Abstract
The main physiological actions of the biologically most active metabolite of vitamin D, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), are calcium and phosphorus uptake and transport and thereby controlling bone formation. Other emergent areas of 1α,25(OH)2D3 action are in the control of immune functions, cellular growth and differentiation. All genomic actions of 1α,25(OH)2D3 are mediated by the transcription factor vitamin D receptor (VDR) that has been the subject of intense study since the 1980’s. Thus, vitamin D signaling primarily implies the molecular actions of the VDR. In this review, we present different perspectives on the VDR that incorporate its role as transcription factor and member of the nuclear receptor superfamily, its dynamic changes in genome-wide locations and DNA binding modes, its interaction with chromatin components and its primary protein-coding and non-protein coding target genes and finally how these aspects are united in regulatory networks. By comparing the actions of the VDR, a relatively well-understood and characterized protein, with those of other transcription factors, we aim to build a realistic positioning of vitamin D signaling in the context of other intracellular signaling systems.
Keywords: Chromatin, Gene regulation, Genome-wide view, Nuclear receptor, Vitamin D, Vitamin D receptor
1. Introduction
The micronutrient vitamin D is essential for maintenance of health [1]. The most abundant form of vitamin D is 25-hydroxyvitamin D3 (25(OH)D3), the serum concentrations of which indicate the vitamin D status of a human individual [2]. The most biologically active vitamin D metabolite is the secosteroid 1α,25(OH)2D3, which acts as a pleiotropic endocrine hormone and influences many physiological processes [3]. For example, severe vitamin D deficiency leads to rickets, as 1α,25(OH)2D3 is essential for adequate Ca2+ and Pi absorption from the intestine and hence for bone formation [4].
An appreciation of the 1α,25(OH)2D3 endocrine system precedes the isolation of the VDR by well over 400 years as rickets was first described in the beginning of the 17th century. However, the molecular etiology for rickets remained unresolved until the beginning of the 20th century, when it was discovered that the dietary deficiency that caused rickets could be ameliorated by fish oil extracts and that the active ingredient was identified as vitamin D3 [1]. Moreover, it was found that rickets could be cured by exposure to UV radiation. The analysis of 1α,25(OH)2D3 metabolism and the identification of 25(OH)D3 in the 1960’s [4] was followed by the identification of vitamin D-binding proteins in the 1970’s [5,6] and the cloning of the VDR (also referred to as NR1I1 in the generic nuclear receptor terminology) in 1988 [7]. All this leads to a functional understanding of the vitamin D endocrine system.
In the subsequent decades remarkable strides have been made in describing the diverse biology that the VDR participates in. Researchers accommodated this diversity of biological actions by separating functions into the so-called “classical” actions, i.e. the regulation of serum calcium levels [8], and “non-classical” actions, i.e. everything else that includes control of metabolism, cellular growth and immune functions [9]. In particular, immuno-regulatory properties of 1α,25(OH)2D3 may be important, as low 25(OH)D3 levels are associated with poor immune function and increased disease susceptibility [10]. Perhaps now these views are beginning to be consolidated into more unified views of the actions of the VDR.
Although a number of rapid and non-genomic actions of 1α,25(OH)2D3 have been described [11], the vast majority of the effects of the hormone are mediated by the VDR, which is the only protein that binds 1α,25(OH)2D3 effectively at sub-nanomolar concentrations [12]. This simplifies the understanding of vitamin D signaling, since the physiological effects of the hormone largely overlap with the actions of the transcription factor VDR.
Taken together, the VDR system can be viewed as a comprehensively understood transcription factor in terms of both mechanistic insight and phenotypic consequences. In this review, we therefore focus on VDR and its actions from multiple perspectives. We will (i) illuminate VDR as a transcription factor and member of the nuclear receptor superfamily, (ii) describe VDR’s genome-wide locations and DNA-binding modes, (iii) analyze VDR’s dynamic interactions with chromatin modifiers and other nuclear co-factors, (iv), address VDR’s primary protein-coding and non-protein coding target genes and (v) delineate these roles and actions of VDR as a modular component in a regulatory network. Finally we will consider these regulatory networks integrated with the actions of other transcription factors, and thereby position the VDR, and its ligand 1α,25(OH)2D3, into the complex signaling system of human tissues and cell types.
2. Perspective 1: VDR is a member of a transcription factor family
In humans there are approximately 1900 classical transcription factors, i.e. proteins that sequence-specifically contact genomic DNA [13]. VDR is one of these DNA-binding transcription factors, but has an important additional property, which it shares only with some other members of the nuclear receptor superfamily: VDR can get specifically activated by low nanomolar concentrations of a small lipophilic molecule in the approximate size and molecular weight of cholesterol [14]. This property is shared with the nuclear receptors for the steroid hormones estradiol (ERα and ERβ), testosterone (AR), progesterone (PR), cortisol (GR) and mineralocorticoids (MR), for the vitamin A derivative all-trans retinoic acid (RARα, RARβ and RARγ) and for the thyroid hormone triiodothyronine (TRα and TRβ). Moreover, also a number adopted orphan members of the nuclear receptor superfamily, such as retinoid X receptors (RXRs) α, β, and γ, peroxisome proliferator-activated receptors (PPARs) α, δ, and γ, liver X receptors (LXR) α and β and farnesoid X receptor (FXR), show a similar mode of action, but their natural ligands, for example, 9-cis retinoic acid, fatty acids, oxysterols and bile acids, respectively, to date have not been considered as classical endocrine hormones and are in most cases bound by their respective receptors with far lower affinity and specificity [15].
The 48 human members of the nuclear receptor superfamily are characterized by a highly conserved DNA-binding domain (DBD) and a structurally conserved ligand-binding domain (LBD) [16]. The lower part of the LBD of all ligand-activated nuclear receptors contains a ligand-binding pocket of 400–1400 Å3 in volume, in which the respective ligands are specifically bound [17]. The interior surface of these pockets is formed by the side chains of mostly non-polar amino acids and thereby complements the lipophilic character of the ligands [18].
All nuclear receptors have a similar mode of action. Therefore, a number of mechanisms that were identified, for example with ERs, apply also for the VDR. For example, ligand specificity is achieved through a limited number of stereo-specific polar contacts that include the so-called anchoring points and the actual shape of the pocket. Nuclear receptors that bind their specific ligand with high affinity, such as VDR and ERs, have a relatively small ligand-binding pocket, which is filled to a high percentage by ligand, while adopted orphan nuclear receptors, such as PPARs and LXRs, have a significantly larger ligand-binding pocket, which is filled to a far lower percentage by their ligand molecules [17].
As observed with other transcription factors, the DBD of the VDR cannot contact more than six nucleotides within the major groove of genomic DNA. Binding sites of monomeric nuclear receptors are therefore hexameric sequences and most members of the superfamily share consensus on the sequence RGKTSA (R = A or G, K = G or T, S = C or G). However, the DNA-binding affinity of monomeric VDR is insufficient for the formation of a stable protein–DNA complex and therefore the VDR has to complex with a partner protein, in order to achieve efficient DNA binding. The predominant partner of VDR is the nuclear receptor RXR [19].
Steric constraints allow dimerization of nuclear receptor DBDs only on DNA-binding sites that contain properly spaced hexameric binding motifs; these sequences are also referred to as response elements (REs). An asymmetric, direct repeat arrangement of two motifs spaced by three nucleotides (DR3) provides an efficient interface of the DBDs of VDR and RXR (Fig. 1A, top). This fits with the so-called “3-4-5 rule” of Umesono et al. [20], in which VDR–RXR heterodimers show optimal binding to DR3-type REs, while other nuclear receptors, reflecting different structures and steric contraints, prefer altered spacing, such as DR4 for TRs and DR5 for RARs.
Fig. 1.
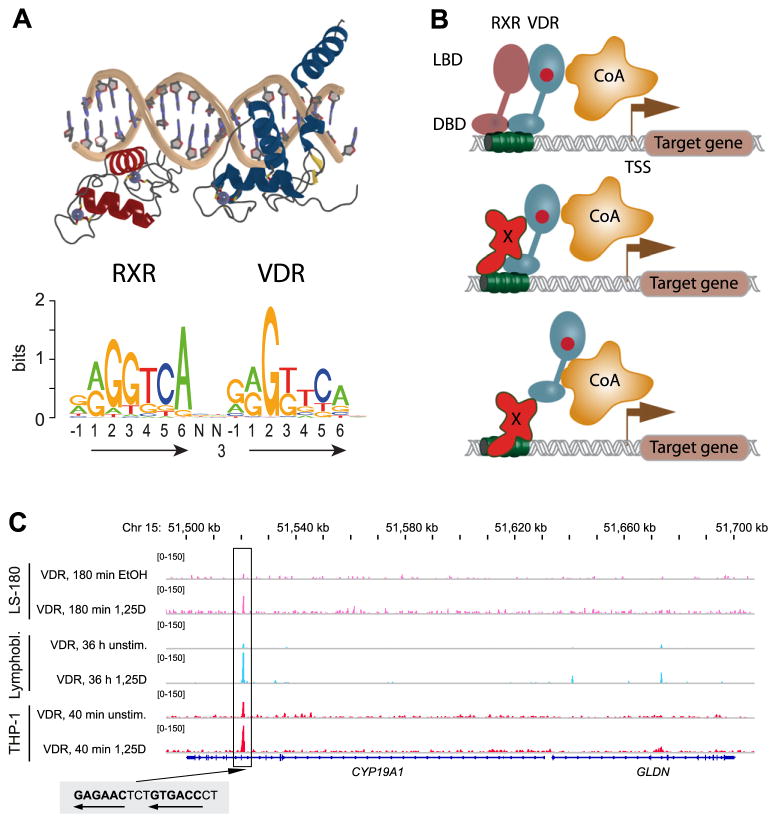
VDR binding sites and target genes. (A) The crystal structure (protein data bank identifier 1YNW [112]) of the heterodimer of the DBDs of VDR (blue) and RXR (red) bound to a DR3-type RE (top) is aligned with the de novo DR3-type sequence motif found below 742 of 2340 VDR peaks (31.7%) in THP-1 cells [35] (bottom). (B) Three modes of VDR regulating its primary target genes are indicated: VDR–RXR heterodimers preferentially binding to a DR3-type RE (top), VDR partnering with undefined protein X bound to DNA (middle) and VDR tethering undefined protein X bound to DNA (bottom). In all three cases it is assumed that the contact of ligand (red)-activated VDR leads to an association with CoA proteins and the activation of primary target genes. (C) The genome view of one primary VDR target gene, CYP19A1, is shown. The peak tracks on top show data from VDR ChIP-seq in LS-180 cells (pink [36]), lymphoblastoids (blue [34]) and THP-1 cells (red [35]) comparing genomic VDR binding at the CYP19A1 locus in unstimulated or vehicle-stimulated cells with that after 1α,25(OH)2D3 (1,25D) treatment for indicated times. The structure of CYP19A1 gene and its direct neighbor GLDN is shown in blue and the sequence of the DR3-type VDRE at the summit of the VDR ChIP-seq peak is indicated.
Genome-wide analyses for VDR binding sites (see Section 4) confirmed the preferential binding of VDR to DR3-type REs (Fig. 1A, bottom), but only for approximately one third of all genomic binding sites. Therefore, there must be additional mechanisms for how the VDR can associate with genomic loci, in order to control its primary target genes. These mechanisms include partnering with presently undefined partner proteins (Fig. 1B, middle) or the tethering to other DNA-binding transcription factors (Fig. 1B, bottom). Independent of the exact mechanism, the VDR recruits to these regions in complexes that include positively and negatively regulating proteins, referred to as co-activators (CoAs) [21] and co-repressors (CoRs) [22], respectively. CoA proteins build a bridge to the basal transcriptional machinery, which is assembled on the transcription start site (TSS) of the primary VDR target gene, and stimulate in this way the transcription of the target gene (more details in Section 4). This process is known as transactivation.
In contrast, transrepression is a process whereby transcription factor actions include gene repression. In the context of nuclear receptors this may include direct mechanism associated with co-repressor recruitment or repression of the activity of a second transcription factor through a protein–protein interaction, such as tethering (Fig. 1B, bottom). With nuclear receptors ligand-dependent transrepression is well established for PPAR and LXR [23], and appears to apply also for other members of the superfamily, such as VDR. The net result of transrepression is a down-regulation of gene transcription and is considered as one of the mechanisms by which VDR down-regulates some of its primary target genes.
The cell specificity of the actions of VDR and its ligand 1α,25(OH)2D3 can be explained in part by VDR’s recognition mode for its genomic binding sites (see Section 4) and the tissue-specific differences in the expression of VDR and its key co-factors. The VDR gene shows highest expression in metabolic tissues, such as kidneys, bone and intestine, but at least low to moderate expression is found in nearly all other of the approximately 250 human tissues and cell-types [24]. Moreover, in contrast to GR and AR, the VDR can bind its genomic targets also in the absence of ligand, i.e. in this respect the functional profile of the VDR is larger than that of its ligand [25]. This relates to both repression and activation events and involves the action of CoAs and CoRs (more details in Section 5). Such a phenotype is also displayed by other members of the nuclear receptor superfamily, such as RARs and TRs [26].
3. Perspective 2: Genome-wide binding of VDR
For a detailed analysis of enhancer and promoter regions of primary transcription factor target genes in living cells, the method of chromatin immunoprecipitation (ChIP) [27] became very popular. This technique uses mild chemical cross-linking, for example, with 1% formaldehyde, to fix nuclear proteins to genomic DNA in living cells or tissues at any chosen time point. After sonication of chromatin into fragments of 200–400 bp in size, immunoprecipitation with an antibody against the chosen nuclear protein, such as the VDR, enriches those chromatin regions that had been in contact with the protein at the moment of cross-linking. After a reverse cross-linking reaction, the resulting chromatin fragments can either be amplified by quantitative PCR using primers specific for the chosen genomic region (ChIP-qPCR) or are directly applied to massive parallel sequencing (ChIP-seq). When a significant enrichment in relation to a control (which mostly is ChIP with unspecific IgGs) is observed for a given genomic region, this is taken as an indication that the nuclear protein had been in contact with the investigated genomic region. For example, by ChIP-qPCR approximately 10 kb of the regulatory regions of the primary VDR target genes CYP24A1 [28], CYP27B1 [29], CCNC [30] and CDKN1A (also called p21) [31,32] were screened for genomic VDR-binding sites and per gene 2–4 specific sites were identified. Alternatively, the complete human ALOX5 gene sequence (some 85 kb) was first screened in silico for regions comprising putative vitamin D response elements (VDREs) and then studied by ChIP-qPCR [33]. From 22 investigated regions, two were shown to be functional in living cells, one of which is located far downstream (+42 kb) of the TSS of the ALOX5 gene.
To date, three VDR ChIP-seq studies have been published. In human lymphoblastoids, which were treated for 36 h with 1α,25(OH)2D3, Ramagopalan et al. [34] reported 2776 genomic VDR-binding sites. In human monocytes (THP-1), Heikkinen et al. [35] observed after 40 min ligand stimulation 1820 VDR ChIP-seq peaks, 1171 of which occur only in the presence of 1α,25(OH)2D3. For comparison, in the absence of ligand in lymphoblastoids and monocytes only 623 and 520 genomic VDR sites were found. Finally, in human colorectal cells (LS180), which were stimulated for 180 min with 1α,25(OH)2D3, Meyer et al. [36] showed that 1674 VDR-binding sites co-locate with those of the VDR partner protein RXR. Importantly, the ChIP-seq studies confirmed a number of previously reported VDR-binding sites on known primary 1α,25(OH)2D3 targets, such as that of the genes MYC [37], VDR [38], CCNC [30] and ALOX5 [33]. In addition, they reported some extra sites for known 1α,25(OH)2D3 target genes and also indicated a large number of previously unknown targets of VDR.
Despite different cellular models and large differences in ligand treatment times, the three ChIP-seq studies revealed a comparable number of VDR-binding sites of approximately 1600–2700 specific peaks. However, only 20% of these genomic sites are identical in all three investigated cell lines, such as in the case of CYP19A1 gene (Fig. 1C). The latter case codes for the estrogen synthesizing enzyme aromatase, which was previously established to be an up-regulated 1α,25(OH)2D3 target gene [39]. Interestingly, the VDR-binding site of this gene is located within an intron some 110 kb downstream of the TSS. In all three cellular models it is bound in a ligand-dependent fashion by the VDR.
Although the majority of VDR binding across the genome is both time and cell background specific, it can reasonably be anticipated that the shared 20% of VDR-binding sites are conserved and represent important functions in all VDR expressing tissues. This implies that data, such as shown in Fig. 1C, may be extrapolated to other human cell types.
Another result, on which the three VDR ChIP-seq studies are in accordance with findings of the ENCODE project [40], is that the distribution of the VDR binding sites has a Gaussian shape, i.e. VDR binding sites are found both up- and downstream of the TSS region of the primary target genes. The likelihood of detecting a functional VDR binding site decreases by distance from the TSS, but there is no maximal distance limiting the interaction between a VDR carrying enhancer region and a TSS region. However, the functionality of the most newly identified genomic VDR binding sites needs to be validated by assays that monitor the three-dimensional interaction of genomic regions, ideally by a genome-wide method, such as chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) [41].
Furthermore, most of the ChIP-seq studies with other members of the nuclear receptor superfamily indicated some 5000–10,000 genome-wide binding sites [42,43], i.e. the numbers reported for VDR is relatively low. However, nuclear receptor binding appears modest compared to other transcription factors, such as FOXA1, for which up to 80,000 ChIP-seq peaks were found [44]. Transcription factors that show such a high number of genomic binding sites are assumed to have greater binding promiscuity and/or diversity of interactions. In this manner they may act more as “pioneer factors”, i.e. as transcription factors that bind regulatory genomic regions at first and start the opening of these loci via the interaction with chromatin modifying enzymes. This then allows “following factors” to bind a subset of these accessible regions and to execute their regulatory actions. Viewed in this manner it is most likely that the VDR is most likely a following than a pioneer factor.
Single gene studies support this model whereby modulation of VDR-binding appears determined by the transcription factors AP1 [45] or RUNX2 [46] suggesting that there are pioneer processes that influence and determine VDR function. So far, however, no genome-wide study of possible pioneer factors cooperating with VDR has been published. However, although a negative result, Meyer et al. [36] showed that in human colorectal cells the transcription factor TCF7L2 does not act as a pioneer factor for VDR. Nevertheless, in analogy to studies with ERα [47], it can be assumed that ubiquitously expressed transcription factors, such as FOXA1, AP1, SPI1 or SP1, may act as pioneer factors for the VDR.
4. Perspective 3: Genomic DNA-binding modes of the VDR
Central to ChIP-seq data studies is the analysis of the sequences below the identified peaks (mostly within ±100 bp of the peak summit) for any enriched sequence motif, the idea being that this sequence will reflect a transcription factor-binding site. In all three VDR ChIP-seq studies [34–36], such agnostic binding site searches identified the well-established DR3-type RE consensus sequence for VDR–RXR heterodimers as being the most highly enriched (Fig. 1A, bottom). Strikingly, using the narrow observation window of ±100 bp either side of the peak height, only 31.7% (742) of all 2340 VDR peak summits in monocytes include one or more DR3-type REs [35]. Similar numbers apply for the datasets from lymphoblastoids and colorectal cancer cells.
When focusing only on 1α,25(OH)2D3-dependent VDR peaks and plotting the percentage of DR3-type RE content over the quality of the VDR ChIP-seq peak, the three ChIP-seq datasets provide similar results. That is, the higher the fold enrichment/value of a VDR peak, the higher is the chance that it contains a high-quality DR3-type RE [48]. In contrast, from the 520 genomic VDR-binding locations that uniquely occur in monocytes in the absence of ligand, only 14% contain a DR3-type VDRE [35]. This observation suggests that after ligand activation, the VDR shifts from genomic regions without a DR3-type RE to those with a DR3-type RE. This suggests that either the VDR becomes more specific in focusing upon its regulated genomic targets, or the binding sites associated with the basal state are more nuanced and less well explored. An intriguing implication of this discovery is that the non-DR3 locations may serve as a nuclear store of VDR to be utilized rapidly upon the introduction of the ligand, partly substituting for the need to transport VDR into the nucleus from outside.
The processes that drive the VDR to re-distribute to these locations remain unresolved. The lack of a DR3-type RE consensus sequence, even in the ligand stimulated state, in the majority of the VDR ChIP-seq peaks suggests that VDR either (i) has far more promiscuous or relaxed DNA binding specificities than previously assumed, probably by forming a complex with a presently undefined transcription factor (Fig. 1B, middle) or (ii) tethers to another DNA-binding transcription factor, such as a pioneer factor, rather than directly contacting DNA (Fig. 1B, bottom). Searches for other VDRE types with either different spacing or relative orientations of the core binding motifs have not provided any statistically significant enrichment within ±100 bp of the peak summit. Although it is still possible that a few individual regions carry such alternative VDRE types, in the published datasets there is no genome-wide evidence for their widespread use.
5. Perspective 4: VDR in dynamic interactions with chromatin components
The complex of genomic DNA and nucleosomes, referred to as chromatin, per se prevents access of transcription factors to their genomic targets [49]. This intrinsic repressive potential of chromatin is essential for long-lasting regulatory decisions, such as terminal differentiation of cells [50]. However, the epigenetic landscape can also be highly dynamic and lead to short-lived states, such as a response of chromatin to extra- and intracellular signals, for example, an exposure to 1α,25(OH)2D3 [51]. One major component of epigenetic changes is the reversible post-translational modification of histone proteins, such as acetylation and methylation, that is directed by a large group of chromatin modifying enzymes, with either histone acetyltransferase (HAT), histone deacetylase (HDAC), histone methyltransferase (HMT) or histone demethylase (HDM) activity [52]. Some of these histone modifications are associated with genes that are actively transcribed, whereas others are a sign of repressed genes [53], i.e. the post-translational modifications of histones correlate with either active or inactive chromatin regions. A second class of nuclear enzymes have ATP-dependent chromatin remodeling activity and induces plasticity of chromatin by rearranging the organization of nucleosomes [54].
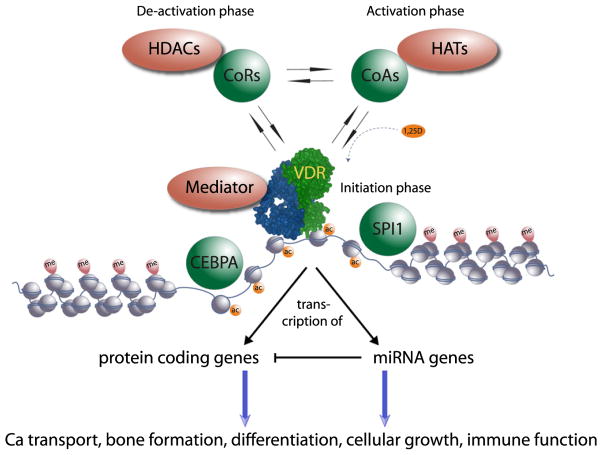
Nuclear receptors in general and the VDR in particular are amongst the first and most well described examples of the dynamic nature of transcriptional regulation in the context of chromatin [55–58]. In the so-called deactivation phase, i.e. in the absence of ligand, nuclear receptors that have a nuclear location, including the VDR, interact with CoR proteins, which in turn associate with HDACs leading to a locally more compact chromatin packaging [59]. In the activation phase, ligand binding induces the dissociation of CoRs and the association of CoAs [60]. Some CoAs have HAT activity or are complexed with proteins harboring such activity, which in net effect results in local chromatin relaxation [61]. In the initiation phase, nuclear receptors interact with another class of CoAs, which are members of the Mediator complex, that build a bridge to the basal transcriptional machinery and initiate a burst of mRNA synthesis by RNA polymerase II [62] (Fig. 2, top). In this way, gene activation by a nuclear receptor, such as VDR, can be separated into three phases, in each of which the transcription factor interacts with a different class of nuclear proteins.
Fig. 2.
Integration of VDR actions. Together with the pioneering factors the VDR is the central part of a differentiation module. Putative pioneer factors such as CEBPA and SPI1 appear to help the VDR to access to its genomic binding sites, but may not be found at all VDR binding loci. At these genomic VDR binding regions there is a cyclical alternation of proteins representing the deactivation phase (for example, CoRs and HDACs), the activation phase (for example, CoAs and HATs) and the initiation phase (for example, VDR and Mediator proteins). The outcome of the dynamic interaction of VDR with its binding sites and partner proteins is the modulation of the transcription of its primary target genes. The latter are either protein coding genes or non-coding genes, such as miRNA genes. Some of the miRNAs are involved in the fine-tuning of the mRNA expression of the protein-coding genes. Together with secondary target genes they mediate the physiological actions of 1α,25(OH)2D3 and its receptor VDR.
Using time-resolved ChIP, Shang et al. [63] demonstrated that several CoA proteins were recruited in a cyclical fashion to an estrogen responsive chromatin region of the human TFF1 gene. Metivier et al. [64] showed on the same genomic region the sequential and ordered recruitment of ERα, RNA polymerase II and many chromatin-associated proteins, such as CoAs, CoRs, HATs, HDACs and HMTs. Similar observations were made with AR on the human KLK3 gene [65], with TRs on the human DIO1 gene [66] and with VDR on the human genes CYP24A1 [28,67], CDKN1A [32], IGFBP3 [68] and MYC [37]. All these examples show cyclical association of co-regulator proteins and, in part, also of the respective nuclear receptor with a periodicity of 30–60 min. Interestingly, the more recently published reports on CDKN1A and IGFBP3 also demonstrate the cycling of mature mRNA [32,68] or even protein [58]. Cycling in the abundance of mature mRNA can be observed only with those genes, whose half-life of the induced mRNA transcript is shorter than the periodicity of cyclical association of transcription factors and their co-regulators, i.e. in average 60 min or less. It is only under this condition that there is enough mRNA degradation within one transcription cycle in order to observe cycling of transcript levels [69]. This reduces the list of genes that show transcriptional cycling to those that encode short-lived regulatory proteins, such as transcription factors and kinases.
The cellular basis for this control most likely reflects the fact that transcriptional dynamics allows a better control of protein expression than controlling protein stability. A gene can be silenced far quicker, when it has to confirm every 60 min, if its transcription is still required. For example, pulsatile exposure of cells with cortisol stimulates transcriptional dynamics of GR [70], but these dynamics are not observed, when the synthetic GR ligand dexamethasone is used. The latter stabilizes the receptor for longer periods than the natural ligand cortisol. A similar observations was made with the synthetic VDR agonist Gemini, which failed to induce transcriptional dynamics of the human IGFBP3 gene, while 1α,25(OH)2D3 does [68]. These observations may have implications for the therapeutic application of synthetic nuclear receptor ligands and may explain some of their side effects.
6. Perspective 5: Primary VDR target genes
Each eukaryotic gene is under the control of a large set of transcription factors that bind up- and downstream of its TSS. An essential prerequisite for a direct modulation of transcription by 1α,25(OH)2D3 is the interaction of activated VDR with the basal transcriptional machinery. This is achieved through the specific binding of VDR to a genomic binding site, which via DNA looping gets into vicinity of a core promoter region of a primary 1α,25(OH)2D3 target gene [71]. The effect of 1α,25(OH)2D3 on gene expression, i.e. 1α,25(OH)2D3-induced changes of the transcriptome, has been investigated by multiple mRNA microarrays and more recently also by miRNA microarrays [72] in various cellular models (either established cell lines or primary cells) or in in vivo models (mostly rodents). However, there is a large variation in the microarray platforms used for these transcriptome studies and also the experimental conditions, such as treatment time and ligand concentration, have been rather divergent. Moreover, the application of a next-generation sequencing technology method for the detection of RNA transcripts, called RNA-seq, has not yet been reported for VDR target genes. Similar to ChIP-seq, this technique is based on the sequencing of all RNA transcripts of all cells and is supposed to be more sensitive than hybridization-based microarrays [73].
Some studies focused on the identification of primary VDR target genes and used rather short incubations with the ligand (2–6 h), while others were more interested in the overall physiological or consequential effects of 1α,25(OH)2D3 and used far longer treatment times (24–72 h). In the past, cDNA arrays with an incomplete number of genes were used and rather short lists of VDR target genes from colon [74], prostate [75–78], breast [79] and osteoblasts were obtained [80,81]. However, despite these limitations many genes appear to respond to 1α,25(OH)2D3 activation. For example, in squamous cell carcinoma cells more than 900 genes responded within 12 h to a stimulation with 1α,25(OH)2D3 [82]. Unfortunately, the results of many of the earlier microarray studies with 1α,25(OH)2D3 were not placed in public data repositories, such as the Gene Expression Omnibus (GEO) of NCBI [83], which made a direct comparison of the results difficult.
Also more recent microarray analyses in various tissues and cells from different species have suggested long lists of VDR target genes. For example, in human monocytes (THP-1) 638 genes responded to a 4 h treatment with 1α,25(OH)2D3 [35], while a 36 h stimulation of human lymphoblastoids let only 229 genes move [34]. However, the overlap between these two 1α,25(OH)2D3 target gene lists is only 5.6%. This confirms the overall impression that most VDR target genes respond to 1α,25(OH)2D3 in a very tissue- and time-specific fashion and some of them show only a rather transient response to the ligand. Although a number of these genes may not be primary VDR targets, they nevertheless contribute to the physiological effects of 1α,25(OH)2D3. Although there are far fewer studies to date on VDR regulation of miRNAs, the numbers regulated and the time-dependent patterns appear comparable to mRNA targets in terms of the proportion of the total number regulated and the kinetics [58,72].
The combination of 1α,25(OH)2D3 microarray data with VDR ChIP-seq data from the same cellular model allows a more detailed exploration of the mechanisms of VDR target gene regulation. This was possible in particular for the study in monocytes [35], where a 40 min ligand stimulation for VDR location mapping and a 4 h 1α,25(OH)2D3 treatment for mRNA expression studies was used. Due to the short stimulation time most of the 638 regulated genes can be assumed to be primary 1α,25(OH)2D3 targets, i.e. that their mRNA expression changes are a direct consequence of the binding of VDR to genomic regions looping to their respective core promoter region. Plotting the positions of the 1α,25(OH)2D3-stimulated VDR ChIP-seq peaks in relation to the TSS of the 1α,25(OH)2D3 target genes showed a clear peak at the TSS region and symmetrical decline towards both the upstream and downstream flanking regions [48]. This emphasizes again that VDR binds as likely upstream as downstream of the core promoter region of its target genes. This fits with insights of the ENCODE project [84] and indicates that the pre-genomic focus on the upstream region only addressed half of the regulatory regions of a gene.
The gene regulatory scenarios of up-regulated VDR target genes vary considerably. In monocytes there are only about 20 genes, such as SP100 or CAMP, where VDR binds close to their core promoter region [35]. More common are situations where one target gene has multiple VDR-binding sites in various distances to its TSS region. Alternatively, a pair of closely located VDR target genes share one or more VDR-binding sites, as shown for the members of the IGFBP gene family [85]. From the 638 1α,25(OH)2D3 target genes in monocytes, 408 are up-regulated and for 93 of the latter (22.8%) the largest 1α,25(OH)2D3-stimulated VDR peak is within 30 kb from their TSS. For another 201 genes (49.3%), the most prominent VDR-binding site is in a distance of 30–400 kb from the core promoter region. For comparison, in pre-genomic studies a distance of 30 kb between a VDRE and the TSS was already considered large [71], while 400 kb was practically unimaginable.
Interestingly, only 99 (43.0%) out of the 230 down-regulated genes in monocytes have a 1α,25(OH)2D3-stimulated VDR peak in the ±400 kb region [35]. This observation emphasizes that the mechanisms of down-regulation of VDR target genes seem to be different from that of up-regulation. They may require gene-specific investigations as demonstrated for the genes CYP27B1 [29] and MYC [37]. In the case of the CYP27B1 gene, the repressive function of VDR results from indirect interaction with genomic DNA, via transcription factor 3, also known as VDR-interacting repressor [86].
Another mechanism of gene regulation is de-repression, which was first described for the nuclear receptors TR and LXR [87,88]. In this regulatory process the nuclear receptor actively represses genes via the interaction with CoR and HDAC proteins. The addition of ligand induces a dissociation of the nuclear receptor from its binding site and a release of the repression. In monocytes, only six up-regulated genes meet the de-repression criteria that they have a VDR peak in the unstimulated sample and no peak in the 1α,25(OH)2D3-treated sample [35]. An additional 21 up-regulated genes can be called dominantly de-repressed, since their main peak is found only in the unstimulated sample. This indicates that for some 10% of all up-regulated 1α,25(OH)2D3 target genes, a de-repression mechanism may apply.
Nevertheless, for some 25% of the up-regulated and more than the half of the down-regulated 1α,25(OH)2D3 target genes in monocytes the ChIP-seq approach does not identify any VDR binding within 400 kb of their core promoter region, i.e. for these genes there is no obvious explanation for their regulation by VDR [35]. However, gene regulation by VDR is a very dynamic process (see Section 5) with rapid changes of VDR-binding site occupancy [32,37,68], which a single, short time point at 40 min may have not fully captured. The time points chosen in each study represent only snap-shots of the actions of the VDR and it is likely that without time-course data, a considerable proportion of transient VDR-binding sites remain unknown.
7. Perspective 6: VDR as a module component
Much of the activity of a cell depends on gene regulatory networks, which are built of interacting regulatory pathways, also referred to as modules. A module is represented by a set of co-regulated genes (both protein and non-protein coding) that respond to different conditions [89]. In such modules, transcription factors and epigenetic modifications serve as inputs, while the output is a gene expression pattern representing a physiological situation, such as a differentiation stage. Transcription factors show two different types of inputs, as they determine the expression of the target genes and serve as functional drivers, which come into play only during specific situations during development or cell fate decisions. Additionally, the regulation of chromatin structure and nuclear organization also play a role in determining and controlling the function of these modules, for example, by regulating the amplitude and magnitude of gene expression periodicity.
Understanding the central control of architectural modules in these gene circuits may yield insight into predicting cellular responses and thus therapeutic targets. For example, nuclear receptors regulate CYP enzymes in negative feedback loops that degrade ligand and signal output [15]. These metabolic enzymes are frequently altered in expression, and equally provide therapeutic targets in various syndromes.
In this context, the regulation of miRNA genes by VDR may be of special importance. After processing of its precursor the active part of a miRNA is a single-stranded RNA molecule of 21–23 nt in length, which associates with cytosolic proteins that use the miRNA for a sequence-specific recognition of the 3′-UTR of mRNA molecules and their consequent degradation [90]. In this way miRNAs control the half-life of their target mRNAs and regulate the level of translated proteins. Like transcription factors, each miRNA can have up to hundred targets [91], i.e. the regulation of a miRNA gene by VDR may have larger impact than the regulation of, for example, a metabolic enzyme. Some VDR regulated modules include feed forward loops that are crucial for the precise regulation of target genes, in terms of signal amplitude and magnitude. These loop motifs often include roles for miRNAs to fine-tune transcriptional signals [92] (see also Fig. 2, bottom). Studies with VDR combined with an emerging literature [93,94] suggest that these motifs are common in normal human biology and disrupted in cancer. For example, VDR regulates the MCM7 gene that encodes the MIR106b cluster. VDR also regulates CDKN1A that in turn is targeted by MIR106b. These members thereby form a VDR feed forward loop that governs cell cycle progression in human prostate epithelial cells [58]. The balance of these interactions appear disrupted in cancer cells compared to non-malignant models with selective attenuation and repression of VDR transcriptional responses of target genes such as CDKN1A. The suppressed transcriptional responses in PC-3 human prostate cancer cells were associated with gene-specific VDR-induced enrichment of the CoR NCOR1 leading to gene silencing. Other cyclin-dependent kinase inhibitors appear to be regulated in a similar manner. VDR represses MIR181a, which targets the CDKN1B gene (encodes for p27) and thereby establish another feed forward loop that promotes hematopoietic differentiation [95].
The architecture of these modules also appears to provide enough flexibility and information to generate spatial and temporal patterns of gene expression, for example, during cellular differentiation. Again, this can be studied best in the hematopoietic system. Hematopoiesis is believed to be controlled by a hierarchy of a relatively small number of critical transcription factors that are sequentially expressed, are largely restricted to a specific lineage and can interact directly to mediate and reinforce cell fate decisions [96]. However, genome-wide studies suggest amore complex architecture in regulatory circuits involving larger numbers of transcription factors that control different combinations of modules of co-expressed genes [97,98].
Novershtern et al. [99] measured the transcriptome profiles of a large number of hematopoietic stem cells, multiple progenitor states and terminally differentiated cell types. They found distinct regulatory circuits in both stem cells and differentiated cells, which implicated dozens of new regulators in hematopoiesis. They identified 80 distinct modules of tightly co-expressed genes in the hematopoietic system. One of these modules is expressed in granulocytes and monocytes and includes genes encoding enzymes and cytokine receptors that are essential for inflammatory responses. Major players in this module are VDR together with the pioneer factors CEBPA and SPI1 (Fig. 2). Further contributors are the proteins ATF3, CREB5, PPARGC1A, VENTX and MYCL1. This indicates that VDR works together with this small set of transcription factors, in order to regulate granulocyte and monocyte differentiation.
These findings also fit with previously obtained information about potent effects of 1α,25(OH)2D3 both on the innate and the adaptive immune system. For example, 1α,25(OH)2D3 enhances the differentiation of monocytes into functional macrophages with increased phagocytic capacity and altered cytokine-secreting capacity, but impairs the differentiation of monocytes into dendritic cells [100]. The main 1α,25(OH)2D3 targets in differentiating monocytes are anti-microbial peptides, such as cathelicidin, co-stimulatory molecules, such as CD14 [35], and cytokines, such as interleukins 10 and 12b [101,102]. The new insight of the dominant role of VDR in the granulocyte/monocyte module now allows more specific investigations on the functional interplay of VDR with its partner transcription factors, for example with the pioneer factors CEBPA and SPI1.
These provocative studies also reflect a very powerful light on much earlier and translational studies on the role of 1α,25(OH)2D3 and its analogs to drive so-called differentiation therapy in myeloid malignancies [103–107]. However, clinical exploitation of these studies was ultimately equivocal and perhaps required more accurate analyses of individual patient responsiveness to such therapies. The new modular understanding of the VDR may ultimately provide this insight.
8. Conclusions
The different perspectives presented here for the VDR reflect the pleiotropic molecular actions of the receptor and its natural ligand 1α,25(OH)2D3. In this context the parameter time has emerged to be very critical due to the dynamic response of tissues and cell types, especially in the early phase of their treatment with 1α,25(OH)2D3. Therefore, further time-course experiments for VDR ChIP-seq and 1α,25(OH)2D3 microarrays will provide a more detailed understanding of this aspect.
Genome-wide the actions of VDR and 1α,25(OH)2D3 to date have best been understood in cells of the hematopoietic system. Modular studies have started to demonstrate with which other partner transcription factors VDR forms integrated units that offer up windows of potent transcriptional actions to determine cell fate. These modular actions may also shed light on the targeted effects of the VDR in physiology. Part of this range of targeted effects and sensitivity is in part determined by the intrinsic epigenetic states and shared expression of co-factors and histone modifying complexes. For example, VDR is important for the differentiation of mesenchymal stem cells to bone and fat cells. The large datasets obtained from genome- and transcriptome-wide investigations on VDR and on related transcription factors and epigenetic modifications provide new insight and will allow the integration of the actions of VDR with that of other signaling systems, such as that of other nuclear receptors or of pioneer factors, such as CEBPA and SPI1. This will allow a more generalized understanding of VDR and 1α,25(OH)2D3 in the control of the whole body’s physiology.
This may also illuminate the discrepancies observed on responsiveness of the VDR in disease states, such as cancer, where responsiveness of cells towards VDR actions, ranging from sensitivity to recalcitrance. Given that miRNA regulation by the VDR appears common, this can be exploited to define individual cell or patient responsiveness to the vitamin D-based therapies. Tumor-specific miRNA patterns are emerging as highly attractive biomarkers, for example, of cancer risk and progression. Given miRNAs are secreted into body fluids [108] and can be reliably extracted and measured [109], they offer significant clinical potential as highly sensitive serum-borne prognostic indicators [110,111]. Using serum-borne miRNAs as prognostic markers is highly attractive for several reasons. First, they can overcome the limitations of inaccurate sampling for the presence of cancer. Second, they can encapsulate the effects of heterotypic cell interactions within the tumor microenvironment. Third, they form a non-invasive test procedure. Therefore understanding miRNA regulation, within critical VDR modules, offers up the real opportunity of tailoring and monitoring vitamin D therapies to the individual.
Acknowledgments
C.C. thanks the Academy of Finland and the Juselius Foundation for support. M.J.C. acknowledges the Biotechnology and Biological Sciences Research Council (UK) and support in part from National Institute of Health Grants R01 CA095367-06 and 2R01-CA-095045-06. M.J.C. also acknowledges support, in part, of the NCI Cancer Center Support Grant to the Roswell Park Cancer Institute. C.C. and M.J.C. acknowledge the support of NucSys, an European Community FP6 Marie Curie Research Training Network and CanSys, an Atlantis EU-US training program. C.C. thanks Drs. S. Heikkinen and F. Molnár for bioinformatic support in the preparation of the figures.
Abbreviations
- 1α, 25(OH)2D3
1α,25-dihydroxyvitamin D3
- 25(OH)D3
25-hydroxyvitamin D3
- ALOX5
arachidonate 5-lipoxygenase
- AR
androgen receptor
- CAMP
cathelicidin anti-microbial peptide
- CCNC
cyclin C
- CDKN1A
cyclin-dependent kinase inhibitor 1A
- CoA
co-activator
- CoR
co-repressor
- ChIP
chromatin immunoprecipitation
- ChIP-seq
ChIP coupled with massive parallel sequencing
- CYP
cytochrome P450
- DBD
DNA-binding domain
- DIO1
thyroxine deiodinase type I
- DR3
direct repeat spaced by 3 nucleotides
- ER
estrogen receptor
- FXR
farnesoid X receptor
- GLDN
gliomedin
- GR
glucocorticoid receptor
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HDM
histone demethylase
- HMT
histone methyltransferase
- IGFBP
insulin-like growth factor binding protein
- KLK3
kallikrein 3
- LBD
ligand-binding domain
- LXR
liver X receptor
- miRNA
micro RNA
- MR
mineralocorticoid receptor
- PR
progesterone receptor
- RAR
retinoic acid receptor
- RE
response elements
- RXR
retinoid X receptor
- SP100
SP100 nuclear antigen
- TFF1
trefoil factor 1
- TR
thyroid hormone receptor
- TSS
transcription start site
- VDR
vitamin D receptor
- VDRE
vitamin D response element
References
- 1.Holick MF. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am J Clin Nutr. 2004;80:1678S–88S. doi: 10.1093/ajcn/80.6.1678S. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–81. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 3.Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78:1193–231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 4.Renkema KY, Alexander RT, Bindels RJ, Hoenderop JG. Calcium and phosphate homeostasis: concerted interplay of new regulators. Ann Med. 2008;40:82–91. doi: 10.1080/07853890701689645. [DOI] [PubMed] [Google Scholar]
- 5.Tsai HC, Norman AW. Studies on calciferol metabolism. 8. Evidence for a cytoplasmic receptor for 1,25-dihydroxy-vitamin D3 in the intestinal mucosa. J Biol Chem. 1973;248:5967–75. [PubMed] [Google Scholar]
- 6.Brumbaugh PF, Hughes MR, Haussler MR. Cytoplasmic and nuclear binding components for 1α 25-dihydroxyvitamin D3 in chick parathyroid glands. Proc Natl Acad Sci USA. 1975;72:4871–5. doi: 10.1073/pnas.72.12.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA. 1988;85:3294–8. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouillon R, Carmeliet G, Verlinden L, van Etten E, Verstuyf A, Luderer HF, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29:726–76. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80:1689S–96S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- 10.Hart PH, Gorman S, Finlay-Jones JJ. Modulation of the immune system by UV radiation: more than just the effects of vitamin D? Nat Rev Immunol. 2011;11:584–96. doi: 10.1038/nri3045. [DOI] [PubMed] [Google Scholar]
- 11.Haussler MR, Jurutka PW, Mizwicki M, Norman AW. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2 vitamin D3: genomic and non-genomic mechanisms. Best Pract Res Clin Endocrinol Metab. 2011;25:543–59. doi: 10.1016/j.beem.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Haussler MR, Haussler CA, Jurutka PW, Thompson PD, Hsieh JC, Remus LS, et al. The vitamin D hormone and its nuclear receptor: molecular actions and disease states. J Endocrinol. 1997;154(Suppl):S57–73. [PubMed] [Google Scholar]
- 13.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 14.Molnár F, Peräkylä M, Carlberg C. Vitamin D receptor agonists specifically modulate the volume of the ligand-binding pocket. J Biol Chem. 2006;281:10516–26. doi: 10.1074/jbc.M513609200. [DOI] [PubMed] [Google Scholar]
- 15.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 16.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–24. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engström O, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–8. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 19.Carlberg C, Bendik I, Wyss A, Meier E, Sturzenbecker LJ, Grippo JF, et al. Two nuclear signalling pathways for vitamin D. Nature. 1993;361:657–60. doi: 10.1038/361657a0. [DOI] [PubMed] [Google Scholar]
- 20.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–66. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 22.Burke LJ, Baniahmad A. Co-repressors 2000. FASEB J. 2000;14:1876–88. doi: 10.1096/fj.99-0943rev. [DOI] [PubMed] [Google Scholar]
- 23.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, et al. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARγ. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verstuyf A, Carmeliet G, Bouillon R, Mathieu C. Vitamin D: a pleiotropic hormone. Kidney Int. 2010;78:140–5. doi: 10.1038/ki.2010.17. [DOI] [PubMed] [Google Scholar]
- 25.Polly P, Herdick M, Moehren U, Baniahmad A, Heinzel T, Carlberg C. VDR–Alien: a novel, DNA-selective vitamin D3 receptor–corepressor partnership. FASEB J. 2000;14:1455–63. doi: 10.1096/fj.14.10.1455. [DOI] [PubMed] [Google Scholar]
- 26.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–23. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 27.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 28.Väisänen S, Dunlop TW, Sinkkonen L, Frank C, Carlberg C. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1α,25-dihydroxyvitamin D3. J Mol Biol. 2005;350:65–77. doi: 10.1016/j.jmb.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 29.Turunen MM, Dunlop TW, Carlberg C, Väisänen S. Selective use of multiple vitamin D response elements underlies the 1α,25-dihydroxyvitamin D3-mediated negative regulation of the human CYP27B1 gene. Nucleic Acids Res. 2007;35:2734–47. doi: 10.1093/nar/gkm179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinkkonen L, Malinen M, Saavalainen K, Väisänen S, Carlberg C. Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res. 2005;33:2440–51. doi: 10.1093/nar/gki502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saramäki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 2006;34:543–54. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saramäki A, Diermeier S, Kellner R, Laitinen H, Väisänen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1α,25-dihydroxyvitamin D3. J Biol Chem. 2009;284:8073–82. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seuter S, Väisänen S, Radmark O, Carlberg C, Steinhilber D. Functional characterization of vitamin D responding regions in the human 5-lipoxygenase gene. Biochim Biophys Acta. 2007;1771:864–72. doi: 10.1016/j.bbalip.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–60. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011;39:9181–93. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26:37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toropainen S, Väisänen S, Heikkinen S, Carlberg C. The down-regulation of the human MYC gene by the nuclear hormone 1α,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J Mol Biol. 2010;400:284–94. doi: 10.1016/j.jmb.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 38.Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 2010;24:128–47. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jakob F, Homann D, Seufert J, Schneider D, Köhrle J. Expression and regulation of aromatase cytochrome P450 in THP 1 human myeloid leukaemia cells. Mol Cell Endocrinol. 1995;110:27–33. doi: 10.1016/0303-7207(95)03512-6. [DOI] [PubMed] [Google Scholar]
- 40.ENCODE-Project-Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Gen Dev. 2008;22:2953–67. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, et al. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Gen Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schüle R, Umesono K, Mangelsdorf DJ, Bolado J, Pike JW, Evans RM. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 46.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, et al. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol Cell Biol. 2003;23:3339–51. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlberg C, Seuter S, Heikkinen S. The first genome-wide view of vitamin D receptor locations and their mechanistic implications. Anticancer Res. 2012;32:271–82. [PubMed] [Google Scholar]
- 49.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–8. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohn F, Schübeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–36. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 52.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 53.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 54.Hager GL, Nagaich AK, Johnson TA, Walker DA, John S. Dynamics of nuclear receptor movement and transcription. Biochim Biophys Acta. 2004;1677:46–51. doi: 10.1016/j.bbaexp.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 55.Metivier R, Reid G, Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO Rep. 2006;7:161–7. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265–266:162–7. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.George AA, Schiltz RL, Hager GL. Dynamic access of the glucocorticoid receptor to response elements in chromatin. Int J Biochem Cell Biol. 2009;41:214–24. doi: 10.1016/j.biocel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorne JL, Maguire O, Doig CL, Battaglia S, Fehr L, Sucheston LE, et al. Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Res. 2011;39:2045–56. doi: 10.1093/nar/gkq875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–7. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 60.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 61.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Gen Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 62.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Näär AM, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–8. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 63.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 64.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor a directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 65.Kang Z, Pirskanen A, Jänne OA, Palvimo JJ. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J Biol Chem. 2002;277:48366–71. doi: 10.1074/jbc.M209074200. [DOI] [PubMed] [Google Scholar]
- 66.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci USA. 2002;99:7934–9. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–17. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 68.Malinen M, Ryynänen J, Heinäniemi M, Väisänen S, Carlberg C. Cyclical regulation of the insulin-like growth factor binding protein 3 gene in response to 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2011;39:502–12. doi: 10.1093/nar/gkq820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carlberg C, Seuter S. Dynamics of nuclear receptor target gene regulation. Chromosoma. 2010;119:479–84. doi: 10.1007/s00412-010-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, et al. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat Cell Biol. 2009;11:1093–102. doi: 10.1038/ncb1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlberg C, Polly P. Gene regulation by vitamin D3. Crit Rev Eukaryot Gene Expr. 1998;8:19–42. doi: 10.1615/critreveukargeneexpr.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- 72.Wang WL, Chatterjee N, Chittur SV, Welsh J, Tenniswood MP. Effects of 1α,25 dihydroxyvitamin D3 and testosterone on miRNA and mRNA expression in LNCaP cells. Mol Cancer. 2012;10:58. doi: 10.1186/1476-4598-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon-Cardo C, Munoz A. Genetic signatures of differentiation induced by 1α,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799–806. [PubMed] [Google Scholar]
- 75.Krishnan AV, Shinghal R, Raghavachari N, Brooks JD, Peehl DM, Feldman D. Analysis of vitamin D-regulated gene expression in LNCaP human prostate cancer cells using cDNA microarrays. Prostate. 2004;59:243–51. doi: 10.1002/pros.20006. [DOI] [PubMed] [Google Scholar]
- 76.Khanim FL, Gommersall LM, Wood VH, Smith KL, Montalvo L, O’Neill LP, et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23:6712–25. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 77.Peehl DM, Shinghal R, Nonn L, Seto E, Krishnan AV, Brooks JD, et al. Molecular activity of 1,25-dihydroxyvitamin D3 in primary cultures of human prostatic epithelial cells revealed by cDNA microarray analysis. J Steroid Biochem Mol Biol. 2004;92:131–41. doi: 10.1016/j.jsbmb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Ikezoe T, Gery S, Yin D, O’Kelly J, Binderup L, Lemp N, et al. CCAAT/enhancer-binding protein delta: a molecular target of 1,25-dihydroxyvitamin D3 in androgen-responsive prostate cancer LNCaP cells. Cancer Res. 2005;65:4762–8. doi: 10.1158/0008-5472.CAN-03-3619. [DOI] [PubMed] [Google Scholar]
- 79.Swami S, Raghavachari N, Muller UR, Bao YP, Feldman D. Vitamin D growth inhibition of breast cancer cells: gene expression patterns assessed by cDNA microarray. Breast Cancer Res Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- 80.Eelen G, Verlinden L, Van Camp M, Mathieu C, Carmeliet G, Bouillon R, et al. Microarray analysis of 1α,25-dihydroxyvitamin D3-treated MC3T3-E1 cells. J Steroid Biochem Mol Biol. 2004;89–90:405–7. doi: 10.1016/j.jsbmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 81.Eelen G, Verlinden L, van Camp M, van Hummelen P, Marchal K, de Moor B, et al. The effects of 1α,25-dihydroxyvitamin D3 on the expression of DNA replication genes. J Bone Miner Res. 2004;19:133–46. doi: 10.1359/JBMR.0301204. [DOI] [PubMed] [Google Scholar]
- 82.Wang TT, Tavera-Mendoza LE, Laperriere D, Libby E, MacLeod NB, Nagai Y, et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol. 2005;19:2685–95. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 83.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.ENCODE-Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matilainen M, Malinen M, Saavalainen K, Carlberg C. Regulation of multiple insulin-like growth factor binding protein genes by 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2005;33:5521–32. doi: 10.1093/nar/gki872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murayama A, Kim MS, Yanagisawa J, Takeyama K, Kato S. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 2004;23:1598–608. doi: 10.1038/sj.emboj.7600157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley BW. Interaction of human thyroid hormone receptor β with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–6. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–93. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, et al. Module networks: identifying regulatory modules and their condition – specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 90.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 91.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 92.Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–45. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cohen EE, Zhu H, Lingen MW, Martin LE, Kuo WL, Choi EA, et al. A feed-forward loop involving protein kinase Calpha and microRNAs regulates tumor cell cycle. Cancer Res. 2009;69:65–74. doi: 10.1158/0008-5472.CAN-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, et al. P53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8:736–41. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iwasaki H, Akashi K. Hematopoietic developmental pathways: on cellular basis. Oncogene. 2007;26:6687–96. doi: 10.1038/sj.onc.1210754. [DOI] [PubMed] [Google Scholar]
- 97.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–63. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.FANTOM-Consortium Riken Center. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–62. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Curr Opin Pharmacol. 2010;10:482–96. doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 101.Matilainen JM, Husso T, Toropainen S, Seuter S, Turunen MP, Gynther P, et al. Primary effect of 1α,25(OH)2D3 on IL-10 expression in monocytes is short-term down-regulation. Biochim Biophys Acta. 2010;1803:1276–86. doi: 10.1016/j.bbamcr.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 102.Gynther P, Toropainen S, Matilainen JM, Seuter S, Carlberg C, Väisänen S. Mechanism of 1α,25-dihydroxyvitamin D3-dependent repression of interleukin-12B. Biochim Biophys Acta. 2011;1813:810–8. doi: 10.1016/j.bbamcr.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 103.Reichel H, Koeffler HP, Tobler A, Norman AW. 1α,25-Dihydroxyvitamin D3 inhibits gamma-interferon synthesis by normal human peripheral blood lymphocytes. Proc Natl Acad Sci USA. 1987;84:3385–9. doi: 10.1073/pnas.84.10.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tobler A, Gasson J, Reichel H, Norman AW, Koeffler HP. Granulocyte-macrophage colony-stimulating factor. Sensitive and receptor-mediated regulation by 1,25-dihydroxyvitamin D3 in normal human peripheral blood lymphocytes. J Clin Invest. 1987;79:1700–5. doi: 10.1172/JCI113009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elstner E, Lee YY, Hashiya M, Pakkala S, Binderup L, Norman AW, et al. 1α,25-Dihydroxy-20-epi-vitamin D3: an extraordinarily potent inhibitor of leukemic cell growth in vitro. Blood. 1994;84:1960–7. [PubMed] [Google Scholar]
- 106.Studzinski GP, Bhandal AK, Brelvi ZS. Potentiation by 1α,25-dihydroxyvitamin D3 of cytotoxicity to HL-60 cells produced by cytarabine and hydroxyurea. J Natl Cancer Inst. 1986:641–8. doi: 10.1093/jnci/76.4.641. [DOI] [PubMed] [Google Scholar]
- 107.Studzinski GP, Bhanda AK, Brelvi ZS. Cell cycle sensitivity of HL-60 cells to the differentiation-inducing effects of 1α,25-dihydroxyvitamin D3. Cancer Res. 1985;45:3898–905. [PubMed] [Google Scholar]
- 108.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids – the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 110.El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50:564–73. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 111.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shaffer PL, Gewirth DT. Structural analysis of RXR–VDR interactions on DR3 DNA. J Steroid Biochem Mol Biol. 2004;89–90:215–9. doi: 10.1016/j.jsbmb.2004.03.084. [DOI] [PubMed] [Google Scholar]