Abstract
The G4C2 hexanucleotide repeat expansion mutation (HREM) in C9ORF72, represents the most common mutation associated with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). Three main disease mechanisms have been proposed to date: C9ORF72 haploinsufficiency, RNA toxicity, and accumulation of dipeptide repeat proteins. Pure GC content of the HREM potentially enables the formation of various non-B DNA structures such as G-quadruplexes and i-motifs. These structures are proposed to act as promoters and regulatory elements affecting replication, transcription and translation of the surrounding region. G-quadruplexes have already been shown on the G-rich sense DNA and RNA strands (G4C2)n, the structure of the anti-sense (G2C4)n strand remains unresolved. Similar C-rich sequences may, under acidic conditions, form i-motifs consisting of two parallel duplexes in a head to tail orientation held together by hemi-protonated C+-C pairs. We show that d(G2C4)n repeats do form i-motif and protonated hairpins even under near-physiological conditions. Rather than forming a DNA duplex, i-motifs persist even in the presence of the sense strand. This preferential formation of G-quadruplex and i-motif/hairpin structures over duplex DNA, may explain HREM replicational and transcriptional instability. Furthermore, i-motifs/hairpins can represent a novel pharmacological target for C9ORF72 associated ALS and FTLD.
The G4C2 hexanucleotide repeat expansion mutation (HREM), located in the first intron or promoter region of the C9ORF72 gene on chromosome 9p21 represents the most common mutation associated with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) in populations of European origin1,2,3. Individuals without the disorder possess on average 2 repeats with some individuals having up to 19, while patients with ALS or FTLD may present with up to 5000 repeats2,3,4,5,6,7.
Three main disease mechanisms have been proposed to date: C9ORF72 haploinsufficiency, RNA toxicity, and accumulation of dipeptide repeat proteins (DPR)8. C9ORF72 haploinsufficiency can arise from changes in the transcription and RNA processing rates of the mutation carrying allele2. On the other hand, the repeat RNA may be toxic as the accumulating RNA foci may sequester important RNA binding proteins9 a mechanism that has already been suggested for myotonic dystrophy10. Finally, the RNA transcripts can undergo repeat-associated non-ATG (RAN) translation, mediated by stable hairpin structures rather than an ATG codon. The resulting DPRs form toxic aggregates in various cell and animal models11,12,13. The complexity surrounding the C9ORF72 disease mechanisms is increased even further as the HREM region undergoes transcription also in the anti-sense direction giving rise to anti-sense RNA foci and additional DPRs11.
The pure GC content of both strands of HREM potentially enables the formation of various non-B DNA structures and the G-rich sense DNA and RNA strands (G4C2)n have been shown to form G-quadruplex structures14,15,16. The structure of the C-rich (G2C4)n anti-sense strand has not been defined to date, however other C-rich sequences have been suggested to form i-motifs, in acidic conditions, that consist of two parallel duplexes in a head to tail orientation held together by hemi-protonated C+-C pairs17,18. Like G-quadruplexes, i-motifs can act as promoters and regulatory elements affecting replication, transcription and translation of the surrounding region19,20,21.
Here we show that d(G2C4)n repeats can form i-motif as well as protonated hairpin structures even under conditions approaching physiological relevance and in the presence of the complementary strand. This property may contribute to replicative instability of HREM as well as affect the local transcription. Finally, i-motifs/hairpins may prove a novel drug target for C9ORF72 associated ALS and FTLD, as similar structures have already been shown to be susceptible to targeting with small molecules22.
Results
CD spectra indicate formation of i-motifs and hairpins
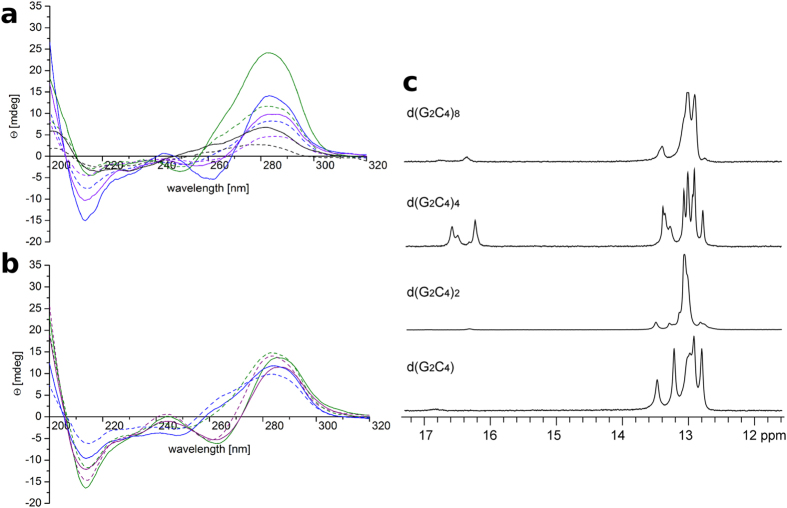
In order to get an indication of the structures formed by the repeats, CD spectra of d(G2C4), d(G2C4)2, d(G2C4)4, and d(G2C4)8 were measured in deionized H2O at 37 °C (Fig. 1a). In case of d(G2C4)4 and d(G2C4)8 a positive peak at 285 nm and a negative peak at 265 nm, characteristic of i-motifs20, were clearly visible. CD spectra measured at 5, 25, and 37 °C showed no differences in peak shapes and respective wavelengths (Supplementary Fig. 1). On the other hand the CD spectra of all four investigated oligonucleotides were pH dependent. Spectra of d(G2C4) did not show any i-motif characteristics at any of the tested pH values (Supplementary Fig. 2), while spectra of d(G2C4)2 and d(G2C4)8 showed a shift toward i-motif characteristic peaks up to pH 5.0 (Supplementary Fig. 3 and 5, respectively) and d(G2C4)4 up to pH 6.5 (Supplementary Fig. 4). However, characteristics of CD spectra as a function of pH could not be entirely explained by a decrease in the population of i-motif structures upon increasing of pH. At pH values higher than 6.5, an additional positive peak at around 260 nm as well as a negative peak at 245 nm were observed while the peak at 285 nm remained. Such observation could be explained by the presence of additional structures, such as hairpins23,24. The occurrence of additional structures was further substantiated with UV melting (Supplementary Table 1 and Fig. 6, respectively) and native PAGE (Supplementary Fig. 7).
Figure 1. CD and NMR spectra indicate the presence of protonated structures.
(a) CD spectra of d(G2C4) (black), d(G2C4)2 (green), d(G2C4)4 (blue), and d(G2C4)8 (purple) immediately after dilution in H2O (dashed line) and after incubation (full line). Positive and negative peaks, at 285 and 265 nm, respectively, are associated with the formation of i-motifs. (b) Comparison of CD spectra of d(G2C4)4 in water (dashed lines) and under molecular crowding conditions in 40% w/v PEG8000 (full lines) at pH 6.5 (purple), 7.0 (green), 7.2 (blue). Peaks characteristic for i-motifs persist up to pH 7.0 in 40% w/v PEG8000. All spectra were acquired at 37 °C. (c) Imino region of 1H NMR spectra of d(G2C4), d(G2C4)2, d(G2C4)4, and d(G2C4)8 after incubation in H2O at pH 6.0 and 5 °C. Signals between δ 12.5 and 13.5 ppm are indicative of Watson-Crick hydrogen bonding, while signals between δ 16.0 and 17.0 ppm correspond to imino protons from hemi-protonated C+-C base pairs.
The formation of non-B DNA structures could be affected by the crowded conditions within the nucleus. Because water availability influences the formation of such structures, CD spectra of d(G2C4)4 were then measured under molecular crowding conditions in 40% w/v PEG800025,26 at 37 °C and different pH. Under these conditions, CD spectra of d(G2C4)4 exhibited i-motif characteristic peaks up to pH 7.0 (Fig. 1b).
NMR reveals coexistence of protonated hairpins and i-motifs
Imino region of 1H NMR spectra of d(G2C4), d(G2C4)2, d(G2C4)4, and d(G2C4)8 acquired at pH 6.0 and 5 °C was examined in order to obtain deeper insights into secondary structure formation (Fig. 1c). All four oligonucleotides exhibit signals between δ 12.5 and 13.5 ppm indicative of Watson-Crick hydrogen bonding, which is to be expected given the high GC content and therefore high intra and intermolecular Watson-Crick binding ability of the oligonucleotides. In addition, signals between δ 16.0 and 17.0 ppm were observed, corresponding to imino protons from hemi-protonated C+-C base pairs (Fig. 1c). The observed chemical shifts are shifted downfield significantly with respect to previously reported values for protonated cytosines within i-motifs27 suggesting formation of other protonated structures.
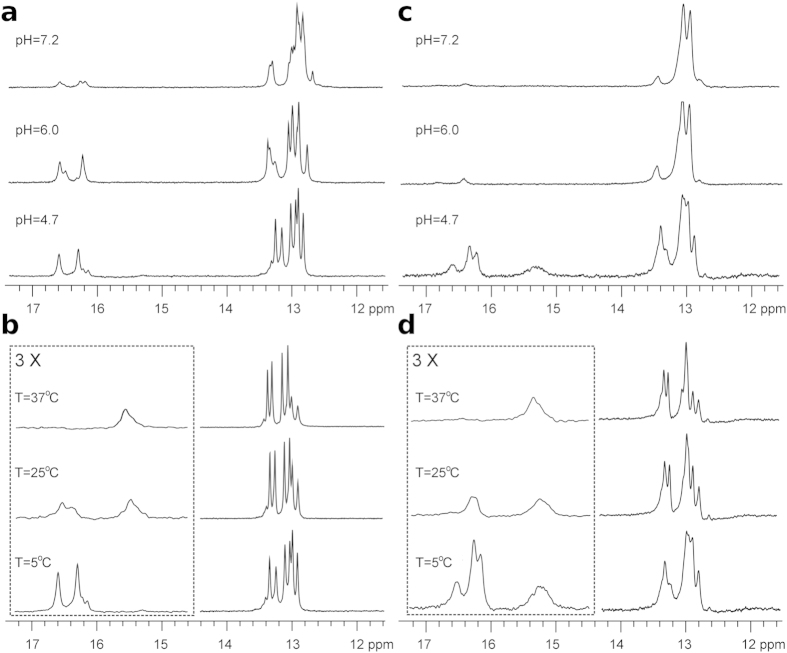
1H NMR spectra of d(G2C4)4 and d(G2C4)8 at a constant temperature of 5 °C and pH ranging from 4.7 to 7.2 showed signals between δ 16.1 and 16.8 ppm corresponding to hemi-protonated C+-C base pairs as well as signals indicative of Watson-Crick hydrogen bonding (Fig. 2a,c). With increasing pH, imino signals from hemi-protonated C+-C base pairs decreased while signals corresponding to GC-base pairs remained constant. Furthermore, at pH 4.7, d(G2C4)8 exhibited a broad signal at δ 15.3 ppm, which is indicative of i-motif structure formation (Fig. 2c)18. In the case of d(G2C4)4 at pH 4.7, a broad signal at δ 15.5 ppm was first observed at 25 °C (Fig. 2b). For both oligonucleotides signals corresponding to i-motifs further increased in intensity up to 37 °C while signals between δ 16.1 and 16.8 ppm gradually decreased at higher temperatures and eventually vanished at 37 °C (Fig. 2b,d). Signals indicative of GC-base pairs persisted under all tested temperatures.
Figure 2. Coexistence of i-motifs and protonated hairpins.
(a) Imino region of 1H NMR spectra of d(G2C4)4 at 5 °C and pH 4.7, 6.0, and 7.2. (b) Imino region of 1H NMR spectra of d(G2C4)4 at 5, 25, and 37 °C at pH 4.7. The vertical scale of the spectral region indicated by the dotted box has been enlarged three-fold. (c) Imino region of 1H NMR spectra of d(G2C4)8 at 5 °C and pH 4.7, 6.0, and 7.2. (d) Imino region of 1H NMR spectra of d(G2C4)8 at 5, 25, and 37 °C at pH 4.7. The vertical scale of the spectral region indicated by the dotted box has been enlarged three-fold.
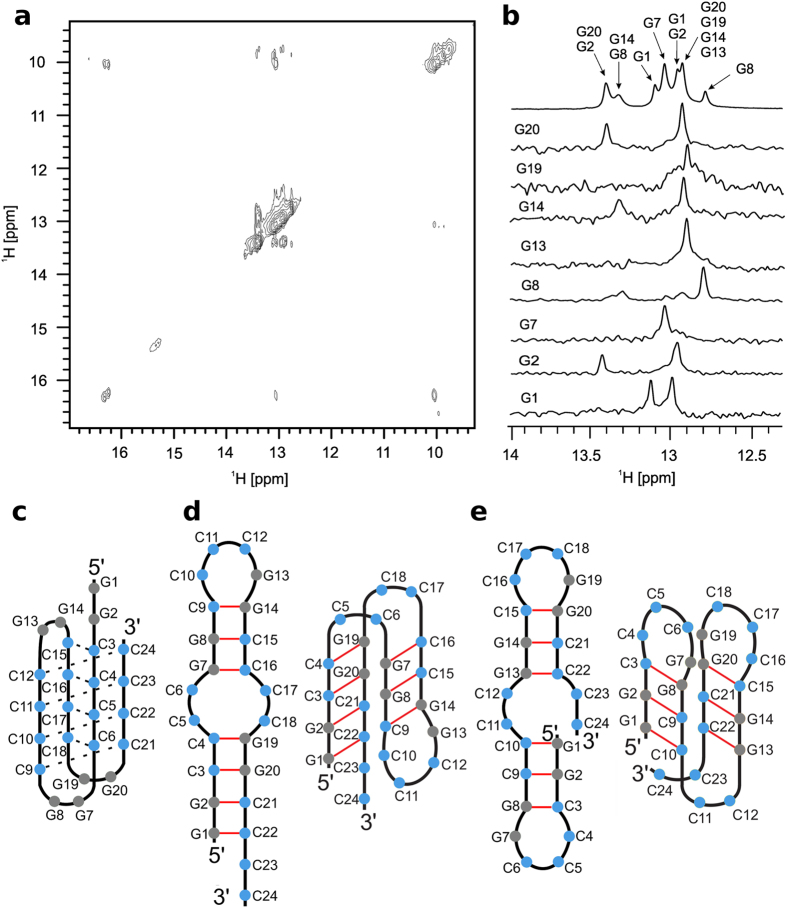
Intra or intermolecular nature of structures adopted by d(G2C4)4 and d(G2C4)8 was established by measuring translation diffusion coefficients (Dt) in 90% H2O/10% 2H2O at 25 °C. The observed Dt values range between 1.3–1.4 and 0.8–0.9 × 10−6 cm2 s−1 for d(G2C4)4 and d(G2C4)8, respectively. The corresponding hydrodynamic dimensions are consistent with a unimolecular nature of structures adopted by both d(G2C4)4 and d(G2C4)8 in solution28,29. Analysis of 2D NOESY NMR spectrum of d(G2C4)4 acquired at pH 4.7 and 5 °C (Fig. 3a) showed that signals corresponding to hemi-protonated C+-C base pairs exhibit NOE correlations with signals of GC-base pairs. Therefore, C+-C and GC-base pairs are in proximity within a single structure. On the contrary, no NOE correlations between signals at δ 15.5 ppm and signals of GC-base pairs were observed, which suggested presence of another distinct structure. Observed NOE correlations together with translation diffusion coefficient value of d(G2C4)4 indicate the coexistence of i-motifs with guanine residues in loops (Fig. 3c) and protonated hairpins involving GC and C+-C base pairs. 1D 15N-edited HSQC NMR spectra on partially 15N-residue-specific labelled d(G2C4)4 were used to determine which guanine residues were involved in GC-base pairs within protonated hairpins30. NMR data clearly showed that all guanine residues were within GC-base pairs since signals of their imino protons appear in the region indicative of Watson-Crick hydrogen bonds (Fig. 3b). It is interesting to note that single imino signals were observed for guanine residues G7, G13 and G19 in 1D 15N-edited HSQC NMR spectra, while two imino signals per guanine residue were observed for the others (Fig. 3b). The presence of two imino signals for some of guanine residues could be explained only by the coexistence of two different protonated hairpins, with a difference in the involvement of guanine residues of their GC-base pairs and their loop region (Fig. 3d,e).
Figure 3. Assignment of guanine imino protons and proposed structures.
(a) Imino-imino region of NOESY spectrum of d(G2C4)4 at mixing time of 150 ms. (b) Assignment of imino protons of guanine residues involved in GC base pairs using 1D 15N-edited HSQC NMR spectra of residue specific (marked on the left side of each spectrum) 15N, 13C-isotopically labelled d(G2C4)4. Top spectrum in B represents imino region of 1H NMR spectrum. All spectra were acquired at pH 4.7 and 5 °C. (c) Proposed structures of i-motif and (d,e) hairpins adopted by d(G2C4)4.
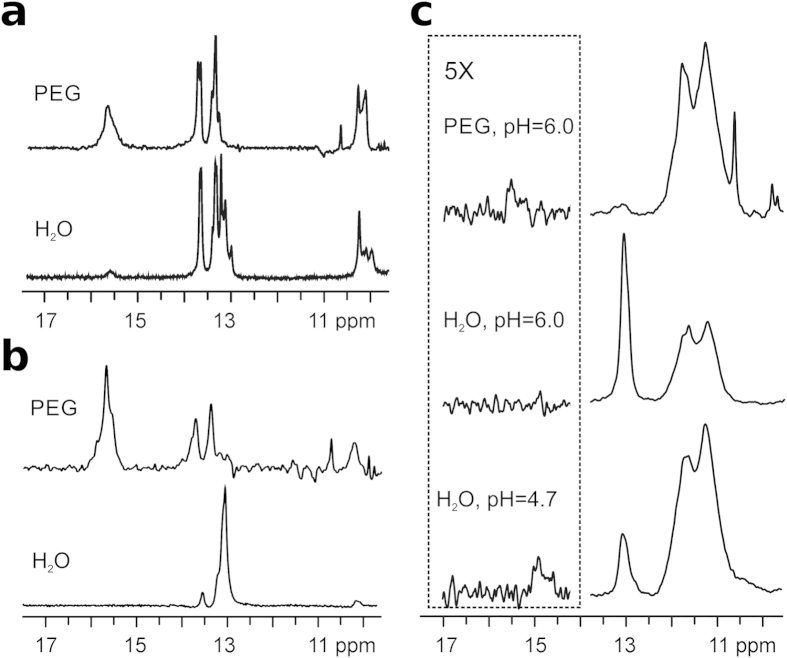
In addition to molecular crowding, other factors including biologically important cations can affect non-B DNA structures (e.g. K+ ion presence promotes formation of G-quadruplexes). In order to simulate intracellular conditions, d(G2C4)4 and d(G2C4)8 were dissolved in 40% w/v PEG8000 with added 100 mM K+ ions and 37 °C. Under these conditions and at pH approaching neutral values, 1H NMR spectra of d(G2C4)4 and d(G2C4)8 exhibit strong broad signals corresponding to i-motifs in addition to signals indicating the presence of hairpins connected through GC-base pairs (Fig. 4a,b). Intensities of signals belonging to hemi-protonated C+-C base pairs within i-motifs were significantly higher under molecular crowding conditions in comparison to the same signals observed for both oligonucleotides dissolved in water alone.
Figure 4. i-motifs persist under molecular crowding and in the presence of the complementary strand.
(a) Imino region of 1H NMR spectra of d(G2C4)4 and (b) d(G2C4)8 in water and in 40% w/v PEG8000, at pH 6.0 with 100 mM K+ ions. (c) Imino region of 1H NMR spectra of equimolar mixture of d(G2C4)8 and d(G4C2)8 in water at pH 4.7 and 6.0, and in 40% w/v PEG8000 at pH 6.0 with 100 mM K+ ions. The vertical scale of the spectral region indicated by the dotted box has been enlarged five-fold. All spectra were recorded at 37 °C.
Since in vivo both G- as well as C-rich strands are present, an equimolar mixture of d(G4C2)8 and d(G2C4)8 was dissolved in water in the presence of 100 mM K+ ions at pH 4.7 and 37 °C. Imino region of the resulting mixture after annealing exhibited signals characteristic of Hoogsteen hydrogen bonding within G-quartets in addition to signals corresponding to GC-base pairs. A weak signal at δ 15.0 ppm, indicating the presence of i-motifs, was also observed at pH 4.7 (Fig. 4c, bottom). Upon the increase of pH to 6.0 this signal disappeared and only signals corresponding to imino protons involved in G-quartets and GC-base pairs were observed (Fig. 4c, middle). However, a weak signal at δ 15.5 ppm indicating i-motifs was observed at pH 6.0 and 37 °C under molecular crowding conditions in the presence of K+ ions and the sense d(G4C2)8 strand (Fig. 4c, top). Therefore, an equimolar mixture of the complementary strands in conditions simulating the intracellular environment forms G-quadruplexes, hairpins and i-motifs, rather than a DNA duplex.
Discussion
Here we have shown that the highly C-rich anti-sense strand of HREM can form protonated hairpins as well as i-motifs and that the formation of these structures takes place at physiological pH, temperature and under molecular crowding conditions. In addition we show that these structures persist even in the presence of the complementary strand, which has important implications for key cellular processes – DNA replication and transcription. The potential separation of the HREM complementary strands independently of the replication/transcription machinery may adversely affect both the stability and rate of the HREM replication and transcription contributing to the proposed disease mechanisms.
Currently, attempts are underway to influence the disease mechanisms through targeting of G-quadruplex structures and their binding proteins on the sense-strand (G4C2)n HREM31,32. Since the transcript of the C9ORF72 mRNA is made on the basis of the anti-sense strand, the latter may also be targeted for regulation of the transcription leading to RNA toxicity and DPR accumulation. So far, several ligands that either stabilize or destabilize specific i-motif/hairpin structures have been developed33. Proof of concept for transcriptional modulation through targeting of i-motif/hairpins was recently demonstrated in the case of BCL2 gene, whose i-motif/hairpin conformation pair was first characterized as part of its promotor region in 200922,34. HREM i-motifs/hairpins could therefore also prove as attractive new target for modulation of all three proposed disease mechanisms in C9ORF72-associated ALS and FTLD at their source.
Methods
Sample preparation
Reverse-phase purified oligonucleotides d(G2C4), d(G2C4)2, d(G2C4)4, d(G4C2)4, and dual HPLC purified oligonucleotides d(G2C4)8 and d(G4C2)8 were purchased from Eurogentec (Seraing, Belgium). Oligonucleotides d(G2C4) and d(G2C4)2 were cleaned using 2M LiCl and dialysed four times against water, and concentrated using an ultra-filtration device (Merck Millipore, Herfordshire, UK) and an ultra-filtration membrane (regenerated cellulose, Millipore). Oligonucleotides d(G2C4)4, d(G4C2)4, d(G2C4)8, and d(G4C2)8 were cleaned using 2M LiCl and passed through an Amicon ultrafilter. Residue specific 10% 15N, 13C-guanine and cytosine labeled samples were synthesized on K&A Laborgeraete GbR DNA/RNA Synthesizer H-8 using standard phosphoramidite chemistry. Deprotection was performed with overnight incubation in 20% aqueous ammonia at 50 °C. 2M LiCl was added prior to purification and concentration of samples using an Amicon ultrafilter.
The samples were lyophilized overnight and diluted in 90% of H2O and 10% of 2H2O. The concentrations of the 300 μL NMR samples were 3.3, 2.7, 1.1, 0.7, 0.4 and 0.3 mM for d(G2C4), d(G2C4)2, d(G2C4)4, d(G4C2)4, d(G2C4)8, and d(G4C2)8 respectively. In case of equimolar mixing experiments, equimolar quantities of d(G2C4)4 with d(G4C2)4 and d(G2C4)8 with d(G4C2)8 were heated to 90 °C prior to annealing. For testing of additional conditions d(G2C4)4, d(G4C2)4, d(G2C4)8, and d(G4C2)8 were diluted to 0.1 mM. For CD measurements, samples d(G2C4), d(G2C4)2 and d(G2C4)4 were diluted 60-fold, while d(G2C4)8 was diluted 30-fold in respective diluents (see CD section). For molecular crowding experiments, samples were prepared by diluting to 0.1 mM in 40% w/v PEG (8000 MW) (Sigma Aldrich, Munich, Germany) in water. 10% residue specific labeled samples were diluted in 90% of H2O and 10% of 2H2O. Concentrations were ranging from 0.4 to 0.7 mM. The pH of samples was adjusted by the addition of LiOH or HCl and measured using the 780 pH Meter (Metrohm, Herisau, Switzerland).
Circular dichroism spectroscopy
CD spectra were recorded on an Applied Photophysics Chirascan CD spectrometer at 5, 25, or 37 °C using a 0.1 cm path length quartz cell. The wavelength was varied from 200 to 320 nm. Three scans were averaged for each CD spectrum. In each case corresponding blanks were used for baseline correction. For CD spectroscopy the initial samples were measured 20 days after dilution at 5, 25, and 37 °C, at pH 6.2, 6.2, 6.5 and 6.5 for d(G2C4), d(G2C4)2, d(G2C4)4, and d(G2C4)8, respectively. The initial CD measurement was performed after denaturation at 90 °C. Measurements at different pH were performed in H2O at 37 °C and pH 4.0, 4.7, 5.0, 6.0, 6.5, 7.0, 7.2, 8.0, and 8.5. In order to model molecular crowding conditions, samples were prepared by diluting d(G2C4)4 and d(G2C4)8 to 15 μM per strand in 40% w/v PEG (8000 MW). The pH of the PEG solutions in water was adjusted using LiOH or HCl. All samples with molecular crowding conditions were measured at 37 °C after 8 days.
UV spectroscopy
Melting experiments of d(G2C4), d(G2C4)2, d(G2C4)4, and d(G2C4)8 were performed on a Varian Cary 100 Bio UV-VIS spectrometer (Varian Inc.) equipped with a thermoelectric temperature controller. UV melting experiments were performed on samples diluted to 2 μM in 100mM K-phosphate buffer at pH 6.0, using 1 cm path-length quartz cells. A combination of mineral oil and a fixed cuvette cap was used to prevent evaporation and sample loss due to high temperatures. A stream of nitrogen was applied throughout the measurements to prevent condensation at lower temperatures. Folding/unfolding processes were followed between 10 and 90 °C by measuring absorbance at 260 nm using scanning rates of 0.5 and 0.1 °C min−1. Temperatures of half transition (T1/2) were determined using the first derivative method.
Native PAGE electrophoresis
Native gel electrophoresis of d(G2C4), d(G2C4)2, d(G2C4)4, d(G4C2)4, d(G2C4)8, d(G4C2)8 and equimolar mixtures of d(G2C4)4 with d(G4C2)4, and d(G2C4)8 with d(G4C2)8, respectively, was performed on a 15% polyacrylamide gel (5 °C at 100 V) in 1xTBE (pH 6.5) buffer with 100 mM KCl. 1nmol of DNA sample was mixed with loading buffer (3 μl 15% ficoll, 2.5 X Tris borate) and diluted to 20 μl with water. The approximate size of the bands was determined by using the GeneRuler Ultra Low range DNA Ladder (Thermo Scientific, Waltham, MA USA). The samples were treated at room temperature overnight before loading, except for the equimolar mixture samples, that were heated to 90 °C and either cooled immediately on ice or cooled slowly at room temperature prior to loading. Following the overnight electrophoresis, the gel was stained with Stains All gel stain solution (Sigma Aldrich) and filmed using the DNR Bio-Imaging Systems instrument.
NMR spectroscopy
All NMR spectra were obtained with Agilent Technologies DD2 600 MHz NMR spectrometer at 5, 25, and 37 °C using a triple resonance cold probe. Standard 1D 1H spectra were acquired with the use of DPFGSE, watergate 3919 or PRESAT solvent suppression. Diffusion coefficient measurements were performed by a spin-echo pulse sequence with PFG gradient strengths between 0.49 and 29.06 G cm−1. NOESY spectra were acquired with mixing times of 80 and 150 ms. Assignment of imino protons of guanine residues was done by 1D 15N-edited HSQC experiments performed on residue specific 10% 15N, 13C-isotopically labeled oligonucleotides. Measurements at different pH were performed at pH 4.7, 6.0 and 7.2. Measurements under molecular crowding conditions were performed using 40% w/v dPEG (8000 MW) (Polymer Source Inc., Dorval, Canada) in water 7 days after annealing. KH2PO4 was used in experiments with 100 mM K+ ions. NMR spectra were processed and analyzed using VNMRJ (Varian Inc.) and Sparky (UCSF) software.
Additional Information
How to cite this article: Kovanda, A. et al. Anti-sense DNA d(GGCCCC)n expansions in C9ORF72 form i-motifs and protonated hairpins. Sci. Rep. 5, 17944; doi: 10.1038/srep17944 (2015).
Supplementary Material
Acknowledgments
This work was supported by the Slovenian Research Agency (grants P1-0242, P4-0127, J3-6789, J3-5502, J1-6733 and Z3-6802). We would like to thank Jean-Marc Gallo, Blaz Koritnik, Dušan Kordiš, and Mateus Webba da Silva for critical reading of the manuscript.
Footnotes
Author Contributions A.K., J.P. and B.R. designed research. M.Z. performed the experiments. P.Š., M.Z. and A.K. analyzed data. J.P. contributed reagents and analytic tools. B.R. provided primary idea and contributed reagents. A.K., M.Z., P.Š., J.P. and B.R. wrote the manuscript with extensive input from all authors.
References
- van der Zee J. et al. A Pan-European Study of the C9orf72 Repeat Associated with FTLD: Geographic Prevalence, Genomic Instability, and Intermediate Repeats. Hum. Mutat. 34, 363–373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J. et al. Large C9orf72 Hexanucleotide Repeat Expansions Are Seen in Multiple Neurodegenerative Syndromes and Are More Frequent Than Expected in the UK Population. Am. J. Hum. Genet. 92, 345–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Tortosa E. et al. C9ORF72 hexanucleotide expansions of 20–22 repeats are associated with frontotemporal deterioration. Neurology 80, 366–370 (2013). [DOI] [PubMed] [Google Scholar]
- Smith B. N. et al. The C9ORF72 expansion mutation is a common cause of ALS+/–FTD in Europe and has a single founder. Eur. J. Hum. Genet. EJHG 21, 102–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M. et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 12, 978–988 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatovec S., Kovanda A. & Rogelj B. Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging 35, 2421.e1–2421.e12 (2014). [DOI] [PubMed] [Google Scholar]
- Haeusler A. R. et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson O. J., Aagaard L., Jensen T. G. & Damgaard C. K. Molecular mechanisms in DM1 - a focus on foci. Nucleic Acids Res. 43, 2433–2441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- Mizielinska S. et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I. et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P. et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep . 2, 1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K., Zamiri B., Stanley S. Y. R., Macgregor R. B. & Pearson C. E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 288, 9860–9866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šket P. et al. Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging 36, 1091–1096 (2015). [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J. L. & Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature 363, 561–565 (1993). [DOI] [PubMed] [Google Scholar]
- Benabou S., Aviñó A., Eritja R., González C. & Gargallo R. Fundamental aspects of the nucleic acid i-motif structures. RSC Adv . 4, 26956 (2014). [Google Scholar]
- Qin Y. & Hurley L. H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90, 1149–1171 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick S. & Hurley L. H. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. Chim. Pure Appl . 82, 1609–1621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks T. A., Kendrick S. & Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 277, 3459–3469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick S. et al. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 136, 4161–4171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypr J., Kejnovská I., Renciuk D. & Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlíčková M., Kejnovská I., Bednářová K., Renčiuk D. & Kypr J. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality 24, 691–698 (2012). [DOI] [PubMed] [Google Scholar]
- Miyoshi D., Karimata H. & Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 128, 7957–7963 (2006). [DOI] [PubMed] [Google Scholar]
- Cui J., Waltman P., Le V. H. & Lewis E. A. The effect of molecular crowding on the stability of human c-MYC promoter sequence I-motif at neutral pH. Mol. Basel Switz . 18, 12751–12767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Ambrus A., Hurley L. H. & Yang D. A Direct and Nondestructive Approach To Determine the Folding Structure of the I-Motif DNA Secondary Structure by NMR. J. Am. Chem. Soc. 131, 6102–6104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusic M., Sket P., Bauer L., Viglasky V. & Plavec J. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 40, 6946–6956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovski M., da Silva M. W. & Plavec J. Unique structural features of interconverting monomeric and dimeric G-quadruplexes adopted by a sequence from the intron of the N-myc gene. J. Am. Chem. Soc. 134, 4132–4141 (2012). [DOI] [PubMed] [Google Scholar]
- Phan A. T. & Patel D. J. A Site-Specific Low-Enrichment 15 N, 13 C Isotope-Labeling Approach to Unambiguous NMR Spectral Assignments in Nucleic Acids. J. Am. Chem. Soc. 124, 1160–1161 (2002). [DOI] [PubMed] [Google Scholar]
- Zamiri B., Reddy K., Macgregor R. B. Jr & Pearson C. E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J. Biol. Chem. 289, 4653–4659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U. S. A. 110, E4530–4539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day H. A., Pavlou P. & Waller Z. A. E. i-Motif DNA: Structure, stability and targeting with ligands. Bioorg. Med. Chem. 22, 4407–4418 (2014). [DOI] [PubMed] [Google Scholar]
- Kendrick S., Akiyama Y., Hecht S. M. & Hurley L. H. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 131, 17667–17676 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.