Abstract
Poly(L-diaminopropionic acid) (PDAP) is one of the four homopoly(amino acid)s that have been discovered in nature. However, the molecular mechanism of PDAP biosynthesis has yet to be described. In this work, the general layout of the PDAP biosynthetic pathway is characterised in Streptomyces albulus PD-1 by genome mining, gene disruption, heterologous expression and in vitro feeding experiments. As a result, L-diaminopropionic acid (L-DAP), which is the monomer of PDAP, is shown to be jointly synthesised by two protein homologues of cysteine synthetase and ornithine cyclodeaminase. Then, L-DAP is assembled into PDAP by a novel nonribosomal peptide synthetase (NRPS) with classical adenylation and peptidyl carrier protein domains. However, instead of the traditional condensation or thioesterase domain of NRPSs, this NRPS has seven transmembrane domains surrounding three tandem soluble domains at the C-terminus. As far as we know, this novel single-module NRPS structure has only been reported in poly(ε-L-lysine) synthetase. The similar NRPS structure of PDAP synthetase and poly(ε-L-lysine) synthetase may be a common characteristic of homopoly(amino acid)s synthetases. In this case, we may discover and/or design more homopoly(amino acid)s by mining this kind of novel NRPS structure in the future.
In recent years, the homopoly(amino acid)s secreted by microorganisms have been extensively investigated because of their unique characteristics. Homopoly(amino acid)s are distinguished from common peptides and proteins because homopoly(amino acid)s are composed of a single type of amino acids1. To date, more than 300 types of amino acids have been discovered. However, only four homopoly(amino acid)s, namely, poly(γ-glutamic acid) (γ-PGA), poly(ε-L-lysine) (ε-PL), poly(γ-L-diaminobutanoic acid) (γ-PAB) and poly(L-diaminopropionic acid) (PDAP) have been found in nature1,2,3 (Fig. 1). The scarcity of homopoly(amino acid)s is probably due to the lack of suitable synthetase that can assemble amino acid monomers into homopoly(amino acid)s. Among the four kinds of homopoly(amino acid)s, γ-PGA and ε-PL have many applications; for example, γ-PGA has been broadly used in foods, cosmetics, medicines, water treatment and agriculture4,5. ε-PL has also been extensively used in foods, medicines, new materials and electronics; furthermore, ε-PL has been approved as a natural food preservative in Japan, the United States, China, and other countries6,7.
Figure 1. Chemical structures of the discovered homopoly(amino acid)s.
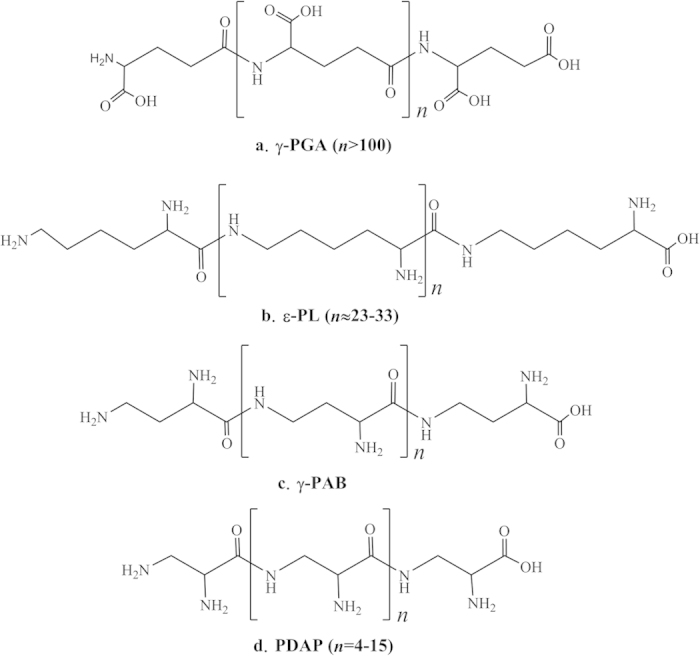
(a) γ-PGA, (b) ε-PL, (c) γ-PAB, and (d) PDAP.
In a previous study, PDAP, the fourth homopoly(amino acid)s, was purified and identified from the culture broth of Streptomyces albulus PD-1, a well-known ε-PL-producing strain. Structurally, PDAP is an L-α,β-diaminopropionic acid (L-DAP) oligomer that links the amine functional group and the carboxylic acid functional group; this substance has a molecular mass distribution ranging from 0.5 to 1.5 kDa3. With a novel polycationic structure, PDAP, similar to ε-PL, can be regarded as a potentially specific and advanced platform for many fields. For example, it can be used as a natural food preservative, as a non-viral gene delivery vector or as a raw material for hydrogels. Despite their similarity, PDAP exhibits a higher charge density than ε-PL. Thus, PDAP is more suitable for some applications than ε-PL. As far as we know, PDAP shows efficient antibacterial activities against various microorganisms and exhibits stronger inhibitory activities against yeasts than ε-PL3. In our previous work, PDAP fermentation was optimised using a pH- and dissolved oxygen-controlled strategy; this optimised process yielded 9.6 g/L PDAP8. However, the genes or enzymes involved in PDAP biosynthesis remain unknown.
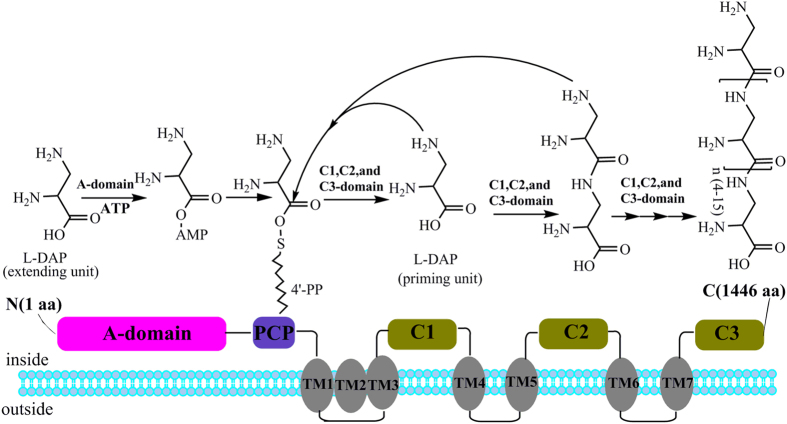
In previous studies, the enzymes involved in the biosynthesis of γ-PGA and ε-PL have already been described to some extent. In γ-PGA biosynthesis, two different assembly modes, the thiotemplate mechanism catalysed by non-ribosomal peptide synthetases (NRPSs) and the amide-ligation mechanism catalysed by amide ligases, have been postulated. In the thiotemplate mechanism, L-glutamic acid is initially activated by ATP; this substance then binds to a sulfhydryl group and isomerises into a D-form, and the isomerised product is subsequently elongated to form the D-homopolymer of γ-PGA9. In the amide-ligation mechanism, ATP is transferred to activate the terminal carboxyl group of a growing γ-PGA chain. Subsequently, a new amide linkage is formed via nucleophilic attack by the amino group of glutamic acid to elongate the γ-PGA chain to the D/L-homopolymer5,10. In ε-PL, L-lysine is catalysed to ε-PL by an unusual NRPS comprised of an adenylation domain (A domain), a peptidyl carrier protein domain (PCP domain) and three special tandem domains surrounding six transmembrane (TM) domains11. In this process, an L-lysine monomer is initiated in the N-terminus by the adenylated A domain and the resulting lysyl-O-AMP is transferred to the PCP domain. Then, the unusual C-terminal tandem domains catalyse peptide bond formation and iteratively extend the biopolymer. Such work raises the following interesting questions: What is the structure of the PDAP synthetase (PDAPs)? How does it assemble L-DAP into a biopolymer? Is there any similarity between the PDAPs and the reported homopoly(amino acid)s synthetases?.
In this study, candidate genes for the de novo biosynthesis of PDAP were predicated from S. albulus PD-1 by sequencing and annotating its genome. The putative genes were identified and characterised using genetic inactivation, heterologous expression and biochemical assays. In particular, the PDAPs was found to be a novel NRPS with a typical A domain, a typical PCP domain and seven TM domains surrounding three tandem soluble domains in the C-terminus. To our knowledge, this novel single-module NRPS structure has only been reported in the poly(ε-L-lysine) synthetase (Pls). Our findings form the basis for genetic and biochemical studies into PDAP biosynthesis. Moreover, the similar NRPS structure of PDAPs and Pls provides us insight into the discovery and/or design of numerous homopoly (amino acid)s in the future.
Results and Discussion
Prediction and identification of the PDAP synthetase
Peptide natural products are commonly synthesised via either ribosomal or non-ribosomal mechanisms1,12. In contrast with the ribosomal machinery and its 21 amino acids building blocks for protein biosynthesis, the non-ribosomal machinery can accept many non-proteinogenic amino acid building blocks as substrates. PDAP is a biopolymer comprised of L-DAP, a novel non-proteinogenic amino acid. Thus this biopolymer is most likely assembled by an NRPS. Recently, with the rapid development of genome sequencing technologies and bioinformatics, an efficient platform for the discovery and identification of new metabolic pathways has been established via genome mining13,14,15,16,17. For example, the genetic variations associated with ε-PL biosynthesis in S. albulus ZPM have been identified through genome sequencing18. A novel xylose isomerase in the xylose utilisation pathway in Bacillus coagulans has also been identified through genome analysis17. Therefore, we initially determined the biosynthetic pathway of PDAP based on the genome data of S. albulus PD-119. We investigated the S. albulus PD-1 genome and found eight NRPS genes. The substrate specificities of these NRPSs were predicted using NRPSpredictor2. The in silico analysis results are listed in Table 1. β-lysine, the substrate of gb|EXU85975.1|, which is an aliphatic amino acid with an amino group at the β position, is most similar to L-DAP in structure (Supplementary Fig. S1). Thus, gb|EXU85975.1| was considered the most likely PDAPs. To verify this hypothesis, we constructed a gene disruption mutant of the putative pdaps gene(Fig. 2A). The pdaps disruption was confirmed by PCR and Southern blot hybridization (Supplementary Fig. S2). Then the culture broth of the disruptant was detected. As shown in Fig. 2B, the disruption of the putative pdaps terminated PDAP production, and this finding provided strong evidence that PDAPs had been correctly identified. Moreover, the mutant strains exhibited poorer spore production activity than wild-type strains.
Table 1. The substrate specificity of those NRPSs in S. albulus PD-1.
| Sequence ID | Substrate cluster description | Nearest neighbor target specificities |
|---|---|---|
| gb|EXU92123.1| | Aliphatic side chain with B-bond donor | glutamine |
| gb|EXU92158.1| | Aliphatic side chain with B-bond donor | lysine |
| gb|EXU92697.1| | Aliphatic side chain with B-bond donor | serine |
| Apolar, aliphatic side chains | glycine | |
| gb|EXU88741.1| | Aliphatic side chain with B-bond donor | asparagine |
| gb|EXU86680.1| | Apolar, aliphatic side chains | phenylalanine |
| gb|EXU85975.1| | Aliphatic, branched hydrophobic side chain | β-lysine |
| gb|EXU91762.1| | Aliphatic side chain with B-bond donor | glutamine |
| gb|EXU90606.1| | hydrophobic-aliphatic | lysine |
Figure 2. Construct used to obtain the loss-of-function mutants.
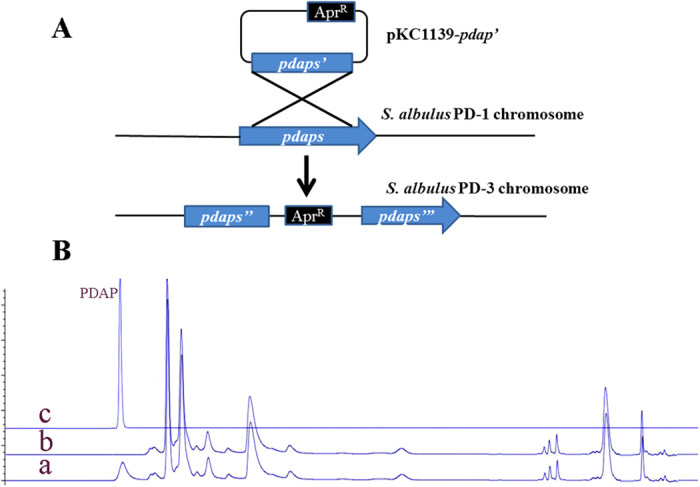
(A) Diagram of putative pdaps disruption. (B) HPLC analysis of the culture of the predicted PDAP-inactivated strain: a, culture broth of the wild-type S. albulus PD-1 strain; b, culture broth of the predicted PDAPs-inactivated strain; c, a PDAP standard.
To further verify the identity of the hypothesised PDAPs, the target gene was expressed in a heterologous host, and an in vitro feeding experiment was conducted. In previous studies, the ε-PL synthetase gene (pls) was expressed via the control of a constitutive promoter; however, neither Pls expression nor ε-PL production was observed. Ultimately, the Pls was successfully expressed in S. albulus and S. lividans under the control of its own promoter20,21. The researchers posited that the Pls expression was probably strictly controlled because Pls is a membrane protein and the constitutive promoter was not suitable for the transcription of Pls. Because similar TM domains also existed in PDAPs, we expressed PDAPs via the control of its own promoter in S. lividans TK24, a model streptomycete commonly used as a host strain for heterologous expression. Positive colonies were selected and cultured in M3G medium supplemented with L-DAP (1 g/L) for one week; subsequently, the supernatant of the culture broth was characterised via high-performance liquid chromatography (HPLC). However, not any significant characteristic peak was detected through HPLC possibly because of the low concentration (<0.05 g/L) of PDAP produced by the recombinant strain. In contrast, the culture broth supernatant tested positive with Dragendorff reagent, which is frequently used to screen alkaloids (Fig. 3A). Thus, the culture broth supernatant from the recombinant strain was concentrated, purified and evaluated with matrix-assisted laser desorption ionisation-time of flight mass spectrometry (MALDI-TOF/MS) using a spectrometer (Autoflex 2, Bruker Daltonics Inc.,USA). The results revealed the presence of a polydisperse PDAP group (Fig. 3B); this group exhibited the same molecular weight as previously reported3. Gene interruption, heterologous expression and in vitro feeding tests rigorously demonstrated that the identified novel NRPS (gb|EXU85975.1|) was indeed the PDAPs.
Figure 3.
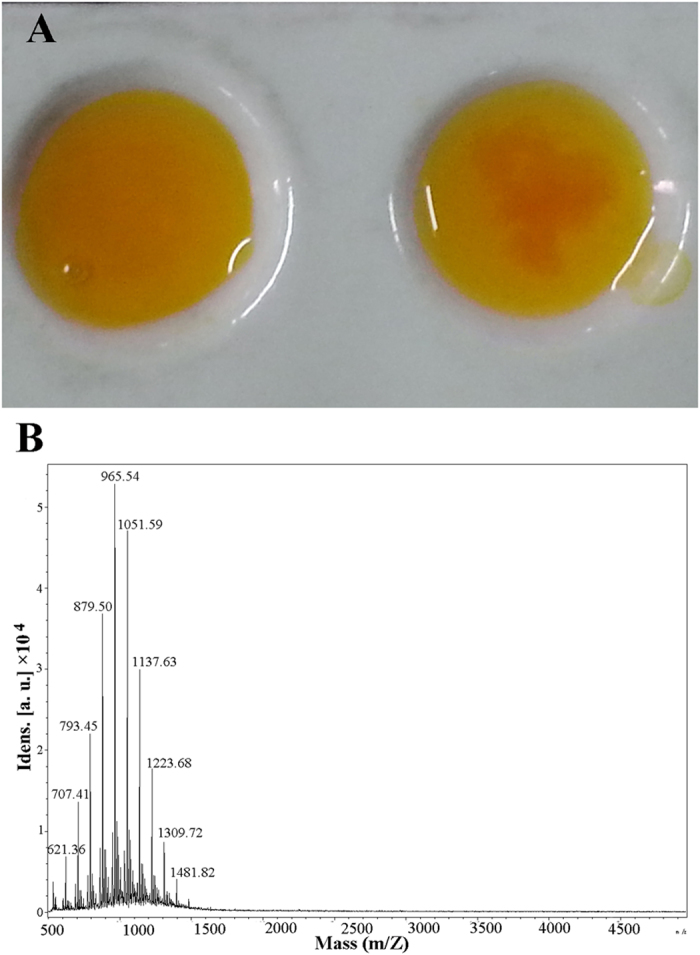
(A) Reaction between culture supernatant and Dragendorff reagent: Culture supernatant of (a) S. lividans TK24 and (b) S. lividans TK24-pdaps. (B) PDAP was purified from the culture broth of S. lividans TK24-pdaps and subsequently analysed through MALDI-TOF/MS. The regular peak at an equal interval is 86.05, which is equal to the molecular of L-DAP (104.07) deducts the molecular of H2O (18.02).
Bioinformatic analysis of the PDAP synthetase
Based on the genome sequence of S. albulus PD-1, the PDAPs is encoded by a single ORF of 4,338 bp. With the nucleotide sequence and protein sequence in hand, the structure and function of the enzyme were investigated. In general, NRPSs contain three core domains. The A domain selects and activates the candidate amino acid as an amino acyladenylate. Then, the PCP domain covalently binds the activated monomer via a phosphopantetheinyl arm. The C domain then catalyses the formation of the peptide linkage between the activated amino acids from two adjacent PCP modules. These three domains constitute the repeating module responsible for the activation and incorporation of an amino acid into the growing peptide chain. Finally, a mature peptide is released from the PCP domain, and this release is catalysed by a C-terminal thioesterase (TE) domain22,23,24. The conserved domains of PDAPs were analysed, and the results revealed that PDAPs exhibited an unusual NRPS structure, with a classical A domain and a classical PCP domain in the N-terminus. However, PDAPs did not contain a domain exhibiting significant sequence similarity to the traditional C domains that are essential for peptide bond formation in NRPSs. Furthermore, PDAPs did not contain a traditional TE domain, which catalyses the release of the final product from NRPS enzymes. Instead, a NRPS terminal domain with an unknown function was found in the C-terminal region of PDAPs. This identified domain was approximately 779 amino acids in length and roughly thrice the size of a traditional TE domain. Physicochemical analysis suggested that seven TM domains were present in this novel domain and this novel domain was divided into three subdomains on the basis of the location of the TM domains (Supplementary Fig. S3). A sequence alignment revealed that the three subdomains displayed substantial sequence similarity (with pairwise identities of 23.1%, 26.1% and 22.6%; Supplementary Fig. S4A). In fact, the structure with three tandem soluble domains surrounded by several TM domains has only been reported in Pls; all three tandem domains were essential for peptide bond formation in ε-PL biosynthesis11,25. Given that these sequences may play the same role in peptide bond formation in PDAPs, we named theses tandem sequences the C1, C2 and C3 domain, similar to those found in Pls. In fact, multiple sequence alignments of the C1, C2, and C3 domains in PDAPs and Pls revealed a few universally conserved amino acids in their structures (Supplementary Fig. S4B). The similar NRPS structures of PDAPs and Pls may be a common characteristic of homopoly(amino acid)s synthetase. For further, this novel type NRPS was mined from the genome database, and many NRPSs with similar structure were discovered, for example, ref|WP_052230086.1|, gb|AJE44528.1|, gb|AHH98491.1|. All of these proteins are presumed to be related to peptide synthesis, but none of them was studied intensively. This is a great resource for homopoly(amino acid)s investigation. As such, we can discover more homopoly(amino acid)s by mining for this novel type of NRPS in the future.
On the basis of the structure of PDAPs, we predicted the mechanism of PDAPs. As shown in Fig. 4, L-DAP is initially activated by an A domain, the active L-DAP monomer is transferred by PCP domain, and three unusual C-terminal tandem domains catalyse peptide bond formation and iteratively extend the polymer. However, several factors remain unknown. For example, a traditional TE domain was not detected in PDAPs; further studies have yet to determine the mechanism by which the release of peptides are catalysed and the conditions that yield a PDAP molecular mass distribution from 0.5 to 1.5 kDa.
Figure 4. Domain architecture and predicted mechanism of PDAPs.
A domain, PCP domain, seven TM domains and three tandem domains (C1, C2 and C3 domains) are shown schematically.
Substrate specificity analysis of the adenylation domain of PDAPs
Given that PDAPs can accept L-DAP as a substrate, we determined whether this novel NRPS could tolerate additional substrate diversity. In NRPS modules, the A domains select and activate the incoming monomeric amino acids26. Hence, the substrate specificity of the A domain of PDAPs was investigated. The amino acid-dependent PPi release assay was conducted to evaluate the substrate specificity of the A domain. As a result, the highest PPi level was detected using L-DAP as a substrate (Fig. 5); no other adenylation activity was observed when L-DAP was replaced with L-lysine, L-alanine, L-valine or L-glycine. Moreover, the adenylation activity was weaker when L-diaminobutyric acid or β-lysine was used as a substrate than when L-DAP was used. However, no significant product was detected via HPLC or Dragendorff reagent when L-diaminobutyric acid or β-lysine was fed to S. lividans TK24-pdaps. Thus, other portions of PDAPs may exert regulatory selection outside of the A domain. In general, the results of the PPi release assay were slightly different from those of the in silico prediction from NRPSpredictor2, which suggested that the preferred substrate of the A domain is β-lysine. As noted before, the NRPS predictor yields suboptimal results when small substrates are used27; thus it is reasonable that the predicted results are slightly inconsistent with our biochemical assay results because L-DAP is a small amino acid.
Figure 5. Substrate specificity of the A domain of PDAPs.
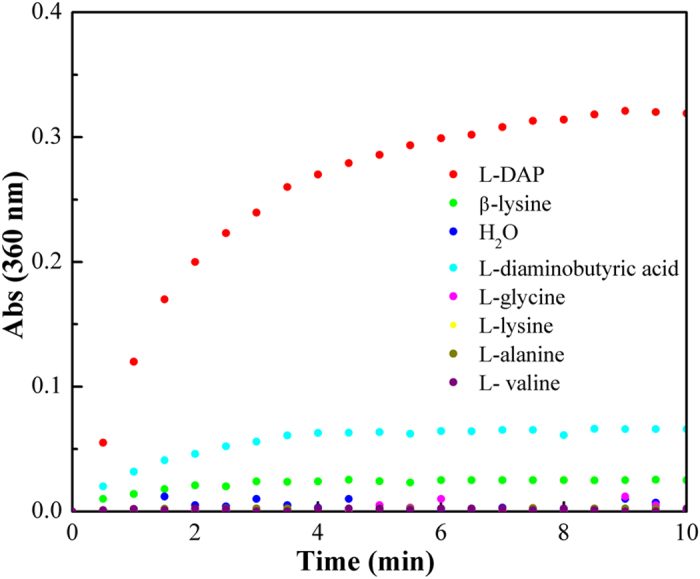
Substrate specificity was indicated by the released PPi level.
Identification of the L-DAP biosynthetic pathway
In the above work, PDAP was shown to be biosynthesised from L-DAP through a novel NRPS in S. albulus PD-1. Then, the biosynthetic pathway of L-DAP was identified in the following work. In previous studies, L-DAP has been reported to be a precursor of some antibiotics, such as viomycin, the capreomycins, and zwittermicin A28,29,30,31. In addition, L-DAP is also a precursor molecule of staphyloferrin B, one of the two siderophores in Staphylococcus aureus32,33. In the past decades, the biosynthetic pathway of L-DAP in some strains has been investigated step by step. Precursor labelling has been applied to the capreomycins and viomycin biosynthetic pathways, and results have demonstrated that L-serine is the precursor of L-DAP28,29. Subsequently, experiments and the analysis of the gene clusters of viomycin, the capreomycins, zwittermicin A and staphyloferrin B suggest that L-DAP biosynthesis is catalysed by the concerted actions of two enzymes30,31,32,33,34. Based on the bioinformatic analysis of these two enzymes, several proposed mechanisms were proposed for L-DAP biosynthesis. Recently, Marek et al. first confirmed O-phospho-L-serine and L-glutamate are the substrates of L-DAP formation in Staphylococcus aureus35. On the basis of these findings, we assumed that L-DAP biosynthesis in S. albulus PD-1 may follow the same pathway and the proteins involved in L-DAP synthesis were mined from the genome data of S. albulus PD-1.
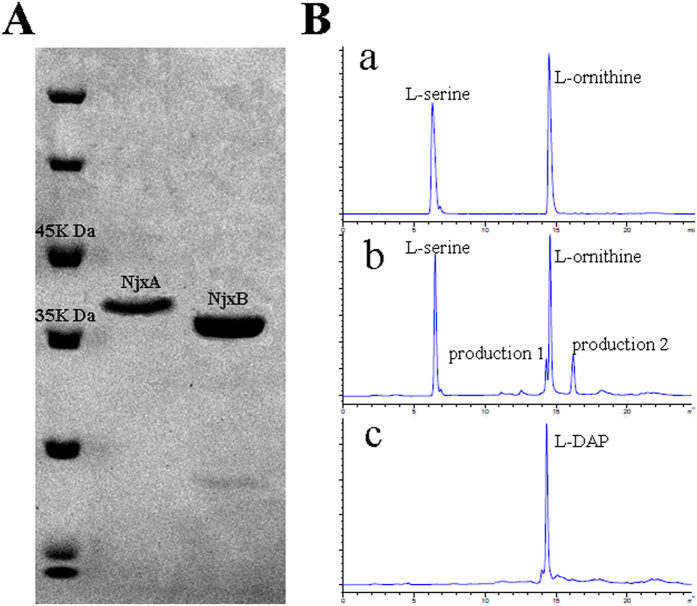
Two proteins encoded by adjacent genes were regarded as the most probable L-DAP synthetase candidantes on the basis of sequence similarity with the reported L-DAP synthetase, namely, NjxA (gb|EXU92816.1|) and NjxB (gb|EXU92121.1|) (Supplementary Table S2). To further confirm the function of these two enzymes, NjxA and NjxB were expressed and purified and then used to jointly perform a catalytic reaction in vitro (Fig. 6). However, when O-phospho-L-serine and L-glutamate were used as substrates, no L-DAP production was detected. These results indicated that L-DAP biosynthesis in S. albulus PD-1 may be somewhat different from that in Staphylococcus aureus. Thus, other substrates proposed in previous studies were added to the reaction mixture30,33. Fortunately, a significant peak of L-DAP was observed when L-serine and L-ornithine were used as substrates (Fig. 6B). These results were little different than those reported by Marek et al. and the L-DAP in S. albulus PD-1 appears to be biosynthesised via the mechanism proposed by Michael et al. In this manner, NjxA acts as the cysteine synthase and serine dehydrate, which first catalyses L-serine to form a pyridoxal phosphate (PLP)-bound α-aminoccrylate intermediate; then it catalyses a β-substitution replacement reaction to generate L-DAP from the α-aminoccrylate intermediate using ammonia as a nucleophile, which is released when NjxB catalyses the cyclisation of L-ornithine (Supplementary Fig. S5). The difference between L-DAP biosynthesis in S. aureus and S. albulus PD-1 may be due to the adaptive evolution of these two different species. In S. aureus, SbnA and SbnB catalyse O-phospho-L-serine and L-glutamate to α-oxoglutarate and L-DAP, which not only provide two of the three precursors required for staphyloferrin B biosynthesis in S. aureus, but also lessen dependence on tricarboxylic acid (TCA) cycle metabolites for staphyloferrin B biosynthesis during iron deprivation. For S. albulus PD-1, a powerful TCA cycle was detected, and thus more metabolic precursors may be available for L-DAP and PDAP biosynthesis from the TCA cycle36,37. Thus, we can conclude that S. albulus PD-1 synthesises PDAP in the following manner: L-DAP is initially synthesised from L-serine and L-ornithine and subsequently assembled into PDAP by a novel NRPS. Although the genes responsible for many secondary metabolites are usually clustered together in the genome38,39, the genes implicated in this L-DAP biosynthesis pathway are not located close to pdaps. In this study, the biosynthetic pathway of PDAP in S. albulus PD-1 was elucidated in detail and it is an important supplement to the basic metabolic network in S. albulus PD-1(Fig. 7). Our findings provide a solid basis for the production of PDAP and ε-PL by S. albulus PD-1. In particular, the novel structure of PDAPs is a relevant reference for the discovery and design of numerous homopoly(amino acid)s in the future.
Figure 6. (I) SDS-PAGE analysis of purified NjxA and NjxB.
M, protein molecular weight marker; 1, purified NjxA 2, purified NjxB. (II) HPLC for the recombinant enzymatic activities of NjxA and NjxB: a, the reaction mixture without NjxA and NjxB; b, reaction mixture with NjxA and NjxB; c, a L-DAP standard.
Figure 7. The basic metabolic pathway of S. albulus PD-1.
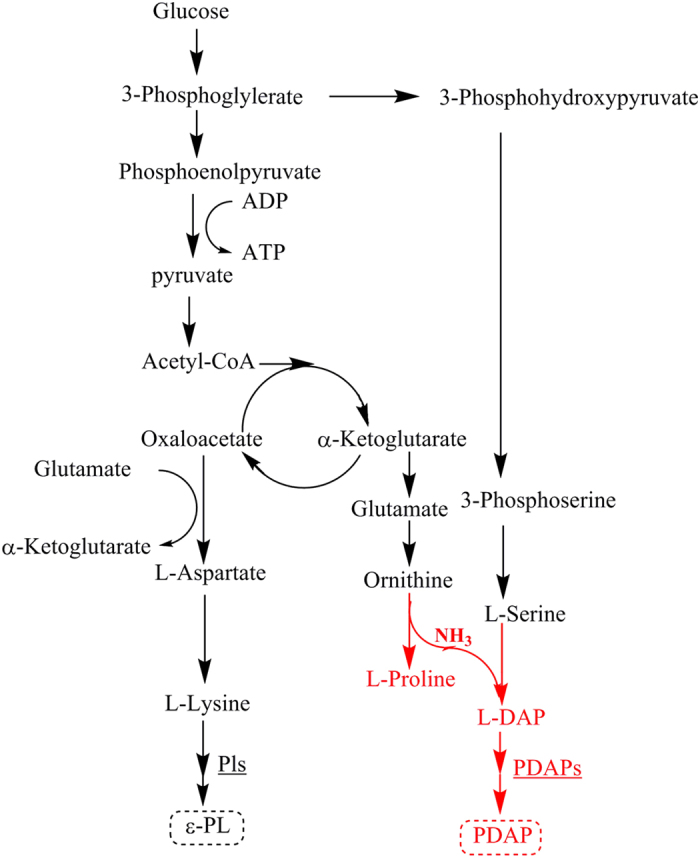
The red part indicates the pathway involved in PDAP biosynthesis.
Methods
Strains, plasmid and media
S. albulus PD-1 (deposited in the China Centre for Type Culture Collection with the strain number M2011043), a well-known ε-PL- and PDAP-producing strain, was used in this study. E. coli ET12567 (pUZ8002) was used as a donor during conjugal transfer. pKC1139 was used to disrupt the putative pdaps gene. pIJ86 was used for heterologous expression of the putative PDAPs. The pET22b plasmid was used to facilitate the expression of the two putative key enzymes involved in L-DAP biosynthesis. The details of the strains, plasmids and media used in this study are listed in Supplementary Table S1.
Gene prediction and bioinformatic analysis
The genome sequence of S. albulus PD-1 was annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/)19. The conserved domains of the candidate proteins were identified using the Conserved Domain Database of NCBI (http://www.ncbi.nlm.nih.gov/cdd/)40. The sequence alignment was performed using the DNAMAN program and the BLAST module in NCBI. The substrate specificities of the NRPS A domains were predicted by using NRPSpredictor2 (http://nrps.informatik.uni-tuebingen.de/Controller? cmd=SubmitJob)41. The TM domains were determined using TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/)42.
Inactivation and heterologous expression of the putative pdaps gene
The target pdaps gene was inactivated through single crossover homologous integration with an apramycin resistance cassette as a selective marker (Fig. 2A). A 3.2-kb homologous fragment of pdaps was amplified from S. albulus PD-1, digested with EcoRI and HindIII and ligated into pre-digested pKC1139 harbouring the apramycin resistance cassette. The resulting plasmid was used for transformation via an optimised conjugal transfer method. The exconjugant was grown at 30 °C on MS medium containing apramycin (50 μg/mL) and nalidixic acid (20 μg/mL). The exconjugant strain was then inoculated into liquid tryptone soy broth (TSB) medium and cultivated in a shake flask at 30 °C and 200 rpm. After 24 h, the culture broth was streaked on an MS agar plate containing 50 μg/mL apramycin and cultivated at 37 °C to obtain the putative gene disruptants.
To confirm whether pKC1139-pdaps was successfully inserted into the chromosomal copy of pdaps, we extracted and analysed the chromosomal DNA from the mutant strains through PCR amplification using start and the end sequences of putative pdaps as primers. In addition, Southern blot was also applied to confirm the disruption of the putative pdaps by using apramycin-resistant gene as a probe. To confirm whether the mutant strains could produce PDAP, we cultivated spores of the recombinants in M3G medium and in a shake flask at 30 °C and 200 rpm for 72 h. The culture filtrates were characterised using HPLC with an Agilent 1200 instrument and according to a previously described method3.
To achieve the heterologous expression of PDAPs in S. lividans TK24, the intact pdaps gene with the corresponding native promoter was amplified with specific primers (Supplementary Table S2). The PCR products were digested with BamHI and HindIII and subsequently inserted into pIJ86 to generate pIJ86-pdaps. pIJ86-pdaps was transformed into E. coli ET12567 (pUZ8002) and then transferred into S. lividans TK24 by the standard method described in Practical Streptomyces Genetics43. The transformants were selected and detected through colony PCR. The spores of the candidate transformants were cultured at 30 °C for 72 h in M3G medium supplemented with 1 g/L L-DAP. The supernatant of the culture broth was tested with Dragendorff reagent and characterised with HPLC for PDAP production.
Analysis of the in vitro substrate specificity of the PDAPs adenylation domain
The A domain-encoding fragment was amplified from S. albulus PD-1 with the primers listed in Supplementary Table S3. The PCR fragment was digested with EcoRI and HindIII and inserted into the corresponding sites of pET28a; as a result, pET28a-pdapsa was generated. The recombinant plasmid was introduced into E. coli BL21 (DE3) to overexpress the corresponding protein. The expressed protein was purified through Ni-chelating affinity chromatography.
In the first step of an NRPS assembly line, a specific amino acid is selected by the A domain. A domains consume ATP to activate amino acid substrates, with concomitant pyrophosphoric acid (PPi) release24. Thus, the released PPi can be used as an indicator of the substrate specificity of an A domain. For PDAPs, the PPi levels were measured through a continuous spectrophotometric assay with the EnzChek pyrophosphate assay kit (Molecular Probes)31. The reactions were conducted at 30 °C in a 500 μL reaction mixture that contained 1 × buffer, 0.2 mM MesG, 10 mM MgCl2, 5 mM ATP, 10 mM amino acid, 0.3 U inorganic pyrophosphatase, 1 U purine nucleoside phosphorylase and 50 μL A domain. The reaction mixture was monitored using a microplate reader (Thermo, Fisher), and readings were conducted every 30 s for 10 min at a wavelength of 360 nm.
Identification of the L-DAP biosynthesis pathway
Two candidate genes responsible for the biosynthesis of L-DAP were identified based on a genetic similarity analysis: njx A and njx B. Both genes were amplified via PCR from the genomic DNA of S. albulus PD-1. The upstream and downstream oligonucleotide primers for njxA and njxB are listed in Supplementary Table S3. Then, the PCR fragments were cloned into the EcoRI/HindIII restriction sites of the pET-22b plasmid. The recombinant plasmid pET-22b-njxA and pET-22b-njxB were verified through restriction enzyme digestions and DNA sequencing (GenScript Biotech Co., China). The recombinant plasmids were transformed into E. coli BL21 (DE3), E. coli Rosetta (DE3) and E. coli Origami (DE3); E. coli Origami (DE3) was selected as the best host for the co-expression of njx A and njx B because more soluble NjxA and NjxB protein could be obtained when expressed in this strain.
Expression studies were conducted in E. coli Origami (DE3). A single colony from freshly transformed cells was inoculated into 5 mL LB medium supplemented with 50 μg/mL ampicillin and grown at 37 °C for 8 to 10 h. Then, 1 mL of the cell suspension was incubated in 50 mL LB supplemented with 50 μg/mL ampicillin and was subsequently incubated at 37 °C. When the OD600 reached 0.6, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 1 mM, and the culture was incubated for 20 h at 15 °C to induce protein expression. The induced cells were harvested through centrifugation and suspended in normal saline. The cells were then disrupted through sonication in an ice bath, and the expressed proteins were purified through Ni-chelating affinity chromatography. The recombinant enzymatic activities were determined by measuring the amount of L-DAP in a reaction mixture. The reaction mixture contained L-serine (1 g/L), L-ornithine (1 g/L), PLP (50 mg/L), NAD+ (50 mg/L), purified NjxA (50 μL) and purified NjxB (50 μL) in a total volume of 200 μL (Tris-HCl, pH 7.5). The reaction mixture was incubated at 37 °C for 30 min and evaluated through HPLC. Online derivatisation was performed using orthophthalaldehyde for the primary amino acids and 9-fluorenylmethyl chloroformate for the secondary amino acids. The amino acid derivatives were analysed using an Agilent 1200 series HPLC system.
Additional Information
How to cite this article: Xu, Z. et al. Systematic unravelling of the biosynthesis of poly (L-diaminopropionic acid) in Streptomyces albulus PD-1. Sci. Rep. 5, 17400; doi: 10.1038/srep17400 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Nature Science Foundation of China (No. 21176123, No. 21476112), the State Key Laboratory of Materials-Oriented Chemical Engineering (ZK201403), the National Basic Research Program of China (973) (No. 2013CB733600), the specialized research fund for the doctoral program of higher education (No.20113221130001), the National High Technology Research and Development Program of China (863) (No. 2015AA020951), the Natural Science Foundation of Jiangsu Province (BK20140933), the Jiangsu Province Agricultural Science and Technology Innovation Program (CX(15)1049).
Footnotes
The authors declare no competing financial interests.
Author Contributions Z.X., S.L., X.F. and H.X. designed the experiments; Z.X., Z.S., and C.C. performed the experiments; Z.X., X.F. and H.X. conceived of the study and wrote the manuscript. All authors read and approved the final manuscript.
References
- Hamano Y., Arai T., Ashiuchi M. & Kino K. NRPSs and amide ligases producing homopoly(amino acid)s and homooligo(amino acid)s. Nat. Prod. Rep. 30, 1087–1097 (2013). [DOI] [PubMed] [Google Scholar]
- Takehara M., Saimura M., Inaba H. & Hirohara H. Poly(γ-L-diaminobutanoic acid), a novel poly(amino acid), coproduced with poly(ε-L-lysine) by two strains of Streptomyces celluloflavus. FEMS Microbiol. Lett. 286, 110–117 (2008). [DOI] [PubMed] [Google Scholar]
- Xia J., Xu H., Feng X., Xu Z. & Chi B. Poly(l-diaminopropionic acid), a novel non-proteinic amino acid oligomer co-produced with poly(ε-l-lysine) by Streptomyces albulus PD-1. Appl. Microbiol. Biot. 97, 7597–7605 (2013). [DOI] [PubMed] [Google Scholar]
- Ogunleye A. et al. Poly-γ-glutamic acid: production, properties and applications. Microbiol. 161, 1–17 (2015). [DOI] [PubMed] [Google Scholar]
- Shih L. & Van Y. T. The production of poly-(γ-glutamic acid) from microorganisms and its various applications. Bioresour. Technol. 3, 207 225 (2001). [DOI] [PubMed] [Google Scholar]
- Bankar S. B. & Singhal R. S. Panorama of poly-ε-lysine. RSC Adv. 23, 8586–8603 (2013). [Google Scholar]
- Shukla S. C., Singh A., Pandey A. K. & Mishra A. Review on production and medical applications of ɛ-polylysine. Biochem. Eng. J. 65, 70–81 (2012). [Google Scholar]
- Xu Z. et al. Economic process to co-produce poly(ε-l-lysine) and poly(l-diaminopropionic acid) by a pH and dissolved oxygen control strategy. Bioresour. Technol. 187, 70–76 (2015). [DOI] [PubMed] [Google Scholar]
- Gardner J. M. & Troy F. A. Chemistry and biosynthesis of the poly (gamma-D-glutamyl) capsule in Bacillus licheniformis. Activation, racemization, and polymerization of glutamic acid by a membranous polyglutamyl synthetase complex. J. Biol. Chem. 14, 6262–6269 (1979). [PubMed] [Google Scholar]
- Ashiuchi M. et al. Enzymatic synthesis of high-molecular-mass poly-γ-Glutamate and regulation of its stereochemistry. Appl. Environ. Microb. 70, 4249–4255 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Maruyama C., Takagi H. & Hamano Y. ε-Poly-L-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase. Nat. Chem. Biol. 4, 766–772 (2008). [DOI] [PubMed] [Google Scholar]
- Maksimov M. O. & Link A. J. Prospecting genomes for lasso peptides. J. Ind. Microbiol. Biothcenol. 41, 233–250 (2014). [DOI] [PubMed] [Google Scholar]
- Hwang K., Kim H. U., Charusanti P., Palsson B. Ø. & Lee S. Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 32, 255–268 (2014). [DOI] [PubMed] [Google Scholar]
- Corre C. & Challis G. L. New natural product biosynthetic chemistry discovered by genome mining. Nat. Prod. Rep. 26, 977–986 (2009). [DOI] [PubMed] [Google Scholar]
- Challis G. L. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiol. 154 1555–1569 (2008). [DOI] [PubMed] [Google Scholar]
- Tang H. et al. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2, 377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F. & Xu P. Genomic analysis of thermophilic Bacillus coagulans strains: efficient producers for platform bio-chemicals. Sci. Rep. 4, 3926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gao C., Tang N., Hu S. & Wu Q. Identification of genetic variations associated with epsilon-poly-lysine biosynthesis in Streptomyces albulus ZPM by genome sequencing. Sci. Rep. 5, 9201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. et al. Genome sequence of Streptomyces albulus PD-1, a Productive Strain for epsilon-Poly-L-Lysine and poly-L-diaminopropionic acid. Genome Announc. 2, e00297–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng W. et al. Cloning of ε-poly-L-lysine (ε-PL) synthetase gene from a newly isolated ε-PL-producing Streptomyces albulus NK660 and its heterologous expression in Streptomyces lividans. Microb. Biotechol. 2, 155–164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K. et al. Development of a recombinant ε-poly-L-lysine synthetase expression system to perform mutational analysis. J Biosci. Bioeng. 111, 646–649 (2011). [DOI] [PubMed] [Google Scholar]
- Marahiel M. A. & Essen L. O. Nonribosomal peptide synthetases: mechanistic and structural aspects of essential domains. Method Enzymol. 458, 337–351 (2009). [DOI] [PubMed] [Google Scholar]
- Finking R. & Marahiel M. A. Biosynthesis of nonribosomal peptides 1. Annu. Rev. Microbiol. 58, 453–488 (2004). [DOI] [PubMed] [Google Scholar]
- Condurso H. L. & Bruner S. D. Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat. Prod. Rep. 29, 1099–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito N. et al. Mutational analysis of the three tandem domains of ε-poly-l-lysine synthetase catalyzing the l-lysine polymerization reaction. J Biosci. Bioeng. 115, 523–526 (2013). [DOI] [PubMed] [Google Scholar]
- Stachelhaus T., Mootz H. D. & Marahiel M. A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 8, 493–505 (1999). [DOI] [PubMed] [Google Scholar]
- Rausch C. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. & Gould S. J. Biosynthesis of capreomycin. 2. incorporation of L-serine, L-alanine, and L-2,3-diaminopropionic acid. J. Org. Chem. 19, 5176–5180 (1993). [Google Scholar]
- Carter J. H. et al. Biosynthesis of viomycin. I. Origin of αβ-diaminopropionic acid and serine. Biochem. 6, 1221–1227 (1974). [DOI] [PubMed] [Google Scholar]
- Felnagle E. A., Rondon M. R., Berti A. D., Crosby H. A. & Thomas M. G. Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Appl. Environ. Microb. 73, 4162–4170 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Song C., Luo Y., Yu Z. & Sun M. L-2,3-diaminopropionate: one of the building blocks for the biosynthesis of zwittermicin A in Bacillus thuringiensis subsp. kurstaki strain YBT-1520. FEBS Lett. 582, 3125–3131 (2008). [DOI] [PubMed] [Google Scholar]
- Cheung J., Beasley F. C., Liu S., Lajoie G. A. & Heinrichs D. E. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74, 594–608 (2009). [DOI] [PubMed] [Google Scholar]
- Beasley F. C., Cheung J. & Heinrichs D. E. Mutation of L-2,3-diaminopropionic acid synthase genes blocks staphyloferrin B synthesis in Staphylococcus aureus. BMC Microbiol. 11, 199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M. G., Chan Y. A. & Ozanick S. G. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Ch. 9, 2823–2830 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobylarz M. J. et al. Synthesis of L-2,3-Diaminopropionic Acid, a Siderophore and Antibiotic Precursor. Chem. Boil. 21, 379–388 (2014). [DOI] [PubMed] [Google Scholar]
- Xia J., Xu Z., Xu H., Feng X. & Bo F. The regulatory effect of citric acid on the co-production of poly(ε-lysine) and poly (L-diaminopropionic acid) in Streptomyces albulus PD-1. Bioproc. Biosyst Eng. 37, 2095–2103 (2014). [DOI] [PubMed] [Google Scholar]
- Xu Z. et al. Effects of oxygen-vectors on the synthesis of ε-poly-lysine and the metabolic characterization of Streptomyces albulus PD-1. Biochem. Eng. J. 94, 58–64 (2015). [Google Scholar]
- Wu Q., Tun H. M., Leung F. C. & Shah N. P. Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Sci. Rep. 4, 1275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci. Rep. 5, 9383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottig M. et al. NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G. & Sonnhammer E. L. L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- Kieser T. Practical Streptomyces genetics. 229–252 (The John Innes Foundation, 2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.