Abstract
Systemic lupus erythematosus (SLE; OMIM 152700) is a genetically complex autoimmune disease characterized by loss of immune tolerance to nuclear and cell surface antigens. Previous genome-wide association studies (GWAS) had modest sample sizes, reducing their scope and reliability. Our study comprised 7,219 cases and 15,991 controls of European ancestry: a new GWAS, meta-analysis with a published GWAS and a replication study. We have mapped 43 susceptibility loci, including 10 novel associations. Assisted by dense genome coverage, imputation provided evidence for missense variants underpinning associations in eight genes. Other likely causal genes were established by examining associated alleles for cis-acting eQTL effects in a range of ex vivo immune cells. We found an over-representation (n=16) of transcription factors among SLE susceptibility genes. This supports the view that aberrantly regulated gene expression networks in multiple cell types in both the innate and adaptive immune response contribute to the risk of developing SLE.
SLE is a clinically heterogeneous disease with a strong genetic component, as demonstrated by the tenfold increase in concordance rates between monozygotic and dizygotic twins1, and familial aggregation (sibling risk ratio, λs = 29)2. Since 2008, the field of SLE genetics has been transformed by GWA3–8 and independent replication studies9,10. However, while the pace of discovery has been unprecedented, providing a richer understanding of lupus genetic etiology, these findings were driven by modestly-sized GWA studies, utilizing 1,800 European patients3,4 and slightly fewer Asian cases5,6; they therefore had limited power to detect loci with relatively low odds ratios and/or minor allele frequencies11. The size of our study, coupled with a meta-analysis and replication study, has greatly increased the power to detect susceptibility loci.
We genotyped 4,946 individuals with SLE and 1,286 healthy controls using the Illumina HumanOmni1-Quad BeadChip. These data were combined with the genotypes of 5,727 healthy controls taken from the University of Michigan Health and Retirement Study (HRS), genotyped using the Illumina HumanOmni2.5 BeadChip. Following quality control (QC) analyses, our data comprised 4,036 SLE cases and 6,959 controls (1,260 controls mainly from southern Europe genotyped using the Omni1-Quad chip and 5,699 controls from the HRS cohort). The final SNP set comprised 644,674 markers that were present on both the Omni1-Quad and Omni2.5 chips (see Online Methods). Four principal components were used as covariates to correct for population structure12,13. The genomic inflation factor14,15 for our data, λ1000, was 1.02, with λGC = 1.16.
Our analysis strategy is described in detail in Online Methods, and is shown schematically in Supplementary Fig. 1. This GWAS identified 25 loci (Table 1 and Supplementary Fig. 2a) of genome-wide significance (P < 5 × 10−08). Three of these associations are novel in SLE: rs6740462 and rs3768792 on chromosome 2p14 and 2q34, respectively and rs7726414 on chromosome 5q31.1.
Table 1. Allelic associations at SLE susceptibility loci following meta–analysis with replication study.
| GWAS | Hom et al. GWAS | Replication study | Post–replication study meta–analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| SNP | Chr | Position (b37) | Locusc | P–value | Odds Ratio | P–value | Odds Ratio | P–value | Odds Ratio | P–value | Odds Ratio | 95% CI |
| rs2476601 | 1 | 114,377,568 | PTPN22 | 8.34E–13 | 1.39 | 9.06E–04 | 1.32 | 6.00E–15 | 1.54 | 1.10E–28 | 1.43 | 1.34 – 1.53 |
| rs1801274 | 1 | 161,479,745 | FCGR2A | 6.05E–11 | 1.21 | 1.78E–02 | 1.13 | 8.38E–03 | 1.10 | 1.04E–12 | 1.16 | 1.11 – 1.21 |
| rs704840 | 1 | 173,226,195 | TNFSF4 | 1.65E–13 | 1.26 | 7.65E–05 | 1.25 | 2.32E–04 | 1.15 | 3.12E–19 | 1.22 | 1.17 – 1.27 |
| rs17849501a | 1 | 183,542,323 | SMG7 NCF2 | 1.63E–59 | 2.24 | 3.96E–05 | 1.58 | 2.84E–30 | 2.08 | 3.45E–88 | 2.10 | 1.95 – 2.26 |
| rs3024505 | 1 | 206,939,904 | IL10 | 2.55E–03 | 1.12 | 3.99E–07 | 1.42 | 4.00E–03 | 1.15 | 4.64E–09 | 1.17 | 1.11 – 1.24 |
| rs9782955 | 1 | 236,039,877 | LYST | 5.58E–04 | 1.12 | 3.93E–06 | 1.33 | 1.38E–03 | 1.15 | 1.25E–09 | 1.16 | 1.11 – 1.22 |
| rs6740462 a | 2 | 65,667,272 | SPRED2 | 2.31E–08 | 1.20 | 9.55E–02 | 1.11 | 4.91E–01 | 0.97 | 2.67E–05 | 1.10 | 1.05 – 1.16 |
| rs2111485 | 2 | 163,110,536 | IFIH1 | 3.44E–06 | 1.15 | 2.97E–03 | 1.17 | 6.52E–05 | 1.16 | 1.27E–11 | 1.15 | 1.11 – 1.20 |
| rs11889341a | 2 | 191,943,742 | STAT4 | 1.17E–65 | 1.75 | 3.70E–13 | 1.54 | 2.16E–48 | 1.79 | 5.59E–122 | 1.73 | 1.65 – 1.81 |
| rs3768792 | 2 | 213,871,709 | IKZF2 | 2.35E–08 | 1.26 | 5.49E–03 | 1.22 | 7.12E–05 | 1.22 | 1.21E–13 | 1.24 | 1.17 – 1.31 |
| rs9311676 | 3 | 58,470,351 | ABHD6 PXK | 5.37E–06 | 1.14 | 7.58E–02 | 1.10 | 1.45E–10 | 1.27 | 3.06E–14 | 1.17 | 1.13 – 1.22 |
| rs564799 | 3 | 159,728,987 | IL12A | 1.15E–06 | 1.15 | 2.83E–01 | 1.06 | 1.78E–04 | 1.15 | 1.54E–09 | 1.14 | 1.09 – 1.18 |
| rs10028805 | 4 | 102,737,250 | BANK1 | 4.50E–10 | 1.21 | 4.68E–01 | 1.04 | 9.84E–11 | 1.28 | 4.31E–17 | 1.20 | 1.15 – 1.25 |
| rs7726414 | 5 | 133,431,834 | TCF7 SKP1 | 9.17E–10 | 1.46 | 2.88E–01 | 1.14 | 3.97E–08 | 1.56 | 4.44E–16 | 1.45 | 1.32 – 1.58 |
| rs10036748 | 5 | 150,458,146 | TNIP1 | 2.83E–18 | 1.32 | 3.36E–07 | 1.35 | 2.53E–24 | 1.50 | 1.27E–45 | 1.38 | 1.32 – 1.45 |
| rs2431697 | 5 | 159,879,978 | MIR146A | 3.23E–14 | 1.25 | 2.22E–03 | 1.18 | 4.16E–14 | 1.32 | 8.01E–28 | 1.26 | 1.21 – 1.31 |
| rs1270942 | 6 | 31,918,860 | MHC class IIId | 1.70E–101 | 2.52 | 6.15E–13 | 1.75 | 7.43E–60 | 2.23 | 2.25E–165 | 2.28 | 2.15 – 2.42 |
| rs9462027 | 6 | 34,797,241 | UHRF1BP1 | 1.80E–05 | 1.14 | 1.47E–01 | 1.09 | 2.42E–04 | 1.15 | 7.55E–09 | 1.14 | 1.09 – 1.19 |
| rs6568431 | 6 | 106,588,806 | PRDM1 ATG5 | 4.33E–12 | 1.22 | 2.29E–03 | 1.17 | No Data | No Data | 5.04E–14 | 1.21 | 1.15 – 1.27 |
| rs6932056a | 6 | 138,242,437 | TNFAIP3 | 1.23E–16 | 1.82 | 8.08E–03 | 1.47 | 1.20E–14 | 1.99 | 1.97E–31 | 1.83 | 1.65 – 2.02 |
| rs849142 | 7 | 28,185,891 | JAZF1 | 3.49E–05 | 1.13 | 4.23E–04 | 1.20 | 2.04E–04 | 1.14 | 8.61E–11 | 1.14 | 1.10 – 1.19 |
| rs4917014 | 7 | 50,305,863 | IKZF1 | 4.10E–05 | 1.14 | 3.25E–03 | 1.19 | 1.49E–09 | 1.27 | 6.39E–14 | 1.18 | 1.13 – 1.24 |
| rs10488631 | 7 | 128,594,183 | IRF5 | 2.66E–44 | 1.79 | 4.50E–17 | 1.93 | 2.86E–52 | 2.12 | 9.37E–110 | 1.92 | 1.81 – 2.03 |
| rs2736340 | 8 | 11,343,973 | BLK | 2.14E–16 | 1.30 | 6.42E–05 | 1.27 | No Data | No Data | 6.28E–20 | 1.29 | 1.22 – 1.37 |
| rs2663052a | 10 | 50,069,395 | WDFY4 | 1.59E–08 | 1.18 | 6.25E–02 | 1.10 | No Data | No Data | 5.25E–09 | 1.16 | 1.10 – 1.22 |
| rs4948496 | 10 | 63,805,617 | ARID5B | 1.17E–06 | 1.15 | 5.76E–01 | 0.97 | 2.76E–08 | 1.22 | 1.04E–10 | 1.14 | 1.10 – 1.19 |
| rs12802200a | 11 | 566,936 | IRF7 | 8.43E–09 | 1.24 | 2.03E–02 | 1.18 | No Data | No Data | 8.81E–10 | 1.23 | 1.15 – 1.31 |
| rs2732549a | 11 | 35,088,399 | CD44 | 1.31E–10 | 1.21 | 1.51E–03 | 1.18 | 1.88E–13 | 1.31 | 1.20E–23 | 1.24 | 1.19 – 1.29 |
| rs3794060 | 11 | 71,187,679 | DHCR7 NADSYN1 | 1.13E–04 | 1.13 | 8.18E–02 | 1.11 | 2.61E–23 | 1.47 | 1.32E–20 | 1.23 | 1.18 – 1.29 |
| rs7941765 | 11 | 128,499,000 | ETS1 FLI1 | 9.82E–07 | 1.15 | 4.64E–03 | 1.17 | 1.55E–03 | 1.12 | 1.35E–10 | 1.14 | 1.10 – 1.19 |
| rs10774625 | 12 | 111,910,219 | SH2B3 | 9.47E–08 | 1.17 | 4.32E–03 | 1.16 | 9.81E–02 | 1.06 | 4.09E–09 | 1.13 | 1.08 – 1.18 |
| rs1059312 | 12 | 129,278,864 | SLC15A4 | 3.20E–06 | 1.14 | 3.97E–03 | 1.16 | 4.14E–07 | 1.20 | 1.48E–13 | 1.17 | 1.12 – 1.21 |
| rs4902562 | 14 | 68,731,458 | RAD51B | 4.85E–05 | 1.13 | 1.49E–02 | 1.14 | 5.78E–05 | 1.16 | 6.15E–10 | 1.14 | 1.09 – 1.19 |
| rs2289583a | 15 | 75,311,036 | CSK | 9.35E–09 | 1.20 | 1.68E–02 | 1.14 | 2.12E–06 | 1.20 | 6.22E–15 | 1.19 | 1.14 – 1.24 |
| rs9652601a,b | 16 | 11,174,365 | CIITA SOCS1 | 3.86E–07 | 1.17 | 4.00E–01 | 1.05 | 2.71E–15 | 1.36 | 7.42E–17 | 1.21 | 1.15 – 1.26 |
| rs34572943a,b | 16 | 31,272,353 | ITGAM | 1.74E–47 | 1.78 | 1.90E–07 | 1.52 | 1.04E–24 | 1.68 | 3.39E–76 | 1.71 | 1.61 – 1.81 |
| rs11644034 | 16 | 85,972,612 | IRF8 | 1.25E–15 | 1.34 | 9.81E–03 | 1.18 | 5.42E–04 | 1.16 | 9.58E–18 | 1.25 | 1.19 – 1.32 |
| rs2286672 b | 17 | 4,712,617 | PLD2 | 5.81E–05 | 1.24 | 2.50E–02 | 1.24 | 2.35E–04 | 1.27 | 2.93E–09 | 1.25 | 1.16 – 1.35 |
| rs2941509 | 17 | 37,921,194 | IKZF3 | 4.32E–06 | 1.41 | 2.34E–01 | 1.16 | 6.27E–04 | 1.35 | 7.98E–09 | 1.35 | 1.22 – 1.49 |
| rs2304256a | 19 | 10,475,652 | TYK2 | 2.34E–12 | 1.26 | 1.51E–02 | 1.16 | No Data | No Data | 3.50E–13 | 1.24 | 1.17 – 1.31 |
| rs7444a,b | 22 | 21,976,934 | UBE2L3 | 1.30E–13 | 1.28 | 1.89E–01 | 1.09 | 3.51E–11 | 1.32 | 1.84E–22 | 1.27 | 1.21 – 1.33 |
| rs887369 a | X | 30,577,846 | CXorf21 | 9.25E–07 | 1.16 | 6.62E–02 | 1.23 | 4.55E–04 | 1.14 | 5.26E–10 | 1.15 | 1.10 – 1.21 |
| rs1734787a | X | 153,325,446 | IRAK1 MECP2 | 2.83E–11 | 1.57 | 8.58E–04 | 1.52 | 9.54E–06 | 1.20 | 1.78E–15 | 1.31 | 1.22 – 1.40 |
Novel SLE associations are shown in bold type.
Imputed data in the Hom et al study. IMPUTE info scores: rs17849501 (0.78), rs6740462 (1.00), rs11889341 (0.99), rs6932056 (0.94), rs2663052 (1.00), rs12802200 (0.90), rs2732549 (1.00), rs2289583 (0.99), rs9652601 (1.00), rs34572943 (0.90), rs2304256 (0.95), rs7444 (1.00), rs887369 (0.83), rs1734787 (0.95).
Imputed controls in the replication study. IMPUTE info scores: rs9652601(0.99), rs34572943 (0.91), rs2286672(0.88), rs7444 (0.99).
For rationale for candidate gene selection at the associated loci see Table 2
For more detailed analysis of MHC see text
To validate these findings, and to search for additional susceptibility loci, we carried out a meta-analysis of our GWAS results and those from an independent European SLE GWAS comprising 1,165 cases and 2,107 controls (the Hom et al.4 study). Each of the 25 loci mapped in the original GWAS had genome-wide significant p-values in this meta-analysis (Supplementary Table 1), and are therefore considered to be associated with SLE. We then designed a replication study, with inclusion based on the meta-analysis of the two GWA studies. At loci with no published association in SLE, we adopted a threshold for inclusion of P < 2.5 × 10−05, while for loci with previously reported associations the threshold was set at P < 1 × 10−04 (see Online Methods for rationale). The 33 SNPs with P-values meeting these criteria were genotyped in our replication study (Supplementary Table 2), using a custom panel that also included 53 ancestry informative markers (see Online Methods). After applying QC measures, the replication data comprised 2,018 cases and 6,925 controls, none of which had been included in either GWAS (see Online Methods).
Finally, we carried out a post-replication meta-analysis of the results of our GWAS, the Hom et al. study and the replication study for those 33 SNPs, again applying the standard measure of genome-wide significance. The 18 SNPs (over and above the 25 already mapped) with P-values < 5 × 10−08 in this meta-analysis were also considered to be associated with SLE (Table 1 and Supplementary Fig. 2b). In addition to the three novel loci mapped in the GWAS, seven further variants, at loci hitherto not showing genome-wide significant association in SLE, were mapped in the overall meta-analysis: rs564799 (3q25.33), rs3794060 (11q13.4), rs10774625 (12q14.1), rs4902562 (14q24.1), rs9652601 (7q32.1), rs2286672 (17p13.2) and rs887369 (Xp21.2). The heritability explained by these 43 validated susceptibility alleles is 19.3% [95% C.I. 14.1–25.5%], where the total heritability of lupus is estimated to be 66%16. This is a large increase on the 8.7% [5.33–12.96%] reported by So et al.17 in 2011 using the same measure.
We imputed both the main GWAS and Hom et al. data to the density of the 1000 Genomes (1KG) study18 and re-analyzed the data (see Online Methods). While no additional loci were identified, we did obtain stronger evidence in support of some loci, for example the signal at the SPRED2 locus, at which the most associated 1KG variant, rs268134, was strongly replicated. In addition, the imputation enabled us to fine map associated loci and to determine whether multiple signals were present (Supplementary Tables 3a and 3b). We identified multiple independent association signals at the TNFSF4, STAT4 and IRF5 loci, as well as five independently associated SNPs at the MHC (see below).
Given that the SNP with the smallest P-value is not necessarily the true causal variant, we considered SNPs from the most associated to a defined cut-off as potentially causal in our subsequent analyses. Specifically, guided by previous work on functional annotation19 (see Online Methods), the cut-off was defined as a Bayes Factor against the most significantly associated SNP equal to 0.34. Any SNPs in this set that were missense variants were considered more likely candidates than the most associated SNP. The results are summarized in Supplementary Tables 3c and 4, listing candidate causal missense variants in PTPN22, FCGR2A, NCF2, TNFAIP3, WDFY4, IRF7, ITGAM and TYK2.
MHC polymorphisms, including SNPs and classical human leukocyte antigen (HLA) alleles, have consistently been observed to be associated with SLE20. We imputed HLA alleles21 in both the main GWAS and Hom et al. data, and incorporated them into our analysis of 1KG imputed data across the MHC (see Online Methods). Of the five MHC SNPs we find to be independently associated with SLE (Supplementary Tables 3a and 3b), the class III SNP in SLC44A4 (rs74290525) is the only association signal that is clearly independent of any HLA alleles. We find that rs74290525 is significantly associated not only when conditioning on each of the HLA genes separately, but even when conditioning on all 199 HLA alleles (see Supplementary Tables 5a–e), and is not in linkage disequilibrium (LD) with any HLA alleles (R2 < 0.1 with each HLA allele). We find that the best model for association includes the HLA class I alleles B*08:01, B*18:01, the class II alleles DQB1*02:01, DRB3*02:00 and DQA*01:02, and the class III SNP rs74290525, consistent with previous findings suggesting multiple SLE associations at the MHC20 (Supplementary Tables 6a and 6b). LD between the five MHC SNPs and HLA alleles on known SLE risk haplotypes can be seen in Supplementary Table 6c.
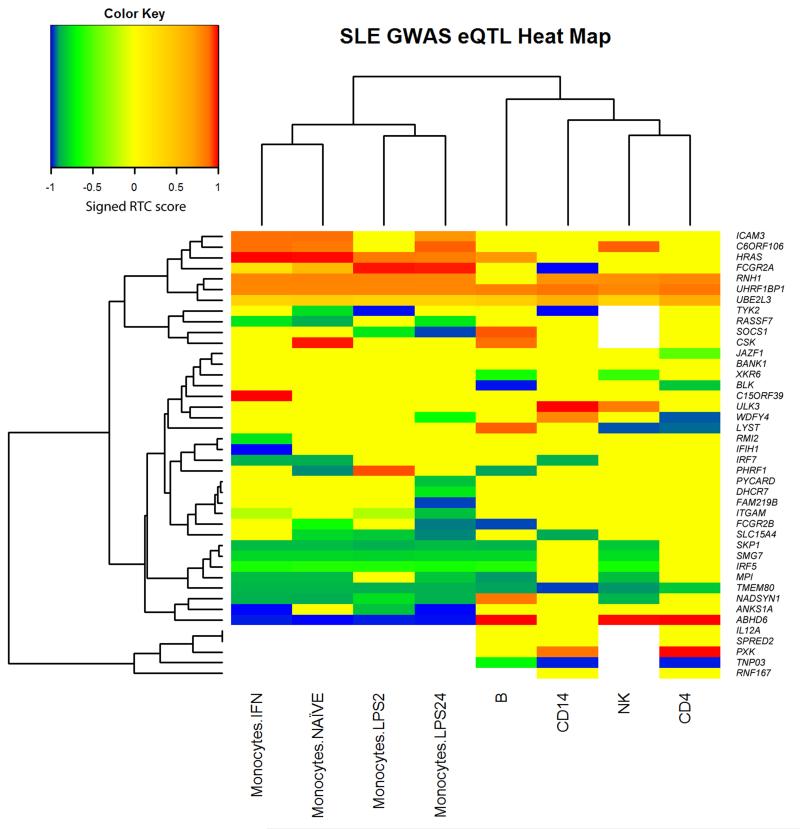
In order to highlight potential causal genes at the susceptibility loci, the associated SNPs at each of the loci were tested for correlation with cis-acting gene expression in ex vivo naïve CD4+ T cells, B cells, natural killer (NK) cells, and stimulated and resting monocytes22–24. Figure 1 displays a heat map across cell types, showing genes exhibiting significant differential expression in relation to the SLE associated alleles. We calculated Regulatory Trait Concordance (RTC) scores25 (see Supplementary Figs. 3a and b) to test the relationship between eQTLs driven by disease-associated alleles, and other, potentially stronger eQTLs, which we identified at each locus. The cis eQTLs were distributed across all cell types tested, some being common to all cell types, such as UBE2L3 and UHRF1BP1, while others are more cell specific: BLK in B cells and JAZF1 in T cells. In general directionality was consistent, although not in all cases: for example ABHD6 showed reduced expression in monocytes and elevated expression in lymphocytes.
Figure 1.
Heat map for cis-acting gene expression RTC scores from ex vivo cells. The heat map includes all genes with evidence of cis-regulatory (+/– 1Mb) action by SLE associated SNPs in at least one cell type. The color represents a signed-RTC-score: a positive score indicates that the associated allele in the GWAS is positively correlated with gene expression; a negative score indicates that the associated allele in the GWAS is negatively correlated with gene expression. We set the RTC score to zero if the P-value for association was > 0.001. Colors represent the RTC-scores as follows: blue, RTC < –0.9 (GWAS risk allele reduces expression); green, RTC < –0.5 (GWAS risk allele reduces expression); yellow –0.5 < RTC < 0.5; orange, RTC > 0.5 (GWAS risk allele increases expression); red, RTC > 0.9 (GWAS risk allele increases expression). A white block indicates that data were not available for this cell type (see Supplementary Figure 4 for results on lymphoblastoid cell lines), either because the probe data failed QC or the probe was not present in the experiment platform. Clustering was performed on cell types, including only genes with data observed for all cell types (i.e., missing data did not inform cell clustering). Genes were clustered using all available data across cells (missing data were not included when determining distance between pairs of genes if eQTL results were not observed for one of the pairs).
We note that some caution must be used when inferring causality, as the RTC score has a uniform distribution and so setting an RTC score threshold of 0.9 for example, sets the type I error rate to be 0.1. Furthermore, some low RTC scores were found in genes (e.g. UBE2L3) where the associated allele resides in a region with strong LD, and the haplotype bearing the associated allele shows robust evidence of functional effects on gene expression26. We suggest that the gene expression analyses provide some support for likely causal genes, but we note that proof of true causality through altered gene expression will only be elucidated by additional experimentation.
We then integrated the results of these eQTL analyses and the coding variant analysis with an in silico survey of murine phenotype data resulting from targeting gene knockouts of genes within the associated SLE loci (Table 2). At some loci, these lines of evidence point to one likely causal gene: examples include IFIH1, LYST, WDFY4 and BANK1. In other instances, we found evidence that supports the role of multiple genes as candidates at a given locus; for example, ABHD6 (an enzyme involved in the endocannabinoid pathway) and PXK (a lymphocyte protein kinase)3 both exhibit correlation of their expression with the associated SNP. Similarly, TCF7 (coding a T cell transcription factor), implicated by the rs7726414 association, has been associated with type 1 diabetes27; however, we show that SKP1 (which encodes a protein involved in the regulation of ubiquitination), within the same LD block exhibits a strong cis eQTL in monocytes and NK cells. rs9652601 resides within CLEC16A, a gene previously reported in association studies in other autoimmune diseases28; we present evidence suggesting that SOCS1 (Suppressor of Cytokine Signaling 1) is a causal gene at this locus in SLE rather than CLEC16A. Our analyses have the advantage of including cis eQTLs based on ex vivo cells, rather than cell line data alone. Nevertheless, we acknowledge the restricted range and activation states of immune cell types available for eQTL analyses and the limited number of murine and other functional studies performed on genes at the loci.
Table 2. Candidate genes at SLE associated loci.
| Associated SNP | Chr | Genes within +/−200kb of SNP | Genes within same LD block as SNPa | Immune phenotype in murine modelb | Coding variant | cis eQTLs with SNP | Functional and/or fine mapping studies and Reference | Likely causal genesc | |
|---|---|---|---|---|---|---|---|---|---|
| rs2476601 | 1 |
MAGI3, PHTF1, RSBN1, PTPN22, BCL2L15
AP4B1, DCLRE1B, HIPK1, OLFML3 |
RSBN1, PTPN22 | PTPN22 | PTPN22 | PTPN22 | 32 | PTPN22 | |
| rs1801274 | 1 |
MPZ, SDHC, C1orf192
FCGR2A, HSPA6, FCGR3A FCGR2B, FCGR2C, FCGR3B, FCRLA |
FCGR2A |
FCGR2A
FCGR2B FCGR3B |
FCGR2A
FCGR2B FCGR3B |
FCGR2A, FCGR2B |
FCGR2A
FCGR2B FCGR3B |
33
34 35 |
FCGR2A
FCGR2B FCGR3B |
| rs704840 | 1 | TNFSF4 | TNFSF4 | TNFSF4 | TNFSF4 | 36 | TNFSF4 | ||
| rs17849501 | 1 | NMNAT2, SMG7, NCF2, ARPC5, RGL1 APOBEC4 | SMG7, NCF2 | NCF2 | SMG7 | NCF2 | 37 | SMG7, NCF2 | |
| rs3024505 | 1 |
RASSF5, EIF2D, DYRK3
MAPKAPK2, IL10, IL19, IL20 IL24, FAIM3, PIGR, FCAMR |
IL10 | RASSF5 MAPKAPK2, IL10 FAIM3, FCAMR | IL10 | 38 | IL10 | ||
| rs9782955 | 1 | LYST, NID1 | LYST | LYST | LYST | LYST | 39 | LYST | |
| rs6740462 | 2 | ACTR2, SPRED2 | SPRED2 | SPRED2 | |||||
| rs2111485 | 2 | DPP4, GCG, FAP, IFIH1, GCA, KCNH7 | IFIH1 | IFIH1 | IFIH1 | IFIH1 | IFIH1 | 40 | IFIH1 |
| rs11889341 | 2 | GLS, STAT1, STAT4, MYO1B | STAT4 | STAT1, STAT4 | STAT4 | 41 | STAT4 | ||
| rs3768792 | 2 | IKZF2 | IKZF2 | IKZF2 | IKZF2 | 42 | IKZF2 | ||
| rs9311676 | 3 |
ABHD6, RPP14, PXK, PDHB, KCTD6
ACOX2, FAM107A, FAM3D |
PXK, PDHB | ABHD6, PXK |
ABHD6
PXK |
43
44 |
ABHD6, PXK | ||
| rs564799 | 3 | SCHIP1, IL12A | IL12A | IL12A | IL12A | IL12A | |||
| rs10028805 | 4 | BANK1 | BANK1 | BANK1 | BANK1 | BANK1 | 45 | BANK1 | |
| rs7726414 | 5 | C5orf15, VDAC1, TCF7, SKP1 | TCF7, SKP1 | TCF7 | SKP1 | TCF7, SKP1 | |||
| rs10036748 | 5 |
IRGM, ZNF300, GPX3, TNIP1, ANXA6
CCDC69, GM2A, SLC36A3 |
TNIP1 | TNIP1 ANXA6 |
TNIP1 | 46 | TNIP1 | ||
| rs2431697 | 5 | C1QTNF2, C5orf54, SLU7, PTTG1, MIR146A, 3142 | intergenic | PTTG1 | MIR146A | 47 | MIR146A | ||
| rs1270942 | 6 | MHCd | |||||||
| rs9462027 | 6 |
C6orf106, SNRPC, UHRF1BP1
TAF11, ANKS1A |
UHRF1BP1 | UHRF1BP1, ANKS1A, C6orf106 | UHRF1BP1 | 48 | UHRF1BP1 | ||
| rs6568431 | 6 |
PRDM1
ATG5 |
intergenic |
PRDM1
ATG5 |
PRDM1
ATG5 |
49
50 |
PRDM1, ATG5 | ||
| rs6932056 | 6 |
TNFAIP3
PERP |
TNFAIP3 |
TNFAIP3
PERP |
TNFAIP3 | TNFAIP3 | 51 | TNFAIP3 | |
| rs849142 | 7 | JAZF1, CREB5 | JAZF1 | JAZF1 | JAZF1 | ||||
| rs4917014 | 7 | ZPBP, C7orf72, IKZF1 | IKZF1 | IKZF1 | IKZF1 | 52 | IKZF1 | ||
| rs10488631 | 7 |
CALU, OPN1SW, CCDC136, FLNC
ATP6V1F, IRF5, TNPO3, TSPAN33 |
IRF5, TNPO3 | IRF5 | IRF5, TNPO3 | IRF5 | 53 | IRF5 | |
| rs2736340 | 8 |
MTMR9, SLC35G5, C8orf12
FAM167A, BLK, GATA4 |
BLK | BLK, XKR6 | BLK | 54 | BLK | ||
| rs2663052 | 10 | WDFY4, LRRC18, VSTM4 | WDFY4 | WDFY4 | WDFY4 | WDFY4 | 55 | WDFY4 | |
| rs4948496 | 10 | ARID5B, RTKN2 | ARID5B | ARID5B | ARID5B | ||||
| rs12802200 | 11 |
B4GALNT4, PKP3, SIGIRR, ANO9, PTDSS2
RNH1, HRAS, LRRC56, C11orf35, RASSF7 PHRF1, IRF7, CDHR5, SCT, DRD4, DEAF1 EPS8L2, TMEM80, TALDO1 |
LRRC56, LMNTD2
RASSF7, MIR210HG PHRF1, IRF7, CDHR5 |
SIGIRR
IRF7 |
IRF7 | IRF7, RNH1, HRAS, RASSF7, PHRF1, and, TMEM80 | IRF7 | 56 | IRF7 |
| rs2732549 | 11 |
APIP, PDHX
CD44, SLC1A2 |
upstream, CD44 | CD44 | CD44 | 57 | CD44 | ||
| rs3794060 | 11 | DHCR7, NADSYN1, KRTAP5 | DHCR7, NADSYN1 | DHCR7, NADSYN1 | DHCR7, NADSYN1 | ||||
| rs7941765 | 11 |
ETS1, FLI1
CUX2 |
intergenic |
ETS1
FLI1 |
ETS1
FLI1 |
58
59 |
ETS1 FLI1 | ||
| rs10774625 | 12 |
FAM109A, SH2B3
ATXN2, BRAP |
SH2B3, ATXN2 | SH2B3 | SH2B3 | 60 | SH2B3 | ||
| rs1059312 | 12 | TMEM132C, SLC15A4, GLT1D1 | SLC15A4 | SLC15A4 | SLC15A4 | SLC15A4 | |||
| rs4902562 | 14 | RAD51B | RAD51B | RAD51B | |||||
| rs2289583 | 15 |
LMAN1L, CPLX3, ULK3, SCAMP2
MPI, FAM219B, COX5A, RPP25 SCAMP5, PPCDC, C15orf39 |
SCAMP5, PPCDC |
CSK, ULK3, MPI,
FAM219B, C15orf39 |
CSK | 61 | CSK | ||
| rs9652601 | 16 |
CIITA, DEXI, CLEC16A, RMI2, SOCS1
TNP2, PRM3, PRM2 |
CLEC16A |
CIITA
SOCS1 |
SOCS1, RMI2 |
CIITA
SOCS1 |
62
63 |
CIITA, SOCS1 | |
| rs34572943 | 16 |
ZNF668, ZNF646, PRSS53, VKORC1, BCKDK KAT8
PRSS8, PRSS36, FUS, PYCARD C16orf98, TRIM72, PYDC1, ITGAM ITGAX, ITGAD, COX6A2, ZNF843, ARMC5 |
ITGAM |
ITGAM
ITGAX ITGAD PYCARD |
ITGAM | ITGAM, PYCARD | ITGAM | 64 | ITGAM |
| rs11644034 | 16 | C16orf74, EMC8, COX4I1, IRF8 | intergenic | IRF8 | IRF8 | 65 | IRF8 | ||
| rs2286672 | 17 |
ALOX15, PELP1, ARRB2, MED11, CXCL16
ZMYND15, TM4SF5, VMO1, GLTPD2 PSMB6, PLD2, MINK1, CHRNE, C17orf107 GP1BA, SLC25A11, RNF167, PFN1, ENO3 SPAG7, CAMTA2, INCA1, KIF1C |
PLD2 |
ALOX15
CXCL16 INCA1 KIF1C PLD2 |
PLD2 | RNF167 | PLD2 | ||
| rs2941509 | 17 |
NEUROD2, PPP1R1B, STARD3, TCAP, PNMT
PGAP3, ERBB2, MIEN1, GRB7, IKZF3, ZPBP2 GSDMB, ORMDL3, LRRC3C, GSDMA |
ERBB2, HER–2, C17orf37
GRB7, IKZF3, ZNFN1A3 ZBPB2, GSDMB |
IKZF3 | IKZF3 | 66 | IKZF3 | ||
| rs2304256 | 19 |
DNMT1, S1PR2, MRPL4, ICAM1, ICAM4 ICAM5
ZGLP1, FDX1L, RAVER1, ICAM3, TYK2, CDC37 PDE4A, KEAP1, S1PR5, ATG4D, KRI1 |
TYK2 |
DNMT1, S1PR2
ICAM1, S1PR5 TYK2 |
TYK2 | TYK2, ICAM3 | TYK2 | 67 | TYK2 |
| rs7444 | 22 |
HIC2, RIMBP3C, UBE2L3, YDJC, CCDC116
SDF2L1, PPIL2, YPEL1, MAPK1 |
UBE2L3
YDJC |
MAPK1 | UBE2L3 | UBE2L3 | 26 | UBE2L3 | |
| rs887369 | X | CXorf21, GK | CXorf21 | CXorf21 | |||||
| rs1734787 | X |
L1CAM, LCA10, AVPR2, ARHGAP4, NAA10
RENBP, HCFC1, TMEM187, IRAK1, MECP2 OPN1LW, TEX28P2, OPN1MW, TEX28P1 OPN1MW2, TEX28, TKTL1 |
ARHGAP4, NAA10
RENBP, HCFC1 TMEM187, IRAK1 MIR718, MECP2 |
IRAK1 |
IRAK1
MECP2 |
68 | IRAK1, MECP2 | ||
The LD block is defined as SNPs showing a correlation (r2) of 0.75 with the associated SNP
The immune phenotype designation is taken from http://www.informatics.jax.org/phenotypes.shtml of genes within +/−200kb of associated SNP
The genes implicated at each locus as potentially causal at each locus
The MHC is not included due to extended LD and gene density at the locus
The 10 previously unmapped SLE loci (shown in bold type in Table 1 and Supplementary Table 3a) encompass genes of diverse function. Those of note include IKZF2 (Helios), which represents the third member of the Ikaros transcription factor family to be associated with SLE (in addition to IKZF1 and IKZF3). The association signal in the phospholipase D2 (PLD2) is a missense variant (R172C), which may alter the function of the enzyme that plays a role in leukocyte migration and apoptosis. The importance of IL12, a cytokine that plays a critical role in the generation of γ-interferon from Th1 T cells and NK cells, is highlighted by the association with IL12A (Table 1), and the suggestive associations at IL12B and the locus encoding the IL12 receptor, IL12RB2 (Supplementary Table 2).
In view of the sexual dimorphism of SLE, the novel X chromosome association revealed by rs887369 is of note. We suggest that the gene CXorf21 is likely to be etiological. While the function of this gene is unknown, it is among a limited set of genes that largely escape X-inactivation29. Sex chromosome dosage has been implicated in the genetic risk of SLE30. We observed an elevated prevalence of Klinefelter’s syndrome31 in male cases in our GWAS compared with the general population (see Online Methods) strengthening the sex chromosome dosage hypothesis. The only other gene close to rs887369 (Table 2) is GK (glycerol kinase) which does not escape X-inactivation, supporting CXorf21 as a candidate gene.
Five other genes (TNIP1, IKZF1, ETS1, WDFY4 and ARID5B) that we mapped are novel in European SLE, but had been previously shown to be associated with SLE in Chinese subjects5,6. SLE is more prevalent in non-European populations – our data suggest that locus heterogeneity among common genetic variants is unlikely to explain this differential prevalence.
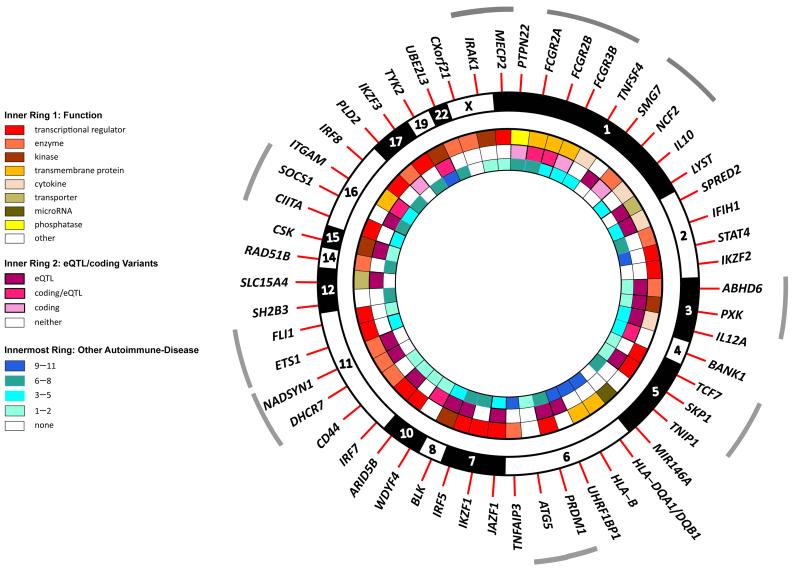
We present all of our principal findings in Fig. 2. This figure indicates ten likely missense coding variants that contribute to SLE risk; these occur largely in genes encoding kinases and other enzymes. It was noted that 16 of the genes shown are transcription factors, an enrichment above the nine expected (P = 2.3 × 10−05, χ2 test). We studied the distribution of the expression of these transcription factors in the ex vivo immune cell types examined for eQTLs; we found no evidence of skewed expression in any cell type. Our results suggest that an important facet in future exploration of SLE pathogenesis will be detailed scrutiny of trans eQTLs and regulatory expression networks in multiple immune cells.
Figure 2.
Summary of functional role of likely causal genes in SLE and other autoimmune diseases. The concentric rings in the figure show several layers of evidence to support the functional annotation of likely causal genes for SLE listed in Table 2. The genes are illustrated clockwise in chromosomal order with the grey arcs delineating those loci for which several genes are implicated. Inner Ring 1 - the gene’s functional category, taken from Ingenuity Pathway Analysis; Middle Ring 2 - the presence of a cis-acting eQTL (Figure 1) and/or coding variant and Innermost Ring 3 - the number of autoimmune diseases (excluding SLE) in Immunobase - Type 1 diabetes (T1D), Celiac disease (CEL), Multiple Sclerosis (MS), Crohn’s Disease (CRO), Primary Billiary Cirrhosis (PBC), Psoriasis (PSO), Rheumatoid Arthritis (RA), Ulcerative Colitis (UC), Ankylosing Spondylitis (AS), Autoimmune Thyroid Disease (ATD), Juvenile Idiopathic Arthritis (JIA), Alopecia Areata (AA), Inflammatory Bowel Disease (IBD), Narcolepsy (NAR), Primary Sclerosing Cholangitis (PSC), Sjögren's Syndrome (SJO), Systemic Scleroderma (SSc), Vitiligo (VIT) - previously reported to be associated with the gene.
ONLINE METHODS
Data: genome-wide association study (GWAS)
We genotyped 4,946 SLE cases and 1,286 healthy controls using the Illumina HumanOmni1-Quad BeadChip (1,140,419 markers). The genotyped controls were mostly from southern Europe, matching our Spanish, Italian and Turkish cases with controls from the same countries. We also used data for 5,727 previously genotyped controls taken from the University of Michigan Health and Retirement Study (HRS). These subjects were genotyped using the Illumina Human2.5M Beadchip (2,443,179 markers).
The clinical features of our GWAS cohort were documented on the basis of standard ACR classification criteria. The experiment was designed to avoid batch effects to the greatest extent possible. All DNA samples were sent to the laboratory at King’s College London, UK, where the integrity of the DNA was checked. The GWAS samples were then genotyped at a single laboratory. All data analysis was carried out in the laboratory at King’s College.
Genotyping for the GWAS was carried out using 82 plates, processed in 13 batches. Duplicate samples taken from HapMap Phase 3 were added to each plate to check genotyping quality. Case-control status and country of recruitment were randomized across plates as far as possible, in order to avoid artifactual differences in genotyping between plates affecting association statistics.
Our final dataset comprised genotyping of 644,674 SNPs for 4,036 SLE cases and 6,959 controls (1,260 controls of mainly southern European ancestry and 5,699 from the HRS).
Data: Hom et al. study
We analyzed data from a previous genome-wide association study of SLE (the Hom et al. study), which comprised 1,165 cases following our QC analysis (see Supplementary Text). We used a further 2,107 previously genotyped controls from the NIH CGEMS study, which were genotyped using the Illumina HumanHap550 chip. Owing to the lower density of genotyping, in some cases data imputed to the density of the 1000 Genomes (1KG) study were used in the analysis of the Hom et al. study and the subsequent meta-analysis. Imputed data are identified in tables.
Data: replication study
A cohort of 2,310 cases not included in any previous genetic study of SLE was genotyped using a custom array. The largest group of samples was from the UK, followed by cohorts from France, the USA, Germany and Canada.
The control data for the replication study comprised 3,672 subjects from the HRS cohort (independent of those used in the GWAS), 3,102 subjects from a study of melanoma and 1,202 subjects from a study of blood clotting. These control data were genotyped using the Illumina 2.5M chip. Following QC procedures (Supplementary Text), the final control dataset comprised 6,925 individuals: 3,668 from the HRS, 2,889 from the melanoma study and 368 from the blood clotting study. The final case dataset consisted of 2,018 samples.
In some cases, SNPs identified by our GWAS as genome-wide significant were not present in the replication control data (owing to absent genotyping in one of the three control sets following QC), and so genotypes for those SNPs were imputed (see below). Again, we identify these SNPs in our results tables.
Ethical approval
The UK subjects with SLE in the study were recruited with the study having obtained ethical approval from the London Ethics Committee (MREC/98/2/06 and 06/MRE02/9). Individuals were invited into the study and given information sheets as well as verbal explanations of what the research entailed. For those individuals willing to participate informed written consent was obtained. The recruitment in continental Europe and Canada were subject to local review and ethical approval. Copies of the relevant supporting documentation were sent to the investigators at King’s College at the commencement of the study.
Quality control
Initial QC analysis of the genotype data was carried out in accordance with Illumina’s Technical Note on Infinium Genotyping Data. In silico QC checks were carried out of:
Individual missingness (3% threshold)
SNP missingness (3% threshold)
Identity-by-descent (IBD, 0.125 threshold)
Population structure
Minor allele frequency (MAF, 0.002 threshold)
Autosomal heterozygosity
X chromosome heterozygosity
Y chromosome calling and homozygosity
Hardy-Weinberg equilibrium (control data only)
IBD analysis included checks both within and across cohorts; no subject in the main GWAS or Hom et al. study is related to any other subject in either cohort. We calculated principal components for the GWAS data using the EIGENSTRAT algorithm12, and derived the empirical genomic inflation factor13,69 for these data. As noted by Price and colleagues14, the definition of genomic control means that λGC is proportional to sample size. We therefore report λ1000, the inflation factor for an equivalent study of 1000 cases and 1000 controls15,70, in the main text, as well as λGC.
For the replication cohort, population structure was estimated using 46 ancestry informative markers (following QC measures on these SNPs). As described in Supplementary Text, we merged these data with HapMap data to help identify non-European samples. Again, principal components were calculated using the EIGENSTRAT algorithm. 120 subjects that clustered with the non-European HapMap populations were removed from the analysis.
Klinefelter’s syndrome
During QC analysis, we identified subjects in our GWAS cohort with abnormal karyotypes, consistent with Klinefelter’s syndrome (47, XXY). Three of the 365 male cases in our main GWAS have clinical and genetic data that confirm their status as Klinefelter’s sufferers (Supplementary Text). Given that the prevalence of Klinefelter’s syndrome in the general population is estimated to be 0.1 – 0.2%31, this estimate suggests an approximately four- to eight-fold increase in prevalence compared with 46, XY males, consistent with Klinefelter’s males and 46, XX females having a similar risk of developing SLE.
Analysis: association
All case-control analysis was carried out using the SNPTEST71,72 algorithm; we use a standard threshold of P = 5 × 10−8 for reporting genome-wide significance throughout. The inverse variance method was used for meta-analysis. All markers were fully genotyped in the main GWAS (i.e, no imputation was carried out). The imputation carried out for the Hom et al. and replication studies, and fine mapping imputation, are described below.
For all SNPs at which we report a novel association with SLE, we compared allele frequencies in the main GWAS controls with those in publically available control cohorts (1KG European samples18, Wellcome Trust Case Control Consortium (WTCCC) genotypes73, TwinsUK samples, HapMap CEU population data, and sample genotypes from the Knight laboratory expression data23). We tested for a statistically significant (α = 0.01) difference in allele frequency between our GWAS and the public controls, using a 1 degree of freedom χ2 test of allele frequencies. One SNP failed this test (rs1439112, MGAT5) and was removed from further analysis. In three further cases, the difference in allele frequency strengthened our observed association. These data are presented in Supplementary Table 7.
Annotation of results
Gene names listed in results tables were identified by overlaying GWAS results onto the UCSC Genome Browser. We adopted a threshold based on linkage disequilibrium: for each SNP, we noted the set of markers with R2 > 0.75 with respect to the SNP of interest (Table 2).
Post hoc QC
Checks carried out following case-control analysis included examination of plots of raw genotype intensity; this was of particular relevance given the increase in the numbers of relatively rare variants due to the higher density of genotyping (as with imputation, genotype calling is by definition more difficult for rarer variants). We checked that the intensity plots showed clusters of genotypes (i.e., homozygotes or heterozygotes) that were compact and well discriminated. This check was also carried out with stratification by QC group. Plots of intensity were examined for each associated SNP, and for all of the SNPs in the replication study.
Analysis: replication study chip design
We selected SNPs for the replication study based on the results of the meta-analysis of the two GWA studies. At loci with no known association in SLE, we adopted a threshold of P = 2.5 × 10−05, while for loci with previously reported associations the threshold was set at P = 1 × 10−04. This followed the methodology used in Box 1 of the WTCCC study of seven common diseases73. This declared SNPs as associated if the posterior odds of association were greater than 10. In that study, the assumption was made that 10 detectable genes were present, so the prior odds of a true association would be in the order of 100,000:1, assuming 1,000,000 independent regions in the genome. Based on the autoimmune genetics literature, we have assumed that there are likely to be as many as 500 genes associated with SLE. We have required posterior odds in favour of a SNP being associated to be >1 (as opposed to >10, which would be advisable if declaring an association rather than choosing SNPs for replication). This gives a P-value threshold of 2.5 × 10−05. For SNPs at loci with previously published SLE associations, we have reduced our threshold for inclusion in the replication study to P = 1 × 10−04. This is because a priori we believe these SNPs are more likely to be at susceptibility loci than those with no evidence of association, increasing the prior odds by at least a factor of 4.
Analysis: 1000 Genomes (1KG) imputation
For imputation, both the main GWAS and the Hom et al. data were pre-phased using the SHAPEIT algorithm74, and then imputed to the density of the 1KG study using IMPUTE71,72 v2.2.3. Only markers with an IMPUTE INFO score > 0.7 were used in analysis. For SNPs identified in our GWAS as genome-wide significant at which data were absent in the replication study controls, we imputed over a +/– 1Mb region around the SNP of interest.
1KG data were used both to fine map loci and to determine whether multiple signals were present. For this analysis, we carried out a meta-analysis of 1KG imputed GWAS and Hom et al. data. Association testing was performed on the 1KG data within a 1 Mb window of the reported SNP. For the MHC, we included the complete 8 Mb region (26–34 Mb) in our analysis. To scan for further independent signals, association tests were performed including the genotype data for the most highly associated SNP as a covariate. If secondary signals were found to be associated by this analysis (with a P-value threshold of 5 × 10−08) and odds ratios were consistent across the single marker and conditional analyses, the secondary signals were reported as independent associations.
In order to address the problem that the most associated (lead SNP, marker with the lowest p-value) variant is not necessarily the best candidate as the true causal variant, we considered markers from the most associated down to a defined cut-off. The cut-off was defined as a Bayes Factor (BF) against the most associated SNP equal to 0.34. This was derived from assuming a prior odds of causality for a non-synonymous SNP equal to 3, taken from an empirical analysis of GWAS annotation19,75. Any SNPs above this BF cut-off that were missense variants were declared as more likely candidates than the most associated SNP: assuming that the prior odds of a missense SNP (being causal) against a non-missense SNP to be equal to 3, any missense SNP with a BF > 0.34 will have a posterior odds > 1 and will therefore have a higher posterior probability than the most associated marker (if the most associated marker is non-missense). Therefore we searched for functional variants within a set of markers where inclusion in this set required a maximum Bayes factor (BF) > 0.34 between the marker and the most associated SNP in the 1KG imputed data. We considered any marker that had a BF > 0.34 with respect to the most associated marker, and noted whether any had functional effects. We calculated an approximate BF following Wakefield76, using a prior distribution on effect size (odds ratio) that was proportional to MAF (as rare variants are believed to have large effects, while common variants are believed to exert small effects). The BF threshold implies that we believe associations with functional variants, such as missense variants, three times more (say) than intergenic variants that do not correlate with gene expression. We then calculated posterior model probabilities following Maller et al.77, but with prior odds of 3 between missense SNPs and non-missense SNPs; Maller et al. use a uniform prior on all model probabilities (all SNPs are considered to have equal weights a priori, and therefore the prior odds are 1). We present these results in Supplementary Table 4 where we also, separately, display SNPs with a BF > 0.1 (as a strict threshold of 0.34 does not reflect the uncertainty in prior odds of causality and BF estimates). We also calculated the BF between SNPs presented in Table 1 and the SNPs listed in Supplementary Table 3a and declared that the marker for association had changed if the BF was greater than 10 (equal to “strong” evidence on the Jeffreys’ scale78). These SNPs are annotated in Supplementary Table 3a.
Analysis: the MHC and HLA alleles
We included imputed HLA alleles in analysis of the MHC, allowing us to determine the most likely model of association within this region. HLA imputation was performed using HLA*IMP V221 using genotyped SNP data. To determine the best model for association within the HLA alleles alone we ran forward stepwise regression. We then tested the five SNPs listed in Supplementary Tables 6a–c for association, conditional on the HLA alleles. To test whether each of the five SNPs was independent of the HLA alleles (rather than just the alleles in the best HLA model), we carried out a test conditional on all alleles (i.e., the HLA alleles were used as covariates) in each HLA gene, and for all HLA alleles over all genes. We used a significance threshold at each stage of the stepwise regression of P = 5 × 10−05, which is a Bonferroni adjustment for 204 tests (199 HLA alleles and 5 SNPs), with a familywise Type I error rate of 0.01.
Analysis: gene expression data
Gene expression data were obtained from three sources: firstly, we obtained data from Fairfax et al.22,23 and unpublished data from Fairfax and Knight for NK cells, naïve monocytes, monocytes stimulated by LPS (harvested after 2 hours and 24 hours), IFN and B cells. Secondly, we interrogated the Genevar database for LCL eQTL results, taking results from the MuTHER resource79. The CD4 (CD4 T cells) and CD14 (CD14/16 Monocytes) data were obtained from a previous study of gene expression in immune related cells24. An adjustment was made for multiple testing using a Bonferroni correction, by counting the number of tests across all loci for genes within +/–1MB of the SLE associated SNP. With a familywise test size of 0.01, the P-value threshold was 1.41 × 10−05.
To test whether observed associations between SNPs and expression levels of cis-acting genes were purely due to chance, we calculated the RTC score25 for all SNP-gene eQTL results displayed in the heat map (Figure 1). This tests the null hypothesis that the GWAS associated SNP and the best eQTL (within a recombination hotspot) are tagging two separate effects, and the observed eQTL is purely due to the LD between the GWAS associated SNP and the “true” eQTL SNP. For our data, we were interested in the distribution of RTC scores, given that eQTL results were generated in multiple cell types. Not all eQTLs were consistently present across all these cells. We therefore plotted the RTC scores against the –log10 P-values supporting each cis eQTL in all cell types (Supplementary Figures 3a and 3b). Supplementary Figures 3a and 3b show that three genes were outlying: ITGAM in two cell types, and UBE2L3 and PLD2 in CD4 cells. However, we have strong a priori evidence of a true causal effect on expression by polymorphisms around UBE2L326. For ITGAM, we note the low RTC scores in Figure 1, which includes all eQTL data for ITGAM given that the results are convincing for the eQTL in LPS stimulated monocytes (P = 2.67 × 10−19 and RTC = 0.85). We have removed the declaration of an eQTL for PLD2. Supplementary Figure 4 displays a heat map for these data using a t-statistic.
Supplementary Material
Acknowledgments
TJV, JDR and MEAR were awarded funding to carry out genotyping and analysis from the George Koukis Foundation and an Arthritis Research UK Special Strategic Award (ref. 19289). MEAR received grants from the Instituto de Salud Carlos III (PS09/00129), co-financed by the FEDER funds of the European Union, the Consejería de Salud de Andalucía (PI0012), the Swedish Research Council of Medicine, and from the European Science Foundation to the BIOLUPUS network. JB was funded by the George Koukis Foundation and the Arthritis Research UK Special Strategic Award. JEW was funded by the Canadian Institutes of Health Research (94825). CLP was funded by a Wellcome Trust grant (ref. 085492). PT is employed by the Biomedical Research Centre. LC was funded by the China Scholarship Council, number 201406380127. The research was funded/supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
The Health and Retirement Study genetic data were obtained from dbGaP under accession phs000187.v1; the study is sponsored by the National Institute on Aging (grant numbers U01AG009740, RC2AG036495, and RC4AG039029) and was conducted by the University of Michigan. The melanoma study data were obtained from dbGaP under accession number phs000187.v1.p1. Research support to collect data and develop an application to support this project was provided by 3P50CA093459, 5P50CA097007, 5R01ES011740, and 5R01CA133996. Funding support for the Genes and Blood Clotting Study was provided through the NIH/NHLBI (R37HL039693). The Genes and Blood Clotting Study is one of the Phase 3 studies as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with genotype cleaning was provided by the GENEVA Coordinating Center (U01 HG004446). Funding support for DNA extraction and genotyping, which was performed at the Broad Institute, was provided by NIH/NHLBI (R37HL039693). Additional support was provided by the Howard Hughes Medical Institute. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession number phs000304.v1.p1. The CGEMS prostate cancer study data were obtained from dbGaP under accession phs000207v1 reference: Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Welch R, Staats BJ, Calle EE, Spencer Feigelson H, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Tucker M, Gerhard DS, Fraumeni Jr. JF, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-Wide Association Study of Prostate Cancer Identifies a Second Locus at 8q24. Nat Genet, 39(5): 645–649, 2007. We thank Genentech Inc. for providing the genotype data from their genome-wide association studies. We thank Vidya Anand and Sophie Ragan for their help in coordinating data collection. We thank Tomas Axelsson, Barbara Fürnrohr, Sophie Ragan and Jennifer Kelly for their help with the replication study.
A large number of people contributed samples or clinical data to the GWAS:
The following samples were obtained via the BIOLUPUS network coordinated by MEAR: Belgium: Bernard Lawerys and Fredric Houssiau (Université Catholique de Louvain) Denmark: Søren Jacobsen (University of Copenhagen), Peter Junker, Helle Laustrup (Odense University Hospital). Germany: Torsten Witte (Medizinische Hochschule Hannover). Greece: Haralampos Moutsopoulos, Etstathia K Kapsogeorgou (National University of Athens). Hungary: Emőke Endreffy and Laszlo Kovacs (Albert Szent-Györgyi Medical University). Iceland: Kristján Steinsson (Landspitali National University Hospital). Italy: Andrea Doria (University of Padova), Pier Luigi Meroni (IRCCS Istituto Auxologico Italiano), Rafaella Scorza (University of Milan), Sandra D’Alfonso (providing samples from Rome, Naples and Siena, Università del Piemonte Orientale). Netherlands: Marc Bijl, Cees Kallenberg (University of Groningen). Portugal: Carlos Vasconcelos (Hospital Santo António, Porto), Berta Martins Silva (University of Porto). Spain: Javier Martín, Ezequiel Martín Rodríguez (Instituto de Parasitología y Biomedicina “Lopez-Neyra”), Ana Suárez (Hospital Universitario Central de Asturias), Iñigo Rua Figueroa (Hospital Dr Negrín, Gran Canaria), Guillermo Pons-Estel (Hospital Clinic, Barcelona). From the GENLES collaboration: Argentina: Bernardo Pons-Estel (Hospital Provincial de Rosario). Other contributors: Canada: Paul Fortin, Joan Wither, Dafna Gladman, Murray Urowitz (Toronto Western Hospital, University Health Network), Ann Clarke, Sasha Bernatsky, Christian Pineau, Joyce Rauch (McGill University), Thomas Hudson (Ontario Institute for Cancer Research), Janet Pope (University of Western Ontario), Christine Peschken, Carol Hitchon (University of Manitoba), John Hanly (Dalhousie University), C Douglas Smith (Ottawa Hospital), Eric Rich, Jean-Luc Senécal (Centre Hospitalier de l’Université de Montréal), Michel Zummer (Maisonneuve-Rosemont Hospital), Gilles Boire (Université de Sherbrooke), Susan Barr (University of Calgary). Germany: Min-Ae Lee-Kirsch (Technische Universität Dresden). Netherlands: Tom Huizinga (Leiden University Medical Center, Dutch and Polish samples). Spain: Josefina Cortés Hernández, Josep Ordi Ros, Jesús Castro Marrero (Vall d’Hebron Research Institute). Turkey: Sule Yavuz (Istanbul Bilim University, Avrupa Florence Nightingale Hospital). UK: Caroline Gordon (University of Birmingham), Katie Vinen (King’s College London), David Isenberg (University College Hospital), Lars Erwig (University of Aberdeen), David D’Cruz (St Thomas’ Hospital, London), AJ Rees (MRC/KRUK GN Biobank), Ian Bruce (University of Manchester). USA: Amr Sawalha (University of Michigan, Turkish samples), Lindsey Criswell (UCSF).
For the replication study, samples were provided by Joan Wither (Toronto Western Research Institute, University Health Network, Canada), Earl Silverman (The Hospital for Sick Children and University of Toronto, Canada), Pat Gaffney (Oklahoma Medical Research Foundation, USA), Ann-Christine Syvänen, Lars Rönnblom (Uppsala Universitet, Sweden), Reinhard Voll, Georg Schett, Barbara Fuernrohr (University of Erlangen-Nuremberg, Germany) and Natalie Costedoat-Chalumeau (Service de médecine interne CHU Paris Centre - Hôpital Cochin, France). Replication genotyping was performed by the SNP&SEQ Technology Platform in Uppsala, which is part of the Swedish National Genomics Infrastructure (NGI) hosted by Science for Life Laboratory.
We thank Towfique Raj and Phil De Jager for contributing gene expression data (CD4 T cells and CD14/16 monocytes). These gene expression data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE56035.
Footnotes
URLs. Department of Twin Research, King’s College London, TwinsUK samples, http://www.twinsuk.ac.uk; Ingenuity Pathway Analysis, http://www.ingenuity.com/; Immunobase, http://www.immunobase.org. Systems Biology and Complex Disease Genetics, http://insidegen.com.
Data access. Summary statistics from the GWAS will be released by deposition in ImmunoBase. All 1KG imputed summary statistics will also be available at http://insidegen.com/insidegen-LUPUS-data.html .
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Deapen D, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 2.Alarcón-Segovia D, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 3.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. New Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 5.Yang W, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J-W, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 7.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada Y, et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 2012;8:e1002455. doi: 10.1371/journal.pgen.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham DSC, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7:e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 12.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bakker PIW, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence JS, Martins CL, Drake GL. A family survey of lupus erythematosus. 1 Heritability. J. Rheumatol . 1987;14:913–921. [PubMed] [Google Scholar]
- 17.So H-C, Gui AHS, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 18.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight J, Barnes MR, Breen G, Weale ME. Using functional annotation for the empirical determination of Bayes Factors for genome-wide association study analysis. PLoS One. 2011;6:e14808. doi: 10.1371/journal.pone.0014808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris DL, et al. Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am. J. Hum. Genet. 2012;91:778–793. doi: 10.1016/j.ajhg.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1118. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis MJ, et al. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am J Hum Genet. 2015;96:221–34. doi: 10.1016/j.ajhg.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noble JA, et al. A polymorphism in the TCF7 gene, C883A, is associated with type 1 diabetes. Diabetes. 2003;52:1579–1582. doi: 10.2337/diabetes.52.6.1579. [DOI] [PubMed] [Google Scholar]
- 28.International Multiple Sclerosis Genetics Consortium (IMSGC) The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10:11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol. Biol. Evol. 2013;30:2588–2601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scofield RH, et al. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanfranco F, Kamischke A, Zitzmann M, Nieschlag E. Klinefelter's syndrome. Lancet. 2004;364:273–283. doi: 10.1016/S0140-6736(04)16678-6. [DOI] [PubMed] [Google Scholar]
- 32.Namjou B, et al. PTPN22 association in systemic lupus erythematosus (SLE) with respect to individual ancestry and clinical sub-phenotypes. PLoS One. 2013;8:e69404. doi: 10.1371/journal.pone.0069404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karassa FB, Trikalinos TA, Ioannidis JPA. Role of the Fcγ receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: A meta-analysis. Arthrit. Rheum. 2002;46:1563–71. doi: 10.1002/art.10306. [DOI] [PubMed] [Google Scholar]
- 34.Floto RA, et al. Loss of function of a lupus-associated FcγRIIb polymorphism through exclusion from lipid rafts. Nat. Med. 2005;11:1056–8. doi: 10.1038/nm1288. [DOI] [PubMed] [Google Scholar]
- 35.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat. Genet. 2007;39:721–3. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manku H, et al. Trans-ancestral studies fine map the SLE-susceptibility locus TNFSF4. PLoS Genet. 2013;9:e1003554. doi: 10.1371/journal.pgen.1003554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob CO, et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. P. Natl. Acad. Sci. USA. 2012;109:E59–67. doi: 10.1073/pnas.1113251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai D, et al. Preferential Binding to Elk-1 by SLE-Associated IL10 Risk Allele Upregulates IL10 Expression. PLoS Genet. 2013;9:e1003870. doi: 10.1371/journal.pgen.1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tchernev VT, et al. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med. 2002;8:56–64. [PMC free article] [PubMed] [Google Scholar]
- 40.Molineros JE, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 2013;9:e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namjou B, et al. High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis Rheum. 2009;60:1085–95. doi: 10.1002/art.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander T, et al. Foxp3+ Helios+ regulatory T cells are expanded in active systemic lupus erythematosus. Ann. Rheum. Dis. 2013;72:1549–58. doi: 10.1136/annrheumdis-2012-202216. [DOI] [PubMed] [Google Scholar]
- 43.Oparina NY, et al. PXK locus in systemic lupus erythematosus: fine mapping and functional analysis reveals novel susceptibility gene ABHD6. Ann. Rheum. Dis. 2015;74:e14. doi: 10.1136/annrheumdis-2013-204909. [DOI] [PubMed] [Google Scholar]
- 44.Vaughn SE, et al. Lupus risk variants in the PXK locus alter B-cell receptor internalization. Front. Genet. 2014;5:450. doi: 10.3389/fgene.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castillejo-López C, et al. Genetic and physical interaction of the B-cell systemic lupus erythematosus-associated genes BANK1 and BLK. Ann. Rheum. Dis. 2012;71:136–42. doi: 10.1136/annrheumdis-2011-200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caster DJ, et al. ABIN1 Dysfunction as a Genetic Basis for Lupus Nephritis. J. Am. Soc. Nephrol. 2013;24:1743–54. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo X, et al. A functional variant in MicroRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Two missense variants in UHRF1BP1 are independently associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2011;12:231–4. doi: 10.1038/gene.2010.66. [DOI] [PubMed] [Google Scholar]
- 49.Kim SJ, Gregersen PK, Diamond B. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J. Clin. Invest. 2013;123:823–33. doi: 10.1172/JCI64712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann. Rheum. Dis. 2011;70:1330–7. doi: 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- 51.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 2011;43:253–8. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kottyan LC, et al. The IRF5-TNPO3 association with systemic lupus erythematosus (SLE) has two components that other autoimmune disorders variably share. Hum. Mol. Genet. 2015;24:582–96. doi: 10.1093/hmg/ddu455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guthridge JM, et al. Two functional lupus-associated BLK promoter variants control cell-type- and developmental-stage-specific transcription. Am. J. Hum. Genet. 2014;94:586–98. doi: 10.1016/j.ajhg.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao H, et al. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes Immun. 2012;13:536–42. doi: 10.1038/gene.2012.33. [DOI] [PubMed] [Google Scholar]
- 56.Heinig M, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–4. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crispín JC, et al. Expression of CD44 variant isoforms CD44v3 and CD44v6 is increased on T cells from patients with systemic lupus erythematosus and is correlated with disease activity. Arthritis Rheum. 2010;62:1431–7. doi: 10.1002/art.27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, et al. Epistatic interaction between genetic variants in susceptibility gene ETS1 correlates with IL-17 Levels in SLE patients. Ann. Hum. Genet. 2013;77:344–50. doi: 10.1111/ahg.12018. [DOI] [PubMed] [Google Scholar]
- 59.Morris EE, et al. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res. Ther. 2010;12:R212. doi: 10.1186/ar3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mori T, et al. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-γ–producing T cells. J. Immunol. 2014;193:1728–36. doi: 10.4049/jimmunol.1303243. [DOI] [PubMed] [Google Scholar]
- 61.Manjarrez-Orduño N, et al. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat. Genet. 2012;44:1227–30. doi: 10.1038/ng.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bronson PG, et al. The rs4774 CIITA missense variant is associated with risk of systemic lupus erythematosus. Genes Immun. 2011;12:667–71. doi: 10.1038/gene.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujimoto M, et al. Inadequate induction of suppressor of cytokine signaling-1 causes systemic autoimmune diseases. Int. Immunol. 2004;16:303–14. doi: 10.1093/intimm/dxh030. [DOI] [PubMed] [Google Scholar]
- 64.Rhodes B, et al. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann. Rheum. Dis. 2012;71:2028–34. doi: 10.1136/annrheumdis-2012-201390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chrabot BS, et al. Genetic variation near IRF8 is associated with serologic and cytokine profiles in systemic lupus erythematosus and multiple sclerosis. Genes Immun. 2013;14:471–8. doi: 10.1038/gene.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun J, Matthias G, Mihatsch MJ, Georgopoulos K, Matthias P. Lack of the transcriptional coactivator OBF-1 prevents the development of systemic lupus erythematosus-like phenotypes in Aiolos mutant mice. J. Immunol. 2003;170:1699–706. doi: 10.4049/jimmunol.170.4.1699. [DOI] [PubMed] [Google Scholar]
- 67.Shaw MH, et al. A natural mutation in the Tyk2 pseudokinase domain underlies altered susceptibility of B10.Q/J mice to infection and autoimmunity. P. Natl. Acad. Sci. 2003;100:11594–9. doi: 10.1073/pnas.1930781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaufman KM. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancxestral groups. Ann. Rheum. Dis. 2013;72:437–44. doi: 10.1136/annrheumdis-2012-201851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freedman ML, et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 70.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet. Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 71.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 72.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 73.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delaneau O, Howie B, Cox AJ, Zagury J-F, Marchini J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 2013;93:687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gagliano SA, Barnes MR, Weale M, Knight J. A Bayesian method to incorporate hundreds of functional characteristics with association evidence to improve variant Prioritization. PLoS One. 2014;9:e98122. doi: 10.1371/journal.pone.0098122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 77.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeffreys H. Theory of Probability. 3rd ed Oxford University Press; Oxford, UK: 1961. [Google Scholar]
- 79.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.