Abstract
Knowledge of global regulatory networks has been exploited to rewire the gene control programmes of the model bacterium Salmonella enterica serovar Typhimurium. The product is an organism with competitive fitness that is superior to that of the wild type but tuneable under specific growth conditions. The paralogous hns and stpA global regulatory genes are located in distinct regions of the chromosome and control hundreds of target genes, many of which contribute to stress resistance. The locations of the hns and stpA open reading frames were exchanged reciprocally, each acquiring the transcription control signals of the other. The new strain had none of the compensatory mutations normally associated with alterations to hns expression in Salmonella; instead it displayed rescheduled expression of the stress and stationary phase sigma factor RpoS and its regulon. Thus the expression patterns of global regulators can be adjusted artificially to manipulate microbial physiology, creating a new and resilient organism.
Gene expression in bacteria is controlled by regulatory factors that include trans-acting DNA binding proteins that operate within hierarchies: the more global the influence of the regulatory protein, the higher its place in the hierarchy1,2,3,4. Any reorganization of the expression pattern of global regulators is likely to alter regulatory network topology and hence cellular physiology3,4,5,6,7. Nucleoid-associated proteins (NAPs) represent a class of global regulatory factor with a very wide range of influences in the genome. NAPs act at the nexus of gene regulation and nucleoid architecture and they typically govern the expression of hundreds of genes, usually in collaboration with other regulatory proteins8. The model bacteria Escherichia coli and its close relative Salmonella enterica serovar Typhimurium have at least 14 NAPs, several of which form paralogous pairs8. Among these are the paralogues H-NS and StpA, proteins that directly or indirectly regulate 13% and 5%, respectively, of the genes in S. Typhimurium in two overlapping regulons9,10.
In S. Typhimurium, the amino acid sequences of H-NS and StpA are 58% identical and the proteins have many functional characteristics in common9,10,11. They also share numerous binding loci in the genome and are involved predominantly in silencing transcription, especially that of genes with a high A + T base composition10,12. Despite these structural and functional similarities, H-NS and StpA are distinguishable. The two proteins have different expression patterns: H-NS is expressed to a high level throughout the growth cycle while expression of StpA is lower and associated principally with the exponential phase of growth13,14. The hns and stpA genes also differ in their transcription regulation: the hns promoter is stimulated by the nucleoid-associated protein Fis15 and the cold-shock protein CspA16, is inhibited by the iron regulatory protein Fur17 and is subject to auto-repression by H-NS18. The stpA promoter is stimulated by osmotic shock and by increases in growth temperature; it shows a sustained expression in minimal medium that depends on the leucine-responsive regulatory protein, Lrp, which is lost upon carbon starvation; transcription of stpA is also silenced by H-NS14,19.
The hns and stpA genes also differ in their genomic locations: hns is in the Ter macrodomain of the chromosome close to the point where chromosome replication ends, making hns among the last genes on the chromosome to be duplicated at completion of the cell cycle2,20. In contrast, the stpA gene is in the non-structured region of the left replichore of the chromosome, mid-way between the origin and terminus of chromosome replication20. Gene location is known to be an important influence on expression in eukaryotes21,22,23 and this has recently been established in bacteria too24. To date, experiments that have investigated the effects of gene location on expression typically have involved moving the promoter of a gene of interest to a series of different locations and monitoring its expression pattern25,26,27,28,29,30,31,32,33. Interestingly, moving the entire hns gene together with its negatively-auto-regulated promoter and upstream regulatory region to different locations on the chromosome did not alter cell physiology in E. coli26. We decided to probe the matter of upstream sequence influences on paralogous gene regulation by taking a much more subtle approach in which just the open reading frames (ORFs) of the two highly-related hns and stpA genes were exchanged reciprocally on the Salmonella chromosome. Since the exchanges began at the translation initiation codons and ended at the translation stop codons, all other components of the participating genes – the transcription regulation, initiation and termination signals – remained in place and unchanged. This allowed us to investigate specifically the effects of ORF exchange while minimizing the number of other variables.
Results
Regulatory re-wiring of the hns and stpA open-reading frames
To investigate the physiological significance of the distinct expression patterns of the hns and stpA genes, their ORFs were exchanged reciprocally on the chromosome of S. Typhimurium strain SL1344 (Methods). In the resulting strain, SL1344RX (RX: reciprocal exchange) the hns ORF was connected to the transcription regulatory inputs of stpA and vice versa (Fig. 1) (Fig S1 and S2). Whole genome sequencing revealed that SL1344RX had not acquired any of the compensatory mutations previously associated with loss of hns expression in SL13449,10 (Table S1, Genome Accession Number CP011233).
Figure 1. The chromosome of Salmonella divided into quarter segments calibrated in minutes and megabasepairs (Mbp).
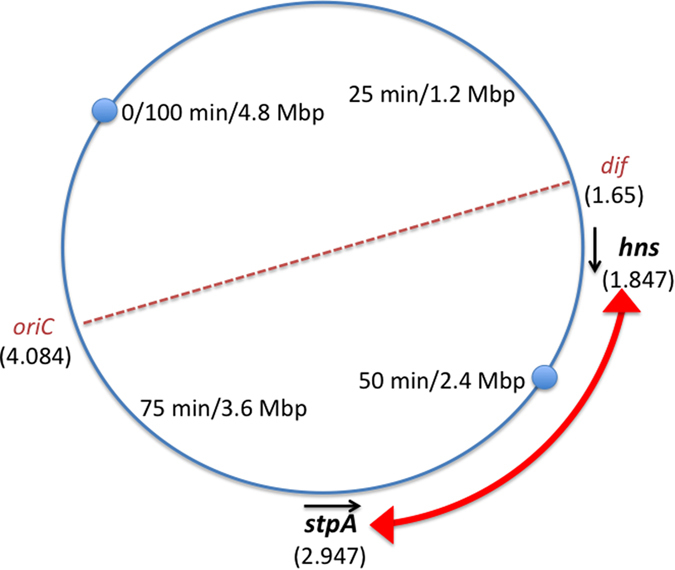
The locations of the origin of chromosome replication (oriC) and the site at the mid-point of the chromosome terminus at which chromosome dimers are resolved by site-specific recombination (dif) are indicated. The locations and directions of transcription of the stpA and hns genes are shown. The numbers in parentheses are the coordinates (in Mbp) of the first nucleotides of their open reading frames; the zero reference point is at 100 minutes (4.8 Mbp). The chromosome is bi-directionally replicated from the origin to the terminus and in Salmonella the stpA and hns genes are both located in the left replichore.
SL1344RX has fitness that is tuneable and can out-compete the wild type
H-NS and StpA regulate genes required for adaptation to environmental stresses, most notably temperature and osmotic shifts10,34,35,36,37. Therefore we assessed the impact of re-wiring the hns and stpA loci on the competitive fitness of S. Typhimurium growing at different temperatures and osmotic pressures. This was done by competing co-cultured SL1344 and SL1344RX at 25 °C, 37 °C or 42 °C (Fig. 2a) and at 37 °C in L broth supplemented with 0 mM, 86 mM or 172 mM NaCl (Fig. 2b).
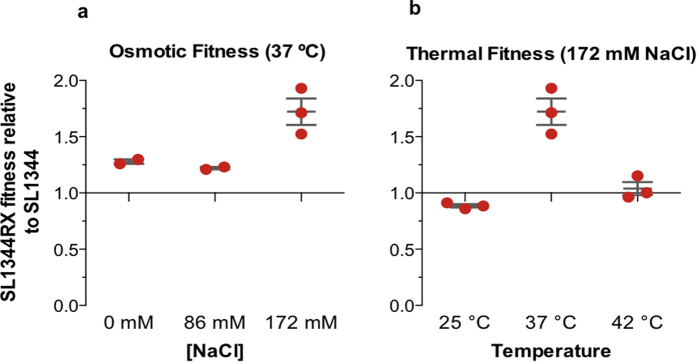
Figure 2. Competitive fitness of SL1344RX relative to SL1344 under different growth conditions.
(a) Fitness of SL1344RX relative to SL1344 grown at 37˚C in a growth medium supplemented with 0 (N = 2), 86 (N = 2) or 172 (N = 3) mM NaCl. Relative fitness values above 1.0 for SL1344RX indicate greater fitness than SL1344; values below 1.0 indicate that SL1344RX is less fit than SL1344. (b) Fitness of SL1344RX relative to SL1344 at three growth temperatures: 25˚C, 37˚C and 42˚C (N = 3). Data points, their mean values and standard deviations are plotted for the biological replicates.
Re-wiring of the hns and stpA genes in SL1344RX altered competitive fitness in a temperature-specific manner. At 25 °C SL1344RX was less fit than the wild type (|s| = 0.88). In strong contrast, at 37 °C SL1344RX significantly out-competed the wild type (|s| = 1.72); no discernable difference in fitness relative to SL1344 was observed at 42 °C (Fig. 2). The fitness enhancement seen in SL1344RX at 37 °C could be tuned by varying the salt concentration of the growth medium: in NaCl-free broth a relative fitness index of 1.27 was recorded for SL1344RX and a marginal decrease in this value was recorded when the strains were competed in 86 mM NaCl (|s| = 1.22); the greatest fitness enhancement was seen at 172 mM NaCl at 37 °C (Fig. 2).
hns and stpA exhibit new expression patterns in SL1344RX
The relocated hns and stpA ORFs did not retain their normal expression patterns: changes in the abundance of hns mRNA and stpA mRNA were observed in SL1344RX when compared to wild-type SL1344 (Fig. 3). When hns mRNA levels were monitored in wild-type SL1344 (Fig. 3a), a characteristic peak was observed during exponential growth after which mRNA levels gradually declined. In SL1344RX, where the stpA promoter drives transcription of the hns ORF, an stpA-like pattern of expression was not conferred upon hns. Instead, a much higher level of hns expression was detected in SL1344RX compared to the wild type (Fig. 3b).
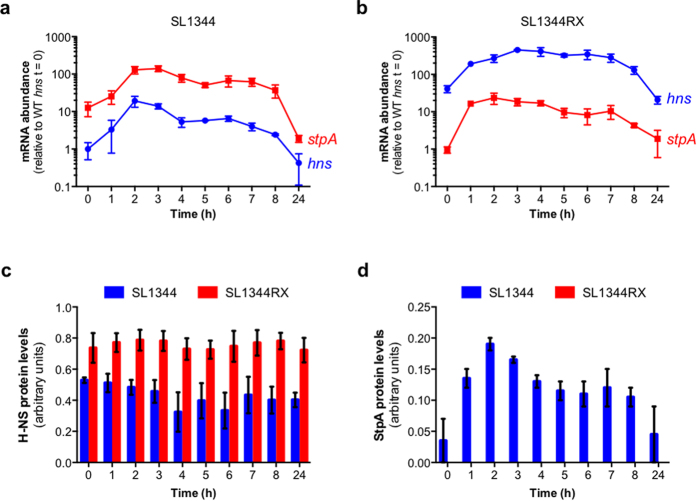
Figure 3. Expression of hns and stpA mRNA and protein in SL1344 and SL1344RX.
Transcripts encoded by the hns (blue) and stpA (red) open reading frames in SL1344 (wild type) (a) and SL1344RX (b) as a function of time were monitored by RT-qPCR. Transcript levels are expressed relative to the level of the hns mRNA transcript in wild type SL1344 at t = 0. H-NS (c) and StpA (d) protein levels in SL1344 (blue) and SL1344RX (red) were determined by Western blotting. Protein levels were measured by densitometry and expressed relative to an internal control in arbitrary units. The mean and standard deviation for mRNA and protein levels are plotted for three biological replicates (N = 3).
The repositioned stpA ORF had also acquired a new pattern of expression in SL1344RX, peaking earlier in the growth cycle than in SL1344 (Fig. 3b). Maximum expression of stpA was sustained for longer in SL1344RX compared to the wild type (Fig. 3a). Despite being transcribed from the hns promoter, stpA mRNA was on average 6-fold less abundant in SL1344RX than in SL1344 throughout the growth cycle.
As the ORF exchanges in SL1344RX altered both the timing and abundance of hns and stpA mRNA expression, it was anticipated that these exchanges could also change the relative sizes of the H-NS and StpA protein populations. Therefore, H-NS and StpA protein expression was monitored as a function of the growth cycle in SL1344 and SL1344RX by western blotting.
In agreement with previously published data18, the abundance of H-NS protein did not change over the growth cycle in wild-type SL1344 (Fig. 3c). H-NS was similarly expressed at a constant level throughout the growth cycle in SL1344RX, despite the new pattern of mRNA expression from the stpA promoter. However, when the levels of H-NS protein in each strain were quantified, it was discovered that H-NS was up to 2-fold more abundant in SL1344RX than in SL1344 (Fig. 3c).
The abundance of StpA protein in wild-type SL1344 agreed with published data13 (Fig. 3d). Contrary to the comparable levels of H-NS in the wild type and SL1344RX, no StpA protein could be detected at any stage of growth in SL1344RX (Fig. 3d). The observation that stpA mRNA levels were lower in SL1344RX than in the wild type raised the possibility that StpA concentrations may have been too low to detect by Western blotting. When total protein from SL1344 and from SL1344RX was analysed by mass spectrometry, StpA-specific peptides were detected in SL1344 but not in SL1344RX (Supplementary Information), consistent with an absence of StpA in SL1344RX.
As mRNA from the stpA ORF was readily detectable in SL1344RX (Fig. 3), transcription silencing by H-NS seemed an unlikely explanation for the lack of StpA protein. Repositioning the hns and stpA open reading frames had the inevitable consequence of creating novel transcripts consisting of the untranslated region (UTR) of one gene fused to the other gene’s ORF. We tested whether these hybrid molecules, 5′-hns[UTR]-stpA[ORF]-3′ mRNA and 5′-stpA[UTR]-hns[ORF]-3′ had different stabilities compared to wild-type transcripts.
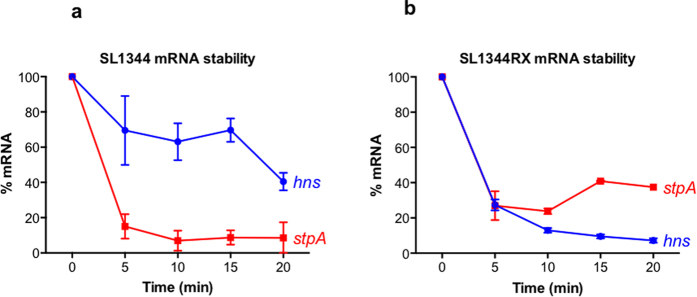
The stabilities of the 5′-stpA[UTR]-hns[ORF]-3′ and 5′-hns[UTR]-stpA[ORF]-3′ hybrid mRNAs in SL1344RX were compared to native transcripts in SL1344 over a 20-min time course by RT-qPCR following transcription inhibition by rifampicin treatment (Methods). The results showed that 5′-stpA[UTR]-hns[ORF]-3′ was less stable than native hns mRNA (Fig. 4a,b) despite generating elevated levels of H-NS in SL1344RX (Fig. 3). In contrast, the hybrid 5′-hns[UTR]-stpA[ORF]-3′ mRNA in SL1344RX was more stable than native stpA mRNA in SL1344 (Fig. 4a,b). Therefore reduced message stability does not account for the absence of StpA protein in SL1344RX. The translatability of the hybrid 5′-hns[UTR]-stpA[ORF]-3′ message and its susceptibility to modulation by RNA secondary structure formation were not measured directly and may underlie the strong reduction in StpA protein in SL1344RX.
Figure 4. Relative stability of mRNA in SL1344 (wild type) and SL1344RX.
The levels of hns mRNA and stpA mRNA were monitored at regular intervals (0, 5, 10, 15 and 20 min) post-rifampicin treatment in SL1344 (a) and SL1344RX (b).
RpoS expression is rescheduled in SL1344RX
The transcriptomes of SL1344RX and SL1344 were compared during mid-exponential growth (OD600 = 0.3) by gene expression microarray analysis. Despite the involvement of H-NS and StpA in regulating so many genes, we identified only 26 differentially expressed genes in SL1344RX relative to SL1344 during exponential growth (Table S2). Twenty-three of these genes were up-regulated and 3 down-regulated in SL1344RX, and 13 were predicted bioinformatically to be RpoS-dependent (Table S3). Further analysis of H-NS, StpA and RpoS regulated genes revealed subtle but significant (P < 0.01) changes in expression in comparison with control gene sets across both the RpoS and StpA regulons but not the H-NS regulon (Fig. S3). Loss of H-NS was shown previously to result in compensatory mutations in the rpoS gene and inactivation of stpA expression to alter the timing of RpoS expression9,10. We detected no compensatory mutations but did find that RpoS was expressed in exponential phase in SL1344RX (Fig. 5) and at a much earlier time than in the stpA knockout derivative of SL134410.
Figure 5. Expression of the RpoS sigma factor in SL1344 (wild type) and in SL1344RX as a function of growth.
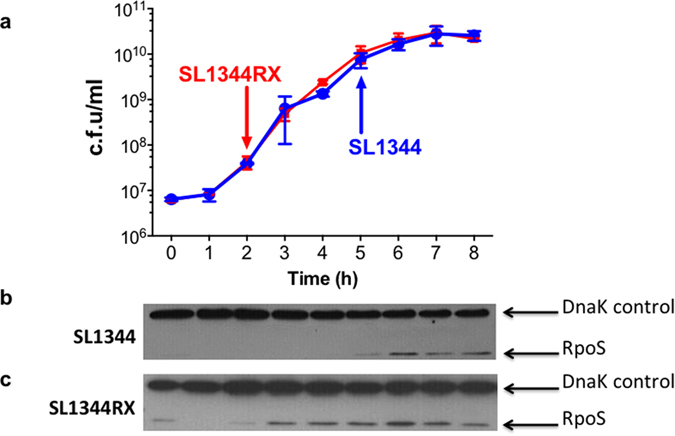
The growth cycles of SL1344 and SL1344RX were determined by colony counts (a). RpoS expression was monitored by Western blotting and was first detected after 2 h and 5 h of growth in SL1344 (b) and SL1344RX (c), respectively.
Many genes within the RpoS regulon are required for adaptation and resistance to environmental stresses such as oxidative, osmotic and thermal stress38,39,40. Up-regulation of RpoS regulon members could explain the enhanced fitness of SL1344RX at 37 °C under different degrees of osmotic pressure (Fig. 2b). Consistent with this model, many of the up-regulated RpoS-dependent genes in SL1344RX were involved in osmotic stress resistance (e.g. osmE, osmY, otsB, wrbA, ybaY, yehY, and ygaU).
Expression of RpoS-regulated genes (and RpoS itself) is normally confined to periods of stress and to stationary phase40,41. Inappropriate expression of the RpoS regulon in E. coli has been described as detrimental to competitive fitness because RpoS competes with other sigma factors for core RNA polymerase resulting in decreased expression of RpoS-independent genes involved in nutrient uptake and metabolism42,43,44,45. The cellular concentration of RpoS is therefore tightly regulated at the level of rpoS transcription, translation and proteolytically at the post-translational level39.
In S. Typhimurium and E. coli, both H-NS and StpA modulate the proteolysis of RpoS by repressing expression of the IraM, IraD and RssC anti-adaptor molecules that prevent degradation of RpoS by the ClpXP protease10,38,42. Since the change in expression of RpoS regulon members in SL1344RX could have been due to a novel pattern of RpoS expression, we monitored RpoS abundance in SL1344 and SL1344RX as a function of growth stage. In the wild-type strain SL1344, RpoS was detectable, as expected, at the onset of stationary phase, 5 h post-inoculation (Fig. 5a,b); however RpoS appeared in SL1344RX in the early stages of exponential growth, 2 h post-inoculation, i.e. 3 h before detection in the wild type (Fig. 5a,c).
SL1344RX did not express measurable levels of StpA protein, although the 5′-hns-stpA-3′ mRNA was detectable. A knockout mutation in the stpA gene in SL1344 causes RpoS to appear in exponentially growing cells10 although not as early as in SL1344RX (Fig. 5c). Further evidence that the observed rescheduling of RpoS expression in SL1344RX was not simply correlated with the absence of StpA came from monitoring RpoS expression in a derivative of SL1344 with two stpA ORFs, one in its native location and one in place of hns; here, RpoS became detectable at 3 h post-inoculation (rather than at 2 h, as in SL1344RX). In yet another derivative of SL1344 that had two hns ORFs, one at its native location and one in place of stpA, RpoS appeared at 4 h post-inoculation (Fig. S4). These findings indicated that the timing of RpoS expression in growing SL1344 was modulated by the overall composition of the StpA and H-NS protein population in the different hns/stpA genotypes and not simply the presence/absence of StpA.
Discussion
Synthetic biology and the creation of artificial gene regulatory circuits are contributing to a deeper understanding of the modular and hierarchical structures of complex regulatory networks5,46. This understanding is being exploited for the rational design and engineering of microbes for use in a wide range of industrial, biotechnological and biomedical processes46,47. Examples include the recent application of re-engineered strains of E. coli as biosensors for the detection of cancer and diabetes in mice48,49. The engineering of stress-tolerant microbes for industrial use has also been a major focus of synthetic biology47.
Our study shows that it is possible to generate a novel derivative of a well-characterized model bacterium that has improved competitive fitness by a simple reciprocal exchange of two paralogous genes that encode global regulators of gene expression. This approach permits retuning of existing gene regulatory networks and is complementary to bottom-up approaches to strain construction and synthetic biology based on the creation of artificial chromosomes. It also informs these approaches by adding to knowledge of the significance of gene position and expression for the preservation of genome integrity50,51. Because compensatory mutations were not detected in the SL1344RX genome sequence, the reciprocal genetic exchange that produced SL1344RX is well tolerated. The enhanced fitness of SL1344RX raises the question of why this genetic arrangement has not occurred in nature, perhaps as a result of horizontal gene transfer? First, it cannot be ruled out conclusively that strains similar to SL1344RX do not exist naturally; they just have not been reported before. Second, our study identified growth conditions (growth at 25 °C) where SL1344RX is outcompeted by the wild type. Thus, the rewired strain may not be capable of thriving in the external environment where it will quickly be eliminated by natural selection.
The RpoS stress and stationary phase sigma factor is expressed prematurely in the growth cycle of SL1344RX. This component of RNA polymerase is central to adaptation to osmotic and thermal stress and is a determining factor in bacterial fitness40,41,42,44,45. Both H-NS and StpA are intimately involved in the regulatory pathways that control RpoS expression and degradation10,38,52, so the early appearance of RpoS in SL1344RX is perhaps not altogether surprising. However, as we have not identified the molecular connection between RpoS and the hns and stpA genes that is responsible for altered RpoS expression in SL1344RX, it is possible that this involves an indirect effect. It is also important to note that since knockout mutations in stpA have obvious phenotypes in Salmonella10 but cause more subtle effects in E. coli19, it is probably unwise to extrapolate our findings even to closely related bacterial species.
The regulatory changes made to hns and stpA in SL1344RX altered the fitness of S. Typhimurium in an environment-specific manner. The variability of fitness outcomes between environmental conditions suggested that altering the relative expression of hns and stpA is one mechanism by which S. Typhimurium manipulates expression of genes within the combined H-NS/StpA regulatory network to adapt to environmental changes. Perhaps unsurprisingly, hns and stpA expression is naturally sensitive to temperature shifts: in E. coli, hns is up-regulated in response to cold-shock while up-regulation of stpA occurs upon an upshift in temperature from 30 °C to 37 °C14,16,39. Expression of stpA in E. coli is also strongly up-regulated in response to osmotic shock16. The ‘unnatural’ changes to the expression of hns and stpA brought about by re-engineering in SL1344RX conferred significant fitness advantages in certain environments, highlighting the potential of S. Typhimurium to evolve to be more competitive through the accumulation of mutations or the acquisition or loss of regulatory inputs that effect the expression of hns and stpA. It is also important to consider the impact of additional hns-like genes that are acquired horizontally; genes of this type are found on many mobile genetic elements that are capable of either self-transmission or of being mobilized53,54.
Conventionally a bacterial species has been viewed as a manifestation of defining phenotypes, of the presence of signature genes, and of the outcome of DNA sequence comparisons with reference strains55. Genomics is enriching this view by revealing that non-coding regions of DNA have a profound influence on species character. For example, orthologous genes with identical open reading frames can differ markedly in their regulatory regions so that the same genes can be used by different species to access different niches56,57. Thus, variation in non-coding regions offers a fruitful route to the evolution of new cellular properties by altering gene expression patterns56. The present study provides a dramatic example of the application of this approach to the construction in the laboratory of a bacterial strain with strikingly novel properties by a simple rewiring of two paralogous global regulators.
Methods
Bacterial strains and growth conditions
Bacterial strains used in this study, their genotypes and source, where applicable, are listed in Table S4; all are derivatives of S. Typhimurium strain SL1344. Unless otherwise stated bacterial strains were cultured with aeration (200 rpm) in Luria broth (LB) at 37 °C. Kanamycin, tetracycline, carbenicillin and chloramphenicol were used at concentrations of 50 μg ml−1, 15 μg ml−1, 50 μg ml−1 and 20 μg ml−1 respectively.
Strain construction and DNA manipulation
All strains generated during this study were constructed using bacteriophage-lambda-mediated recombination58. To construct SL1344RX the neoR and tetRA resistance genes were PCR amplified from plasmids pSUB1159 and pACYC184. These amplicons were integrated onto the SL1344 chromosome 40 bp downstream of the hns and stpA stop codons, respectively. The hns ORF and downstream neoR gene were then PCR amplified and the resulting amplicon was integrated onto the SL1344 chromosome replacing the stpA ORF. This created a strain with two hns ORFs (SL13442Xhns). Similarly the stpA ORF and tetRA cassette were PCR amplified and used to replace the hns ORF, creating a strain with two stpA ORFs (SL13442XstpA). The Phns-stpA promoter-ORF fusion from SL13442XstpA was then introduced into SL13442Xhns by P22 phage-mediated generalised transduction60 to produce SL1344RX. Strain variants of SL1344 and SL1344RX expressing 3X FLAG-epitope-tagged H-NS or StpA were constructed using a modified version of bacteriophage-lambda-mediated recombination60. The DNA oligonucleotides used in this study and their uses are listed in Table S5.
Real-time PCR
The hns and stpA mRNA levels in SL1344 and SL1344RX were determined by RT-qPCR. 0.2 OD600nm units of bacteria were harvested, held on ice and 2/5 volume of stop solution (95% ethanol/5% phenol) was added. Total RNA was extracted using a Promega SV Total RNA Isolation kit and RNA concentration was quantified using a Nanodrop ND-1000. Extracted RNA (200 ng) was reverse transcribed using GoScript™ reverse transcription system (Promega) to generate cDNA pools and the relative abundances of target mRNA molecules were determined by quantitative PCR (qPCR) using gene specific primer pairs (Table S5) and GoTaq qPCR master mix (Promega). Quantification of mRNA was achieved using an internal calibration curve generated from serially diluted genomic DNA of known quantity. For mRNA stability assays, cultures were grown to an OD600 = 0.2 before the addition of rifampicin (250 μg ml−1) to inhibit further transcription. Levels of mRNA were then quantified at 0, 5, 10, 15 and 20 min after rifampicin addition.
Western immunoblotting
Bacteria (0.2 OD600 nm units) were harvested by centrifugation (13,000 rpm, 5 min) and re-suspended in 1× Laemmli sample buffer61. 12% SDS-polyacrylamide gels were used for SDS-PAGE and separated proteins were electroblotted onto 0.22 Protran nitrocellulose membranes (Whatman) for immunodetection as previously described61. Antibodies were used at the following dilutions: anti-FLAG (1/10,000) (Sigma), anti-RpoS (1/5000) (Neoclone) and anti-DnaK (1/100,000) (Abcam), secondary HRP-conjugated goat anti-mouse antibody (1/10,000). The Pierce West Pico Super Signal kit was used for chemiluminescent detection of bound peroxidase conjugate. Blots were probed for the presence of DnaK to ensure equal loading in each lane. Protein levels were quantified relative to a known concentration of Carboxy-Terminal FLAG-BAP fusion protein (Sigma) loaded in each gel.
Competitive fitness assays
The fitness of SL1344RX relative to wild-type SL1344 was calculated for several environmental conditions as described previously62,63. Colony-forming units of each competing strain were enumerated at time zero (t = 0) and after 24 h of co-culturing by selective plating. The fitness/selection coefficient, |s|, of SL1344RX was calculated using the formula:
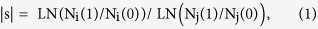 |
where Ni (0) and Ni (1) = initial and final colony counts of SL1344RX, respectively and Nj (0) and Nj (1) = initial and final colony counts of wild type SL1344, respectively63.
RNA extraction, DNA labelling and hybridisation for microarray
OD600 nm units of bacterial cultures were harvested during mid-exponential growth (OD600 nm = 0.3). Total RNA was extracted using a Promega SV Total RNA Isolation kit, quantified using a Nanodrop ND-1000 and checked for purity and degradation by gel electrophoresis. Total RNA samples were converted to double-stranded cDNA pools using a Superscript™ Double-Stranded cDNA synthesis kit (Invitrogen). Three independent RNA samples were extracted for microarray analysis of transcript abundance. Double stranded cDNA (200 ng) was fluorescently labelled with Cy3™ dCTP using the BioPrime® Array CGH Labelling system (Invitrogen). 200 ng of isolated S. Typhimurium SL1344 genomic DNA was labelled with Cy5™ dCTP for use as a co-hybridization control and reference on all arrays. Labelled DNA and cDNA was purified using Micro-spin G50 columns (GE Healthcare) to remove unlabelled nucleotides. Cy3™ and Cy5™ labelled cDNA and reference genomic DNA were mixed in a 1:1 ratio (180 μl of each) and the combined DNA was ethanol precipitated. Precipitated pellets were re-suspended in 100 μl of hybridisation buffer (1 M NaCl, 100 mM MES pH 6.5, 20 mM EDTA, 20% formamide, 20% Triton X-100) at 70 °C for 15 min. Samples were then denatured (100 °C, 10 min), applied to a 44k-probe Salmonella Typhimurium SL1344 microarray slide (Oxford Gene Technologies) and allowed to hybridise for 48 h at 55 °C.
Microarray data acquisition and analysis
After hybridisation, slides were washed and scanned with a GenePix 4000B scanner (Axon Instruments). Quantification of fluorescent spot intensities and local background data was performed using the GenePix 3.0 software supplied. Microarray data files were loaded into ArrayPipe64 for quality assessment and for normalisation with the loess function applied to each sub-grid. Multiple probes per gene were merged into one average value using the median. This was followed by between-array normalisation using the Rquantile method and differential expression analysis through the Bioconductor package Limma65. P-values were corrected for multiple testing through the Benjamini-Hochberg method. To detect subtle regulon-wide changes in gene expression, the expression intensities and Log2 ratios of known H-NS, StpA or RpoS regulated genes were used to generate MA-plots (Fig. S3). Gene expression across each regulon was compared with an equivalent number of control genes. Ten independent comparisons were made using randomly selected control gene sets and analysed by t-test to detect significant changes in expression (P < 0.01). The complete datasets are available at GEO (Accession number GSE52235).
Genome sequencing and analysis
Whole genome sequencing was performed on an Illumina MiSeq platform. Genomic DNA was extracted using a Gentra® Puregene® Yeast/Bact. Kit (Qiagen) and quantified by Qubit dsDNA BR Assay (LifeTechnologies). A Bioruptor Standard (Diagenode, NJ, USA) was used to shear DNA (1 μg in 100 μl Tris-HCl, pH 7) by sonication (low power, 30 s on/90 s off, 15 min) to an optimal fragment size of 600–800 bp. NEBNext Ultra DNA library prep and NEBNext Multiplex Oligo kits for Illumina (New England Biolabs) were used to generate paired–end libraries. Libraries were quantified with the Qubit dsDNA HS assay (LifeTechnologies) and the quality and size distribution was assessed on an Agilent high sensitivity DNA chip with a Bioanalyzer (Agilent Technologies, CA, USA). A final combined 4 nM library was prepared, denatured, diluted to 14 pM and sequenced using a MiSeq Reagent v3 Kit (600 cycle) according to manufacturer guidelines (Illumina Inc., CA, USA).
Paired-end reads were first quality checked with FastQC and then assembled using the velvet de novo assembler, version 1.2.0766. Under default settings we varied k-mer and found k = 227 to produce the highest N50 of 377, 806. This generated 27 contigs of length greater than 1000 bp with the longest at 635, 541 bp. The contigs were ordered and concatenated using ABACAS version 1.3.1.267 providing a full genome of length 4.96 Mbp. This assembly was then aligned to wild type Salmonella Typhimurium SL1344. The Geneious R8.1 package (Biomatters, Auckland, New Zealand)68 was used to align paired-end reads to the SL1344 reference genome (NC_016810, NC_017718, NC_017719, NC_017720). Aligned reads were then searched for variants with a minimum 95% frequency in reads and again at a relaxed stringency of 45% variant frequency. The quality of read alignment at variants was confirmed visually. Only two SNPs were detected and these were in genes (menC and manX) that have no known connections to H-NS, StpA or RpoS.
Additional Information
How to cite this article: Fitzgerald, S. et al. Re-engineering cellular physiology by rewiring high-level global regulatory genes. Sci. Rep. 5, 17653; doi: 10.1038/srep17653 (2015).
Supplementary Material
Acknowledgments
This work was supported by Science Foundation Ireland Principal Investigator Award 13/IA/1875 to CJD and the Saskatchewan Health Research Foundation [Grant 2867] and Natural Sciences and Engineering Research Council of Canada [Discovery Grant 435784-2013] to ADSC.
Footnotes
Author Contributions S.F. designed and performed the experiments, analysed the data, wrote the paper; S.C.D., H.W. and K.H. analysed the data; T.-C.C. performed experiments and analysed the data; A.D.S.C. designed the experiments, analysed the data, wrote the paper; C.J.D. designed the experiments, analysed the data, wrote the paper.
References
- Bhardwaj N., Kim P. M. & Gerstein M. B. Rewiring of transcriptional regulatory networks: hierarchy, rather than connectivity, better reflects the importance of regulators. Sci. Signal. 3, ra79 (2010). [DOI] [PubMed] [Google Scholar]
- Dorman C. J. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11, 349–355 (2013). [DOI] [PubMed] [Google Scholar]
- Lin Z., Zhang Y. & Wang J. Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol. Adv. 31, 986–991 (2013). [DOI] [PubMed] [Google Scholar]
- Yu H. & Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl. Acad. Sci. USA 103, 14724–14731 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M. et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452, 840–845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Tombor B., Albert R., Oltvai Z. N. & Barabási A. L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000). [DOI] [PubMed] [Google Scholar]
- Zwir I. et al. Bacterial nucleoid-associated protein uncouples transcription levels from transcription timing. MBio 5, e01485–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S. C. & Dorman C. J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 (2010). [DOI] [PubMed] [Google Scholar]
- Ali S. S. et al. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 10, e1004500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S., McDermott P., Thompson A. & Hinton J. C. The H-NS-like protein StpA represses the RpoS (sigma 38) regulon during exponential growth of Salmonella Typhimurium. Mol. Microbiol. 74, 1169–1186 (2009). [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Hinton J. C. & Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7, 124–128 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400 (2004). [DOI] [PubMed] [Google Scholar]
- Hinton J. C. et al. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6, 2327–2337 (1992). [DOI] [PubMed] [Google Scholar]
- Free A. & Dorman C. J. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179, 909–918 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi M., Brandi A., La Teana A., Gualerzi C. O. & Pon C. L. Antagonistic involvement of FIS and H-NS proteins in the transcriptional control of hns expression. Mol. Microbiol. 19, 965–975 (1996). [DOI] [PubMed] [Google Scholar]
- La Teana A. et al. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. USA. 88, 10907–10911 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B. et al. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 497–505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi M., Higgins N. P., Spurio R., Pon C. L. & Gualerzi C. O. Expression of the gene encoding the major bacterial nucleotide protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 10, 273–282 (1993). [PubMed] [Google Scholar]
- Sonden B. & Uhlin B. E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 15, 4970–4980 (1996). [PMC free article] [PubMed] [Google Scholar]
- Sobetzko P., Travers A. & Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. USA. 109, E42–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P., Lund E. G. & Oldenburg A. R. Closing the (nuclear) envelope on the genome: how nuclear lamins interact with promoters and modulate gene expression. Bioessays 36, 75–83 (2014). [DOI] [PubMed] [Google Scholar]
- Girton J. R. & Johansen K. M. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv. Genet. 61, 1–43 (2008). [DOI] [PubMed] [Google Scholar]
- Wilson C., Bellen H. J. & Gehring W. J. Position effects on eukaryotic gene expression. Annu. Rev. Cell Biol. 6, 679–714 (1990). [DOI] [PubMed] [Google Scholar]
- Bryant J. A., Sellars L. E., Busby S. J. & Lee D. J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 42, 11383–11392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D. H. S., Hussein R., Liang L. W. & Lim H. N. Regulatory consequences of gene translocation in bacteria. Nucleic Acids Res. 40, 8979–8992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla E. & Sclavi B. Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 (Bethesda) 5, 605–614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. G. & Pritchard R. H. The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol. Gen. Genet. 138, 127–141 (1975). [DOI] [PubMed] [Google Scholar]
- Miller W. G. & Simons R. W. Chromosomal supercoiling in Escherichia coli. Mol. Microbiol. 10, 675–684 (1993). [DOI] [PubMed] [Google Scholar]
- Pavitt G. D. & Higgins C. F. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol. Microbiol. 10, 685–696 (1993). [DOI] [PubMed] [Google Scholar]
- Schmid M. B. & Roth J. R. Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169, 2872–2875 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C., de Lorenzo V. & Cebolla A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology 143, 2071–2078 (1997). [DOI] [PubMed] [Google Scholar]
- Thompson A. & Gasson M. J. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 67, 3434–3439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B.-W., Tsuru S., Seno S., Matsuda H. & Yomo T. Gene expression scaled by distance to the genome replication site. Mol. Biosyst. 10, 375–379 (2014). [DOI] [PubMed] [Google Scholar]
- Lucchini S. et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W. W. et al. Selective silencing of foreign DNA with low GC content by the H-NS protein In Salmonella. Science 313, 236–238 (2006). [DOI] [PubMed] [Google Scholar]
- Rajkumari K., Kusano S., Ishihama A., Mizuno T. & Gowrishankar J. Effects of H-NS and potassium glutamate on sigmaS- and sigma70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J. Bacteriol. 178, 4176–4181 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T. & Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24, 7–17 (1997). [DOI] [PubMed] [Google Scholar]
- Zhou Y. & Gottesman S. Modes of regulation of RpoS by H-NS. J. Bacteriol. 188, 7022–7025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald S. Expression of hns and stpA in Salmonella enterica serovar Typhimurium. University of Dublin (2012).
- Sledjeski D. D., Gupta A. & Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000 (1996). [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ruiz M., Robbe-Saule V., Hermant D., Labrude S. & Norel F. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar typhimurium. J. Bacteriol. 182, 5749–5756 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Majdalani N. & Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacour S. & Landini P. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57, 1–8 (2005). [DOI] [PubMed] [Google Scholar]
- Stoebel D. M., Hokamp K., Last M. S. & Dorman C. J. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 5, e1000671 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C. J., Horwitz A. A., Peisajovich S. G. & Lim W. A. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys 39, 515–537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Zhang Y. & Wang J. Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol Adv 31, 986–991 (2013). [DOI] [PubMed] [Google Scholar]
- Courbet A., Endy D., Renard E., Molina F. & Bonnet J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci Transl Med 7, 289ra83 (2015). [DOI] [PubMed] [Google Scholar]
- Danino T. et al. Programmable probiotics for detection of cancer in urine. Sci Transl. Med. 7, 289ra84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A., Tehranchi A., MacAlpine D. M. & Wang J. D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 6, e1000810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Scolari V. F., Lagomarsino M. C. & Seshasayee A. S. The genome-scale interplay amongst xenogene silencing, stress response and chromosome architecture in Escherichia coli. Nucleic Acids Res. 43, 295–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A., Tsegaye Y. M., Packer D. G., Majdalani N. & Gottesman S. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J. Bacteriol. 194, 2470–2478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M. et al. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315, 251–252 (2007). [DOI] [PubMed] [Google Scholar]
- Takeda T., Yun C. S., Shintani M., Yamane H. & Nojiri H. Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int. J. Evol. Biol. 2011, 685015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C., Alm E. J., Polz M. F., Spratt B. G. & Hanage W. P. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323, 741–746 (2009). [DOI] [PubMed] [Google Scholar]
- Oren Y. et al. Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc. Natl. Acad. Sci. USA. 111, 16112–16117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn H. J., Cameron A. D. & Dorman C. J. Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 10, e1004215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S., Figueroa-Bossi N., Rubino S. & Bossi L. (2001) Epitope tagging of chromosomal genes In Salmonella. Proc. Natl. Acad. Sci. USA 98, 15264–15269 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen. Genet. 119, 75–88 (1972). [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F. & Maniatis T. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory Press, New York, 1989). [Google Scholar]
- Dykhuizen D. Experimental studies of natural selection in bacteria. Annu. Rev. Ecol. Syst. 21, 373–398 (1990). [Google Scholar]
- Lenski R. E. Quantifying fitness and gene stability in microorganisms. Biotechnology 15, 173–192 (1991). [DOI] [PubMed] [Google Scholar]
- Hokamp K. et al. ArrayPipe: A flexible processing pipeline for microarray data. Nucleic Acids Res. 32, W457–W459 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–26 (2004). [DOI] [PubMed] [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa S., Keane T. M., Otto T. D., Newbold C. & Berriman M. ABACAS: Algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25, 1968–1969 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647forma (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.