA).
Pyrosequencing data analysis
Quality control of reads, removal of chimeric sequences and operational taxonomic unit (OTU) clustering were performed using UPARSE (Edgar 2013) according to Pessi et al. (in preparation). Two and zero mismatches were allowed to the primer and barcode sequences, respectively, and reads were required to have a maximum expected error of 0.5 and a length of 370 bp. Quality-filtered sequences were clustered into OTUs at 97.5% similarity, according to Taton et al. (2003). OTUs were classified using CREST (Lanzén et al. 2012) based on the Greengenes database (McDonald et al. 2012), and non-cyanobacterial OTUs were removed from the datasets. Furthermore, a manual inspection of the taxonomic classification was carried out in order to comply with the cyanobacterial taxonomy recently published by Komárek et al. (2014). In this classification, filamentous cyanobacteria from families Pseudanabaenaceae and Leptolyngbyaceae were placed into the order of Synechococcales. The order Chroococcales was considerably reduced, and some species were placed into the order of Synechococcales. The latter has thus become a large order with both unicellular and filamentous types and thus, in this work, we divide it into ‘Synechococcales (coccoid forms)’ and ‘Synechococcales (filamentous forms)’ for a better description of diversity.
The most closely related isolates and uncultured sequences for each OTU (based on the most abundant sequence type inside each OTU cluster) were retrieved using the SeqMatch tool from RDP (Cole et al. 2014). To avoid the inclusion of artefact OTUs, low abundance OTUs that had less than 97.5% similarity with their best SeqMatch isolate hit and were present in only one sample were discarded. A distance tree was constructed with the software package TREECON for Windows 1.3b (Van de Peer and De Wachter 1997), with an alignment of 412 positions obtained with MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar 2004) containing our OTUs, and the two most related sequences of isolates indicated by SeqMatch. The dissimilarity values were corrected for multiple substitutions by the model of Jukes and Cantor (1969) and were used to calculate a distance matrix. A distance tree was constructed by the Neighbour-joining method (Saitou and Nei 1987). Indels were not taken into account. A bootstrap analysis was performed that involved construction of 500 resampled trees. A manual curation based on such a phylogenetic analysis and a BLAST analysis had previously allowed to detect 13 OTUs that were either clusters of sequences of bad quality but related to an existing OTU (‘bad variants’), or seemed to be artefactual based on their isolation and low similarities. OTUs’ representative sequences have been deposited in GenBank under the accession numbers listed in Table S1 (Supporting Information).
To account for uneven sequencing depths across samples, diversity calculations were carried out after rarefying datasets to 1000 sequences. Samples SC3-4, SC3-5, SC3-6, SC4-1, SC4-2, SC4-7 and SC4-8, which consisted of less than 1000 sequences, were thus excluded from downstream analysis. Alpha diversity indices [Good's coverage, richness, Chao1 and Shannon's diversity index (H’)] were calculated using QIIME (Caporaso et al. 2010). Changes in phylotype richness and relative abundances of cyanobacterial orders across sites were assessed by Kruskal–Wallis non-parametric analysis of variance followed by Mann–Whitney posthoc tests using the PAST 3 software. Cyanobacterial community dissimilarities at the phylotype level were examined by non-metrical multidimensional scaling (NMDS) and significant differences in community structure across sites were assessed by a PERMANOVA routine using the Vegan package in R (Oksanen et al. 2013). For this, OTU abundance data were square root transformed and submitted to Wisconsin double standardization, and dissimilarities were calculated based on Bray–Curtis distances. The contribution of environmental parameters to changes in beta diversity was assessed by indirect gradient analyses (Spearman's rank correlation coefficients) using the Vegan package in R. For this, soil physico-chemical parameters were ln+1 transformed (except pH). Differences in OTU relative abundances across the sites were assessed using log-likelihood ratio tests in QIIME.
RESULTS
Soil parameters
Chemical analysis of the studied soil crusts is shown in Table 1. For some parameters, samples were found to represent a gradient corresponding to the level of soil crust development observed macroscopically (Fig. 1b). Total carbon content was lowest in the poorly developed, cyanobacteria-dominated soil crust (site SC1). It increased gradually and reached the highest value in the more developed, lichen-dominated soil crust (site SC4). The same gradient was detected for water content whereas an inverse relationship (from more- to less-developed) was observed for pH. In site SC2, the highest conductivity and Ca2+ concentration were observed. No significant difference was found in N-NH4, N-NO3 and P-PO4 concentrations between sites.
Table 1.
Chemical parameters of the studied soil crusts.
| Parameter | SC1 | SC2 | SC3 | SC4 |
|---|---|---|---|---|
| pH | 8.1 ± 0.01 (a) | 8.0 ± 0.08 (a) | 7.8 ± 0.06 (a,b) | 7.5 ± 0.05 (b) |
| Water content (%) | 19.6 ± 2.4 (a) | 30.8 ± 3.9 (b) | 35.3 ± 3.0 (b) | 34.5 ± 0.1 (b) |
| Conductivity (μS cm−1) | 101 ± 5 (a) | 365 ± 136 (b) | 242 ± 31 (a,b) | 269 ± 34 (a,b) |
| Total organic carbon (%) | 4.8 ± 0.2 (a) | 11.5 ± 2.0 (b) | 15.6 ± 2.7 (b,c) | 16.9 ± 0.8 (c) |
| N-NH4 (μg kg−1) | 3.5 ± 0.6 (a) | 4.0 ± 1.5 (a) | 5.7 ± 1.7(a) | 3.4 ± 0.6(a) |
| N-NO3 (μg kg−1) | 0.7 ± 0.5 (a) | 0.4 ± 0.1 (a) | 0.7 ± 0.1(a) | 4.4 ± 0.8(b) |
| P-PO4 (μg kg−1) | 19.1 ± 0.0 (a) | 15.6 ± 1.1 (a) | 18.3 ± 4.3 (a) | 19.4 ± 0.3 (a) |
| Ca2+ (μg g−1) | 5.8 ± 0.3 (a) | 16.5 ± 1.2 (b) | 7.6 ± 0.2 (c) | 8.2 ± 0.2 (c) |
| Mg2+ (μg kg−1) | 808 ± 39 (a) | 534 ± 54 (b) | 799 ± 40(a) | 417 ± 14(c) |
| K+ (μg kg−1) | 144 ± 5 (a) | 208 ± 16 (b) | 322 ± 40(c) | 221 ± 13 (b) |
| Na+ (μg kg−1) | 36 ± 12 (a,b) | 39 ± 10 (a,b) | 45 ± 4(a) | 18 ± 0.3(b) |
a–cValues are shown as the average (± SD) of the measurements of the three independent samples per site.
Same letters indicate no statistical difference between groups according to one-way ANOVA followed by Tukey's pairwise posthoc tests (P < 0.05).
Cyanobacterial community structure
Pyrosequencing analysis of partial 16S rRNA gene amplicons generated 292210 sequences from the 36 soil crust samples. After bioinformatic analysis and manual removal of spurious sequences, 94 592 cyanobacterial sequences remained, representing 136 OTUs at 97.5% similarity level. A distance analysis shows the large spread of the OTUs along the cyanobacterial radiation, their mutual relationships and to the most related isolates (Fig. S1, Supporting Information).
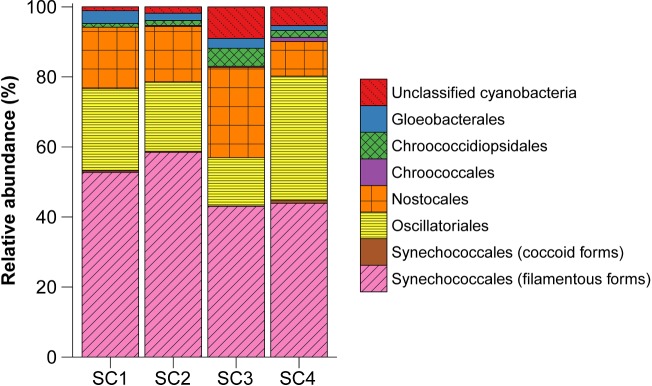
Taxonomic assignment of sequences revealed the presence of six cyanobacterial orders across the four studied sites (Fig. 2; Table S2, Supporting Information). Synechococcales (32 OTUs, 43.1%–58.7% of the sequences) was the dominant order, followed by Oscillatoriales (36 OTUs, 13.8%–35.5% of the sequences), Nostocales (25 OTUs, 9.9%–25.8% of the sequences), Chroococcidiopsidales (8 OTUs, 1.1%–5.1% of the sequences), Gloeobacterales (8 OTUs, 1.5%–3.6% of the sequences) and Chroococcales (14 OTUs, 0.03%–1.2% of the sequences). However, between 1.1% and 9.0% of the sequences (13 OTUs) could not be assigned to any cyanobacterial order by the used bioinformatic tool.
Figure 2.
Taxonomic assignment at the order level of OTUs found in the four soil crust samples.
Differences in the relative abundance of cyanobacterial orders were observed across the sites. Site SC3 presented a considerably lower abundance of Synechococcales and Oscillatoriales and a higher abundance of Nostocales in comparison to the other sites. The highest abundance of filamentous cyanobacteria from the order Synechococcales was found in site SC2. Filamentous cyanobacteria from the order Oscillatoriales were more abundant in site SC4 than in the others (Fig. 2; Table S2, Supporting Information).
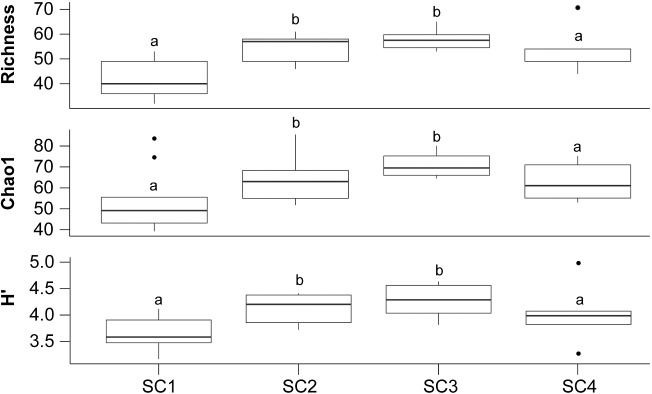
Good's coverage estimates ranged from 98.1% to 99.6%, indicating that the large majority of the cyanobacterial diversity was captured in the analysis. Phylotype richness ranged from 32 to 71 OTUs per sample (Fig. 3). The highest phylotype richness was observed in samples from sites SC2 and SC3. The same trend was observed with the Chao1 and Shannon's diversity index estimates.
Figure 3.
Cyanobacterial richness and diversity estimates calculated for the four soil crust samples. Identical letters indicate that there is no statistical difference between the groups according to a Kruskal–Wallis analysis of variance test followed by Mann–Whitney posthoc tests (P < 0.05, Bonferroni-corrected).
Beta-diversity analysis at the phylotype level showed that samples from the same site harbour cyanobacterial communities more similar to each other than to samples from the other sites (Fig. 4). Moreover, community dissimilarities were explained to some extent by the soil chemical properties, particularly total organic carbon, pH and water content. No influence was observed for Na+, N-NO3 and P-PO4.
Figure 4.
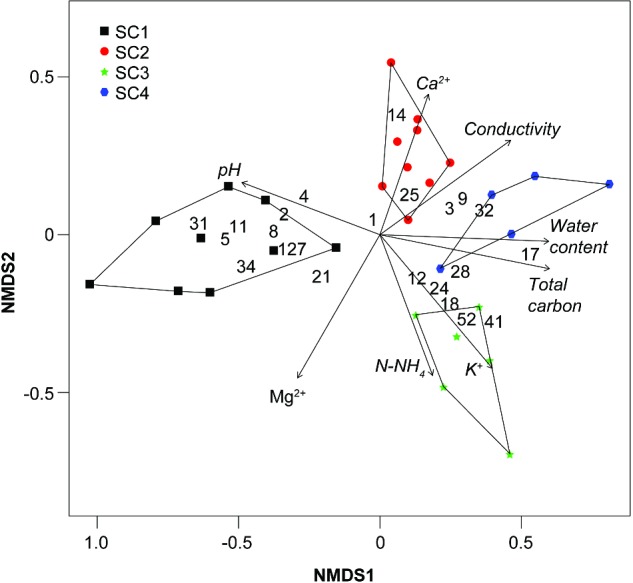
Non-metric multidimensional analysis. Numbers indicate OTUs with statistically different relative abundances across the four sites (Table 2).
Log-likelihood ratio tests identified OTUs with relative abundances that differed statistically across the four sites (Table 2). OTU1 (97.5% identity to Leptolyngbya sp. LLi18) was present in all sites but was the dominant OTU in sites SC2 and SC3 with statistically higher relative abundances. OTU2 (98.9% identity to Leptolyngbya antarctica ANT.L67.1) was the dominant phylotype in site SC1, but was also quite abundant in site SC2. Finally, OTU17 (98.6% similar to Phormidium sp. CYN64) was the dominant OTU in site SC4, but was also found at lower abundances in the other sites. In general, cyanobacterial communities were dominated by phylotypes related to the form-genera Leptolyngbya (94.8%–99.5% similarity), Calothrix (95.1% similarity), Coleofasciculus (95.1% similarity), Oscillatoria (100% similarity), Stigonema (98.9% similarity), Microcoleus (95.1% similarity) and Phormidium (96.4%–98.6% similarity).
Table 2.
List of OTUs with statistically different relative abundances across the four sites.
| Mean relative abundance (%) | |||||
|---|---|---|---|---|---|
| OTU | SC1 | SC2 | SC3 | SC4 | Best SeqMatch isolate hit; accession number (% ID) |
| OTU1 | 13.8 | 21.7 | 21.5 | 12.1 | Leptolyngbya sp. LLi18; DQ786166 (97.5%) |
| OTU2 | 16.4 | 13.2 | 2.8 | 3.0 | L. antarctica ANT.L67.1; AY493572 (98.9%) |
| OTU3 | 0.5 | 7.5 | 8.6 | 9.9 | L. nostocorum UAM 387; JQ070063 (98.4%) |
| OTU4 | 7.6 | 6.8 | 0.6 | 2.4 | Calothrix sp. KVSF5; EU022730 (95.1%) |
| OTU5 | 11.9 | 3.9 | 2.7 | 0.3 | Coleofasciculus chthonoplastes EcFYyyyy00; KC463190 (95.1%) |
| OTU8 | 9.2 | 3.5 | 1.1 | 3.0 | L. subtilissima EcFYyyy700; KC463197 (99.5%) |
| OTU9 | 0.3 | 2.6 | 1.2 | 2.7 | Leptolyngbya sp. CENA112; EF088337 (96.2%) |
| OTU11 | 6.8 | 1.9 | 0.8 | 0.1 | Oscillatoria sp. PCC 7112; AB074509 (100.0%) |
| OTU12 | 1.3 | 2.5 | 6.2 | 0.3 | Stigonema ocellatum SAMA 35; GQ354275 (98.9%) |
| OTU14 | 0.2 | 3.9 | 0.1 | 0.5 | Microcoleus sp. HTT-U-KK5; EF654070 (95.1%) |
| OTU17 | 0.2 | 0.4 | 5.3 | 26.5 | Phormidium sp. CYN64; JQ687330 (98.6%) |
| OTU18 | 0.3 | 0.5 | 6.1 | 3.1 | Unidentified cyanobacterium Ni2-C1; AB275351 (93.4%) |
| OTU21 | 4.8 | 2.9 | 6.5 | 1.5 | Nostoc sp. CCAP 1453/28; HF678493 (99.5%) |
| OTU24 | 0.3 | 0.7 | 3.1 | 0.7 | Chroococcidiopsis sp. CC1; DQ914863 (98.1%) |
| OTU25 | 0.1 | 1.7 | 1.4 | 0.3 | Oscillatoria duplisecta ETS-06; AM398647 (95.6%) |
| OTU28 | 0.2 | 0.7 | 2.5 | 1.5 | L. cavernicola LF-B5; HM748318 (98.6%) |
| OTU31 | 2.1 | 0.7 | 0.1 | 0.1 | G. violaceus PCC 8105; AF132791 (92.9%) |
| OTU32 | 0.0 | 1.2 | 1.2 | 2.3 | Oscillatoriales cyanobacterium EcFYyy200; KC463201 (99.5%) |
| OTU34 | 1.7 | 0.1 | 0.5 | 1.1 | Phormidium sp. LEGE 07317; HM217043 (96.4%) |
| OTU41 | 0.0 | 0.6 | 2.2 | 1.1 | Stigonema ocellatum SAMA 35; GQ354275 (94.2%) |
| OTU52 | 0.1 | 0.2 | 1.5 | 0.8 | Chroococcidiopsis sp. CC1; DQ914863 (94.2%) |
| OTU127 | 7.7 | 1.2 | 2.7 | 2.4 | Lo. tenuis PMC304.07; GQ859652 (99.5%) |
Numbers in bold indicate OTUs with higher relative abundances in specific sites according to log-likelihood test (P < 0.05, Bonferroni-corrected).
DISCUSSION
Cyanobacterial diversity in polar soil crusts may be restricted due to low water availability, high solar radiation, temperature fluctuations and frequent freeze-thawing cycles (Elster 2002). There are not a lot of studies dealing with Arctic soil crusts. However, it has been shown that cyanobacterial morphotype diversity and abundance there are limited compared to tropical and temperate regions (Kaštovská et al. 2005, 2007; Patova and Beljakova 2006). Here, we report the cyanobacterial diversity found in soil crust samples from Petunia Bay, Svalbard (Fig. 1) based on pyrosequencing of partial 16S rRNA gene sequences. Moreover, multiple samples were taken from four sites at different development stages, to take into consideration any patchiness of the cyanobacterial communities and the influence of the environmental conditions. To the best of our knowledge, this is the first indepth assessment of cyanobacterial diversity in Arctic soil crusts based on next-generation sequencing.
The level of soil crust development observed macroscopically in the studied sites was related to chemical parameters of the soil (Fig. 1b, Table 1). Collection of nine soil crust samples in each site allowed us to obtain a more global image of the cyanobacterial diversity present there and assess the influence of pH and nutrient availability on cyanobacterial community composition, as these parameters play a major role in soil crust development (Belnap and Lange 2001). Soil crusts at different stages of development and disturbance have been extensively studied in Australia (Eldridge, Semple and Koen 2000; Eldridge, Freudenberger and Koen 2006; Thompson, Eldridge and Bonser 2006). These studies showed a relation between soil crust components (lichens and mosses) and crust development, where bare soils hosted the most extensive and diverse cryptogamic communities. However, information about soil crusts in polar regions is limited. The most related study using pyrosequencing investigated the global bacterial diversity of High Arctic soil crusts in relation to water pulses (Steven et al. 2013). Their main finding was that Oscillatoriales were more abundant inside the water tracks than outside. Interestingly, the three cyanobacterial sequences that belonged to their 25 most abundant OTUs were either closely related to OTU703 (Nostoc) or to OTU11 (Oscillatoria) and OTU35 (Arthronema) of this study (98%–99% 16S rRNA similarity on the 370 common nucleotides). Colesie et al. (2014) illustrated the importance of water availability for soil crusts in extreme Antarctic terrestrial sites, but the emphasis was on the lichens and mosses and no cyanobacteria were observed in the very dry biotopes of the Diamond Hill. The few studies describing and comparing cyanobacterial communities in Arctic soil crusts at different stages of development, generally, focus on the succession of microbial communities that grow after glacier retreat (Kaštovská et al. 2005, 2007; Schütte et al. 2010; Knelman et al. 2012). However, these studies present whole microbial community without a detailed assessment of cyanobacteria, or the cyanobacterial community is only described using morphological methods.
Taxonomic assignment of the obtained sequences revealed a dominance of the orders Synechococcales, Oscillatoriales and Nostocales across all samples (Fig. 2; Table S2, Supporting Information), based on the most recent cyanobacterial classification (Komárek et al. 2014). Obtained results agreed with previous studies which have shown that Arctic soil crusts are dominated by filamentous cyanobacteria that can survive in extreme conditions due to their motility and mucilage production (Kaštovská et al. 2005; Strunecký, Elster and Komárek 2010). Filamentous cyanobacteria belonging to the Leptolyngbyaceae family were the most abundant in the four studied sites (relative abundances between 42.17%–57.44%). Heterocyst-forming cyanobacteria were mostly represented by Nostoc sp., Calothrix sp., Stigonema sp. Moreover, Nostocales are very important for nitrogen fixation in the Arctic (Zielke et al. 2005). In addition, they are widely distributed around the world in different types of soil crusts (Belnap and Lange 2001; Yeager et al. 2004; Řeháková, Chlumska and Dolezal 2011; Hu, Gao and Whitton 2012; Bastida et al. 2014). Unicellular cyanobacteria (mainly Chroococcales) were rare in all soil crust samples, whereas Gloeocapsa sp. together with Microcoleus sp. were observed in cyanobacterial soil crusts of the Dry Valleys in Antarctica (Colesie et al. 2014). However, Chroococcales were slightly more conspicuous in the well-developed soil crust (site SC4). The dominance of Chroococcales in lichenized soil crusts have been already shown in temperate regions (Bastida et al. 2014). On the other hand, the relative abundance of Oscillatoriales and Synechococcales (coccoid forms) gradually decreased from poorly developed soil crust (site SC1) to mid-developed soil crusts (sites SC2 and SC3, respectively) but greatly increased in the well-developed soil crust (site SC4). The opposite trend was observed for the relative abundance of Chroococcidiopsidales which increased from site SC1 to site SC3, and slightly decreased in site SC4. Based on 16S rRNA clone libraries, Chroococcidiopsis sequences appeared to be abundant in more developed soil crusts (Redfield et al. 2002; Yeager et al. 2004).
At the phylotype level, all soil crust samples were dominated by OTUs related to filamentous cyanobacteria from the genus Leptolyngbya (Table 2; Table S2, Supporting Information), common cyanobacteria in soil crusts around the world (Yeager et al. 2004; Kaštovská et al. 2005; Newsham, Pearce and Bridge 2010; Williams and Eldridge 2011; Strunecký, Elster and Komárek 2012; Osorio-Santos et al. 2014). The widespread filamentous cyanobacteria from the genus Phormidium were most abundant in the well-developed soil crust (site SC4). The sequence of OTU2, dominant in site SC1, was 98.9% similar to L. antarctica ANT.L67.1 isolated from an Antarctic microbial mat (Taton et al. 2006). This morphospecies is ubiquitous in Antarctic lakes (Anagnostidis and Komárek 1988) and has been suggested as endemic to the Antarctic continent (Komárek 2007). However, the sequence of OTU2 has already been found in lake from an Himalayan cold desert (Singh et al. 2014b) and in a freshwater stromatolite sample from Spain (Santos et al. 2010), but had not yet been reported in soil crusts. The OTU28 was related to a strain of Loriellopsis cavernicula isolated from a cave in Spain (98.6% similarity) (Lamprinou et al. 2011), which has likewise never been reported in polar soil crusts. On the other hand, several dominant OTUs (4, 12, 21 and 41), identified as heterocyst-forming cyanobacteria from the order Nostocales, have been previously observed in Arctic soils on the basis of their morphology (Elster et al. 1999; Zielke et al. 2005; Patova and Beljakova 2006; Řeháková, Chlumska and Dolezal 2011).
Relative abundances of OTU2 (98.9% similarity to L. antarctica), and OTU11 (100% similarity to Oscillatoria sp.) decreased with soil crust development. Apparently, these species are able to live in coarse and unstable soil crusts and they even dominated site SC1. This might be due to the reduced competition with other soil cyanobacteria. Interestingly, a sequence very similar to OTU11 has been found amongst the 25 most dominant bacterial OTUs in the biocrusts over permafrost soils of the High Arctic polar desert (Steven et al. 2013). On the contrary, the relative abundance of OTU3 (98.4% similar to L. nostocorum) and OTU17 (98.6% similar to Phormidium sp.) increased with soil crust development. The morphospecies L. nostocorum has already been found and isolated from lichenized well-developed soil crusts in USA (Flechtner, Johansen and Belnap 2008). Similarly, morphotypes related to the Oscillatoriales were more abundant in vegetated soil crusts than in poorly developed ones (Kaštovská et al. 2005).
An important caveat of trying to connect molecular data with morphotypes is that the databases of 16S rRNA gene sequences are still lacunous. If the sequence corresponding to a morphotype is unknown because no culture has yet been sequenced, it might be retrieved from environmental samples but will remain unidentified or loosely affiliated with a known morphotype, though it, in fact, corresponds to a quite different morphology. Another problem is that the strain identification linked to sequences in databases is not always documented with a morphological description.
A surprising result obtained in this study was the presence of OTUs assigned to the order Gloeobacterales in studied soil crusts. Only two species belonging to this order have been described so far: Gloeobacter violaceus, isolated from calcareous rock in Switzerland (Rippka, Waterbury and Cohen-Bazire 1974) and G. kilaueensis, found in epilithic biofilm in a lava cave in Kilauea Caldera, Hawaii (Saw et al. 2013). Gloeobacter-like sequences have been already reported in Ellesmere Island (High Arctic) from mats of Ward Hunt Lake (Jungblut, Lovejoy and Vincent 2010; Lionard et al. 2012), but have never been recorded in Arctic soil crusts. However, given that OTUs obtained in this study had 95.4% or lower identity to the database sequences of Gloeobacterales, their classification within this order cannot be claimed with confidence. Nevertheless, the possibility of Gloeobacterales being present in Arctic soil crusts should not be completely disregarded.
Phylotype richness estimates differed greatly between the four sites (Fig. 3). Site SC1 with poorly developed soil crust had the lowest phylotype richness, probably as a consequence of low concentration of nutrients, which greatly influence abundance of soil microbial community (Housman et al. 2006; Newsham, Pearce and Bridge 2010; Pietrasiak et al. 2013). Phylotype richness estimates were highest in the mid-developed soil crusts (sites SC2 and SC3) consolidated by a mixture of lichens and cyanobacteria, and decreased in site SC4 with stable lichenized soil crust. In lichenized soil crusts, cyanobacteria might compete with lichens for water, nutrients and light. Competition could be an important factor shaping the structure of microbial communities, and might explain the decrease in phylotype richness observed in well-developed soil crust, despite the higher availability of nutrients. However, there is no knowledge how competition of soil crust components influences the cyanobacterial community composition.
Multivariate analysis at the phylotype level showed that the four sites harbour distinct cyanobacterial communities (Fig. 4), mirroring the differences in chemical characteristics (Table 1) and development stage of soil crusts (Fig. 1b). Community dissimilarities were mostly explained by pH, total organic carbon and water content, as it was found by other authors (Li et al. 2010; Ganzert, Bajerski and Wagner 2014). Although a neutral to slightly alkaline pH interval is considered to be the optimum for cyanobacterial growth (Burja et al. 2002; Singh et al. 2014a), apparently, even small fluctuations in pH can influence cyanobacterial community composition. It has been suggested that the abundance of filamentous cyanobacteria increases with increasing pH (Nayak and Prasanna 2007). Our results agree with this observation, given the higher relative abundance of several OTUs related to filamentous cyanobacteria from the orders Synechococcales (OTUs 2, 8, 34 and 127) and Oscillatoriales (OTUs 5 and 11) in samples with higher pH (Table 2). Water availability is an important factor for Arctic soil crusts and positively influences soil crust community growth (Fischer and Subbotina 2014). Lack of water might cause a deceleration of the carbon production processes as already shown at 20% water content in High Arctic soil crusts, whereas a water content of 50% appeared optimal for photosynthesis and respiration rates (Yoshitake et al. 2010). Thus, it seems that soil crusts in this study were at the lower end of the values needed for growth (19.6%–35.3%). We can also infer that the nitrogen-fixation could be potentially more active in the later developmental stages due to a higher water content there (Stewart et al. 2014). In addition, the total organic carbon also increased with crust development (from site SC1 to SC4). Cyanobacteria are physiologically active only when wet (Belnap and Lange 2001) and, thus, it is logical to observe this parallel increase.
CONCLUSIONS
Our results indicate that the cyanobacterial community composition of Arctic soil crusts from the same type of substrate is diverse and changes considerably with the stage of development. Moreover, we have found that pH, total organic carbon and water content were the key parameters shaping the cyanobacterial communities. Lower concentration of total organic carbon, lower water content and higher pH resulted in lower cyanobacterial richness in poorly developed soil crusts where filamentous cyanobacteria were dominant. The increase of water content and concentration of total organic carbon and decrease in pH resulted in higher cyanobacterial richness in mid developed soil crusts, where, despite the dominance of Leptolyngbya sp, other filamentous and heterocyst-forming cyanobacteria were highly abundant. However, in stable well-developed soil crusts, in spite of the higher nutrient availability, the cyanobacterial richness appears to decrease, probably due to the competition with dense populations of lichens.
Supplementary Material
SUPPLEMENTARY DATA
FUNDING
This work was supported by the FRS-FNRS projects BIPOLES and PYROCYANO. IS Pessi is a FRIA PhD fellow from the FRS-FNRS, and A Wilmotte is a Research Associate of the FRS-FNRS of Belgium. The field work in Svalbard was funded by the Ministry of Education, Youth and Sports of the Czech Republic: (i) CzechPolar - Czech polar stations, construction and logistic expenses (LM2010009); and (ii) Establishing of working team and conditions for education in the field of polar ecology and life in extreme environments, (No.CZ.1.07/2.2.00/28.0190). The project is also funded by the European Social Fund and from the governmental budget of the Czech Republic.
Conflict of interest. None declared.
REFERENCES
- Anagnostidis K, Komárek J. Modern approach to the classification system of cyanophytes. 3. Oscillatoriales. Arch Hydrobiol. 1988;80:327–472. [Google Scholar]
- Bastida F, Jehmlich N, Ondono S, et al. Characterization of the microbial community in biological soil crusts dominated by Fulgensia desertorum (Tomin) Poelt and Squamarina cartilaginea (With.) P. James and in the underlying soil. Soil Biol and Biochem. 2014;76:70–9. [Google Scholar]
- Belnap J. Biological crusts. In: Lal R, editor. Encyclopedia of Soil Science. New York: Taylor and Francis Group; 2008. pp. 1–4. [Google Scholar]
- Belnap J, Lange OL. Biological Soil Crust: Structure, Function and Management. Berlin, Germany: Springer-Verlag; 2001. [Google Scholar]
- Boutte C, Grubisic S, Balthasart P, et al. Testing of primers for the study of cyanobacterial molecular diversity by DGGE. J Microbiol Meth. 2006;65:542–50. doi: 10.1016/j.mimet.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Burja AM, Abu-Mansour E, Banaigs B, et al. Culture of marine cyanobacterium, Lyngbya majuscula (Oscillatoriaceae), for bioprocess intensified production of cyclic and linear lipopeptides. J Microbiol Meth. 2002;48:207–19. doi: 10.1016/s0167-7012(01)00324-4. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Fierer N, Lauber CL, et al. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;41:633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colesie C, Gommeaux M, Green ATG, et al. Biological soil crusts in continental Antarctica: Garwood Valley, southern Victoria Land, and Diamond Hill, Darwin Mountains region. Antarct Sci. 2014;26:115–23. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Eldridge DJ, Freudenberger D, Koen TB. Diversity and abundance of biological soil crust taxa in relation to fine and coarse-scale disturbances in a grassy eucalypt woodland in eastern Australia. Plant Soil. 2006;281:255–68. [Google Scholar]
- Eldridge DJ, Semple WS, Koen TB. Dynamics of cryptogamic soil crusts in a derived grassland in south-eastern Australia. Austral Ecol. 2000;25:232–40. [Google Scholar]
- Elster J. Ecological classification of terrestrial algal communities of polar environment. In: Beyer L, Boelter M, editors. GeoEcol of Terrestrial Oases Ecological Studies. Berlin, Heidelberg: Springer-Verlag; 2002. pp. 303–19. [Google Scholar]
- Elster J, Lukešová A, Svoboda J, et al. Diversity and abundance of soil algae in the polar desert, Sverdrup Pass, central Ellesmere Island. Polar Rec. 1999;35:231–54. [Google Scholar]
- Fischer T, Subbotina M. Climatic and soil texture threshold values for cryptogamic cover development: a meta analysis. Biologia. 2014;69:1520–30. [Google Scholar]
- Flechtner VR, Johansen JR, Belnap J. The Biological Soil Crusts of the San Nicolas Island: Enigmatic Algae from a Geographically Isolated Ecosystem. West N Am Naturalist. 2008;68:405–36. [Google Scholar]
- Ganzert L, Bajerski F, Wagner D. Bacterial community composition and diversity of five different permafrost-affected soils of Northeast Greenland. FEMS Microbiol Ecol. 2014;89:426–41. doi: 10.1111/1574-6941.12352. [DOI] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:4. [Google Scholar]
- Housman DC, Powers HH, Collins AD, et al. Carbon and nitrogen fixation differ between successional stages of biological soil crusts in the Colorado Plateau and Chihuahuan Desert. J Arid Environ. 2006;66:620–34. [Google Scholar]
- Hu C, Gao K, Whitton BA. Semi-arid regions and deserts. In: Whitton BA, editor. Ecology of Cyanobacteria II: Their Diversity in Space and Time. Netherlands: Springer Science; 2012. pp. 345–69. [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. Vol. 3. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Jungblut AD, Lovejoy C, Vincent WF. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 2010;4:191–202. doi: 10.1038/ismej.2009.113. [DOI] [PubMed] [Google Scholar]
- Kaštovská K, Elster J, Stibal M, et al. Microbial Assemblages in soil microbial succession after glacial retreat in Svalbard (High Arctic) Microbial Ecol. 2005;50:396–407. doi: 10.1007/s00248-005-0246-4. [DOI] [PubMed] [Google Scholar]
- Kaštovská K, Stibal M, Sabacka M, et al. Microbial community structure and ecology of subglacial sediments in two polythermal Svalbard glaciers characterized by epifluorescence microscopy and PLFA. Polar Biol. 2007;30:277–87. [Google Scholar]
- Knelman JE, Legg TM, O'Neill SP, et al. Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol Biochem. 2012;46:172–80. [Google Scholar]
- Komárek J. Phenotype diversity of the cyanobacterial genus Leptolyngbya in maritime Antarctica. Pol Polar Res. 2007;28:211–31. [Google Scholar]
- Komárek J, Kaštovský J, Mareš J, et al. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014 using a polyphasic approach. Preslia. 2014;86:295–35. [Google Scholar]
- Lamprinou V, Hernández-Mariné M, Canals T, et al. Morphology and molecular evaluation of Iphinoe spelaeobios gen. nov., sp. nov. and Loriellopsis cavernicola gen. nov., sp. nov., two stigonematalean cyanobacteria from Greek and Spanish caves. Int J Syst Evol Micr. 2011;61:2907–15. doi: 10.1099/ijs.0.029223-0. [DOI] [PubMed] [Google Scholar]
- Lanzén A, Jørgensen SL, Huson DH, et al. CREST – classification resources for environ sequence tags. PLoS One. 2012;7::e49334. doi: 10.1371/journal.pone.0049334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Láska K, Witoszová D, Prošek P. Weather patterns of the coastal zone of Petuniabukta, central Spitsbergen in the period 2008–2010. Pol Polar Res. 2012;33:297–318. [Google Scholar]
- Li X-R, He M-Z, Zerbe S, et al. Micro‐geomorphology determines community structure of biological soil crusts at small scales. Earth Surf Proc Land. 2010;35:932–40. [Google Scholar]
- Lionard M, Péquin B, Lovejoy C, et al. Benthic Cyanobacterial Mats in the High Arctic: Multi-Layer Structure and Fluorescence Responses to Osmotic Stress. Front Microbiol. 2012;3:140. doi: 10.3389/fmicb.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moquin SA, Garcia JR, Brantley SL, et al. Bacterial diversity of bryophyte-dominant biological soil crusts and associated mites. J Arid Environ. 2012;87:110–7. [Google Scholar]
- Nayak S, Prasanna R. Soil pH and its role in cyanobacterial abundance and diversity in rice field soils. Appl Ecol Environ Res. 2007;5:103–13. [Google Scholar]
- Newsham KK, Pearce DA, Bridge PD. Minimal influence of water and nutrient content on the bacterial community composition of a maritime Antarctic soil. Microbiol Res. 2010;165:523–30. doi: 10.1016/j.micres.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microb. 1997;63:3327–32. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. Vegan: Community Ecology Package. R package version 2.0-7. 2013. http://CRAN.R-project.org/package=vegan (24 November 2015, date last accessed)
- Osorio-Santos K, Pietrasiak N, Bohunicka M, et al. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. Eur J Phycol. 2014;49:450–70. [Google Scholar]
- Patova EN, Beljakova RN. Terrestrial cyanoprokaryota of Bolshevik island (Severnaya Zemlya Archipelago) Novitates Systematicae Plantarum Non Vascularium. 2006;40:83–91. (in Russian) [Google Scholar]
- Pietrasiak N, Regus JU, Johansen JR, et al. Biological soil crust community types differ in key ecological functions. Soil Biol Biochem. 2013;65:168–71. [Google Scholar]
- Pushkareva E, Elster J. Biodiversity and ecological typification of cryptogamic soil crust in the vicinity of Petunia Bay, Svalbard. Czech Polar Rep. 2013;3:7–18. [Google Scholar]
- Rachlewicz G, Szczucinsky W, Ewertowski M. Post-“Little Ice Age” retreat rates of glaciers around Billefjorden in central Spitsbergen, Svalbard. Pol Polar Res. 2007;28:159–86. [Google Scholar]
- Redfield E, Barns SM, Belnap J, et al. Comparative diversity and composition of cyanobacteria in three predominant soil crusts of the Colorado Plateau. FEMS Microbiol Ecol. 2002;40:55–63. doi: 10.1111/j.1574-6941.2002.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Řeháková K, Chlumska Z, Dolezal J. Soil cyanobacterial and microalgal diversity in dry Mmountains of Ladakh, NW Himalaya, as related to site, altitude, and vegetation. Microb Ecol. 2011;62:337–46. doi: 10.1007/s00248-011-9878-8. [DOI] [PubMed] [Google Scholar]
- Rippka R, Waterbury J, Cohen-Bazire G. A cyanobacterium which lacks thylakoids. Arch Microbiol. 1974;100:419–36. [Google Scholar]
- Rossi F, Potrafka RM, Pichel FG, et al. The role of the exopolysaccharides in enhancing hydraulic conductivity of biological soil crusts. Soil Biol Biochem. 2012;46:33–40. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santos F, Pena A, Nogales B, et al. Bacterial diversity in dry modern freshwater stromatolites from Ruidera Pools Natural Park, Spain. Syst Appl Microbiol. 2010;33:209–21. doi: 10.1016/j.syapm.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Saw JH, Schatz M, Brown MV, et al. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a Lava Cave in Kilauea Caldera, Hawai'i. PloS One. 2013;8:e76376. doi: 10.1371/journal.pone.0076376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütte UME, Abdo Z, Foster J, et al. Bacterial diversity in a glacier foreland of the high Arctic. Mol Ecol. 2010;19:54–66. doi: 10.1111/j.1365-294X.2009.04479.x. [DOI] [PubMed] [Google Scholar]
- Singh SS, Kunui K, Minj RA, et al. Diversity and distribution pattern analysis of cyanobacteria isolated from paddy fields of Chhattisgarh, India. J Asia-Pacific Biodivers. 2014a;7:462–70. [Google Scholar]
- Singh Y, Khattar J, Singh DP, et al. Limnology and cyanobacterial diversity of high altitude lakes of Lahaul-Spiti in Himachal Pradesh, India. J Biosci. 2014b;39:643–57. doi: 10.1007/s12038-014-9458-4. [DOI] [PubMed] [Google Scholar]
- Steven B, Lionard M, Kuske CR, et al. High bacterial diversity of biological soil crusts in water tracks over permafrost in the high Arctic Polar Desert. PLoS One. 2013;8::e71489. doi: 10.1371/journal.pone.0071489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart KJ, Grogan P, Coxson DS, et al. Topography as a key factor driving atmospheric nitrogen exchanges in arctic terrestrial ecosystems. Soil Biol Biochem. 2014;70:96–112. [Google Scholar]
- Strunecký O, Elster J, Komárek J. Phylogenetic relationships between geographically separate Phormidium cyanobacteria: is there a link between north and south polar regions? Polar Biol 2010. 33 1419–28. [Google Scholar]
- Strunecký O, Elster J, Komárek J. Molecular clock evidence for survival of Antarctic cyanobacteria (Oscillatoriales, Phormidium autumnale) from Paleozoic times. FEMS Microbiol Ecol. 2012;82:482–90. doi: 10.1111/j.1574-6941.2012.01426.x. [DOI] [PubMed] [Google Scholar]
- Taton A, Grubisic S, Brambilla E, et al. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (Mc Murdo Dry Valleys, Antarctica): A morphological and molecular approach. Appl Environ Microb. 2003;69:5157–69. doi: 10.1128/AEM.69.9.5157-5169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A, Grubisic S, Ertz D, et al. Polyphasic study of Antarctic cyanobacterial strains. J Phycol. 2006;42:1257–70. [Google Scholar]
- Thompson WA, Eldridge DJ, Bonser SP. Structure of biological soil crust communities in Callitris glaucophylla woodlands of New South Wales, Australia. J Veg Sci. 2006;17:271–80. [Google Scholar]
- Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–30. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- Williams W, Eldridge DJ. Deposition of sand over a cyanobacterial soil crust increases nitrogen bioavailability in a semi-arid woodland. Appl Soil Ecol. 2011;49:26–31. [Google Scholar]
- Yeager CM, Kornosky JL, Housman DC, et al. Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado Plateau and Chihuahuan Desert. Appl Eviron Microb. 2004;70:973–83. doi: 10.1128/AEM.70.2.973-983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake S, Uchida M, Koizumi H, et al. Production of biological soil crusts in the early stage of primary succession on a high Arctic glacier foreland. New Phytol. 2010;86:451–60. doi: 10.1111/j.1469-8137.2010.03180.x. [DOI] [PubMed] [Google Scholar]
- Zielke M, Solheim B, Spjelkavik S, et al. Nitrogen fixation in the high Arctic: role of vegetation and environmental conditions. Arct Antarct Alp Res. 2005;37:372–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.