Abstract
Summary: We report the creation of Drug Signatures Database (DSigDB), a new gene set resource that relates drugs/compounds and their target genes, for gene set enrichment analysis (GSEA). DSigDB currently holds 22 527 gene sets, consists of 17 389 unique compounds covering 19 531 genes. We also developed an online DSigDB resource that allows users to search, view and download drugs/compounds and gene sets. DSigDB gene sets provide seamless integration to GSEA software for linking gene expressions with drugs/compounds for drug repurposing and translational research.
Availability and implementation: DSigDB is freely available for non-commercial use at http://tanlab.ucdenver.edu/DSigDB.
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: aikchoon.tan@ucdenver.edu
1 Introduction
High-throughput genomic technologies enable researchers to analyze tens of thousands of ‘omic’ data points within biological systems. Typically, long lists of interesting candidate genes were generated from these analyses. However, interpreting these gene lists remains a challenge for biomedical researchers. Recognizing that genes act in concert to drive various biological processes, Gene Set Enrichment Analysis (GSEA) was introduced to summarize genomics data using a priori defined gene sets (Mootha et al., 2003, Subramaniam et al., 2005). This seminal article has revolutionized genomic data analyses and interpretation, influenced the development of other analytical enrichment tools. The critical components of these tools are the ‘annotated databases’ used as the gene sets for summarizing high-level biological processes. The popular Molecular Signatures Database (Liberzon et al., 2011), which provides the gene sets for GSEA, currently contains positional, curated, Gene Ontology, motif and computational gene sets. It also contains specialized gene sets for oncogenic and immunologic signatures that were extracted from literatures. GeneSigDB (Culhane et al., 2009), a database of gene signatures extracted and manually curated from the published literature, is another resource for analytical enrichment tools.
Here, we introduce Drug Signatures Database (DSigDB), a collection of drug and small molecule-related gene sets based on quantitative inhibition and/or drug-induced gene expression changes data (see Supplementary Fig. S1). DSigDB differs from the existing resources in the following aspects: (i) DSigDB gene sets were extracted and compiled from quantitative inhibition data of drugs/compounds from a variety of databases and publications. These genes represent the direct targets of the drugs/compounds. (ii) DSigDB gene sets are acquired through both automatic computational methods and manual curation. (iii) Gene sets from DSigDB are explicitly designed to provide seamless integration to GSEA software (see Supplementary Fig. S2). (iv) DSigDB contains the largest number of drugs/compound-related gene sets to date. Here, we will describe the gene set collections of DSigDB, the accompanying online resource, and provide an example illustrating the utility of DSigDB gene sets for GSEA software.
2 DSigDB Collections
2.1 DSigDB collections
DSigDB organizes drugs and small molecules-related gene sets into four collections based on quantitative inhibition and/or drug-induced gene expression changes data.
2.2 D1: approved drugs
This collection of gene sets contains 1202 Food and Drug Administration (FDA) approved drugs covering 1288 target genes. We obtained all the approved drugs from US FDA website, and retrieved bioactivity data for these drugs from PubChem and ChEMBL. Genes with ‘active’ bioassay results recorded in these databases were compiled as the drug target genes (see Supplementary Data for details). The mean gene set size for D1 is 10 (range 1–258).
2.3 D2: kinase inhibitors
The human kinome has been a class of intensely pursued drug targets by the pharmaceutical industry. Kinases are frequently mutated in various cancers. Therefore targeting these kinases with small molecules is an attractive therapeutic approach for personalized cancer treatment. This collection of gene sets contains 1220 kinase inhibitors (1065 unique kinase inhibitors) covering 407 kinases. We collected large-scale in vitro kinase profiling assays from literature and two databases (Medical Research Council Kinase Inhibitor database and Harvard Medical School Library of Integrated Network-based Cellular Signatures database). We considered the kinase a target of a kinase inhibitor if the IC50/Kd/Ki ≤ 1 µM or the Percent of inhibition over Control ≤ 15% from the assays. These target kinases make up the gene sets for the kinase inhibitors. The mean gene set size for D2 is 15 (range 1–315).
2.4 D3: perturbagen signatures
This collection of gene sets was obtained from gene expression profiles induced by compounds. We collected 7064 gene expression profiles from three cancer cell lines perturbed by 1309 compounds from CMap (build 02) (Lamb et al., 2006). For each compound, we compared the treated versus control gene expression profiles for each cell line. Genes with >2-fold change from the control were considered as gene sets (either up or down) for that compound. We defined 1998 gene sets (1154 unique compounds) covering 11 137 genes in this collection. The mean gene set size for D3 is 81 (range 1–3468).
2.5 D4: computational drug signatures
We compiled 18 107 drug signatures extracted from literatures using a mixture of manual curation and text mining approaches. Using manual curation of targets, we compiled 10 830 and 5163 gene sets from the Therapeutics Targets Database (Qin et al., 2014) and the Comparative Toxicogenomics Database (Davis et al., 2013), respectively. For the text mining approach, we used the Biomedical Object Search System (Choi et al., 2012) to acquire 2114 co-occurrences of compounds and genes from PubMed abstracts. In addition, we also retrieved genes with ‘active’ bioactivity data for these drugs from PubChem and ChEMBL as in D1. These genes, with quantitative inhibition data, were integrated with the drug signatures obtained from the source to construct the final gene sets for the drug (see Supplementary Data for details). The mean gene set size for D4 is 28 (range 1–8312).
2.6 Gene set annotations
Each DSigDB gene set consists of a list of target genes of a compound. The current version of DSigDB focuses on human gene sets. We used human Entrez Gene IDs to serve as universal identifiers to map across different databases. We used InChiKey to serve as the universal compound identifiers to map between PubChem and ChEMBL, and to determine the number of unique compounds within DSigDB. As described in the DSigDB collections, these gene sets are collected from several sources and some compounds could appeared multiple times according to their source of collection. DSigDB currently holds 22 527 gene sets, consists of 17 389 unique compounds covering 19 531 genes. Statistics for the gene set size is available in Supplementary Materials.
2.7 File formats
DSigDB gene sets are available to download as GSEA gene set (.gmt), plain text (.txt) or detailed text (_detailed.txt) formats. The .gmt file format can be directly imported into GSEA to execute the program. The gene set results generated from GSEA provide links to the DSigDB online resource for detailed information about the compounds. The plain text format provides a simple list of gene set membership for the compound. The detailed text format provides detailed information of the relations between genes and drug. It contains four columns: Drug, Gene, Type and Source. Every line represents the relation between drug and gene, the type of interactions (either quantitative binding results or qualitative interactions), and the source of the relation (See USER MANUAL for details). We also provide these files (either .gmt, .txt or detailed.txt) for the whole database as downloadable in the Download Page.
3 DSigDB online resource
As a companion to the DSigDB gene sets, we developed an online resource to allow users to search, view and download the compound information and annotated gene sets.
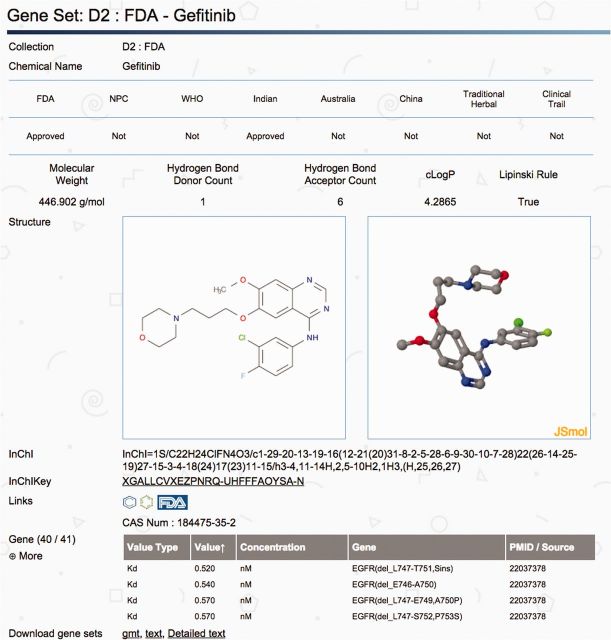
Each gene set and all of its annotations are presented as an individual web page (Fig. 1 ). Each web page contains four parts: (i) top part describes the clinical development of the compound (approved or clinical trials); (ii) middle part indicates the molecular details of the compound including chemical structure (2D and 3D), links to PubChem or ChEMBL; (iii) bottom part lists the gene memberships with embedded links to the source of evidence; (iv) download gene set.
Fig. 1.
An example of the DSigDB gene set web page
The web resource provides support for two search functions: i) compound search and ii) gene search. For ‘compound search’ function, the query compound will return the molecule information of the compound, its gene memberships with links to external sources (Fig. 1). For the ‘gene search’ function, it will return the list of compounds that target the query gene. Both search functions will provide the user to navigate the interactions between drug-gene in DSigDB. All DSigDB data are freely available for download.
4 Use case example
To illustrate an application of DSigDB, we performed GSEA on a previously published non-small cell lung cancer (NSCLC) microarray gene expression data (Coldren et al., 2006) using the D2 gene sets. We selected nine gefitinib (first-generation EGFR inhibitor)-sensitive (IC50 ≤ 2µM) and nine resistant (IC50 > 4µM) EGFR wild-type NSCLC lines for the analysis. Raw microarray gene expression data for these 18 cell lines were normalized, and gene expression profiles were extracted and served as the input for GSEA. GSEA was conducted by comparing gefitinib sensitive versus resistant lines using all of the D2 gene sets. We performed 1000 gene set permutations, and considered gene sets with P < 0.05 as significant. From the results, we observed 16 and 7 gene sets were enriched in the sensitive and resistant groups (P < 0.05), respectively. Notably, the top two gene sets of the sensitive group are CI-1033 and AZD9291, which are newer generation of EGFR inhibitors currently being tested in the clinic for NSCLC patients. According to the kinase inhibition profiles, 15 of the 16 gene sets enriched in the sensitive group inhibited EGFR. Conversely, none of the compounds enriched in the resistant group inhibit EGFR. This is expected as the comparison is between EGFR inhibitor sensitive versus resistant group. Interestingly, RO-3306, a CDK1 inhibitor was identified as enriched in resistant group. From the GDSC website (Yang et al., 2013), two of the gefitinib-resistant lines have lower IC50 as compared with the four gefitinib-sensitive lines, supporting the GSEA results and suggesting that this compound may be useful for EGFR inhibitor resistant lines (see Supplementary Data for details).
5 General applications of DSigDB
More generally, the DSigDB gene sets could be used with other enrichment tools for data analysis. For example, if a user has obtained a list of significant genes from a particular experiment, these genes could be tested for drug-target enrichment or over-representation analysis using the DSigDB gene sets. This will provide a list of candidate drugs/compounds enriched in the gene list for follow-up experiments.
Another application of DSigDB gene sets is to construct drug-target interactions networks for other bioinformatics analyses and discoveries. The gene sets could be easily added in as plug-in modules for visualizing the interactions between chemical–biological networks. This highlights the general usefulness of the DSigDB data in other bioinformatics analyses.
6 Conclusions
In conclusion, we developed DSigDB, a novel collection of gene sets based on quantitative inhibition and/or drug-induced gene expression changes data of drugs and compounds, as a resource for various gene set enrichment tools. We implemented an online resource to allow users to search, view and download the DSigDB gene sets. We believe that DSigDB represents a significant improvement in drugs/compounds related gene sets by quantity and quality (standardized formats for both genes and compounds). Users could seamlessly integrate DSigDB in their gene set enrichment analyses to provide direct links from gene lists to drugs for translational research and drug repurposing studies.
Supplementary Material
Acknowledgements
We thank the Tan Lab members for their useful comments on this manuscript. We also like to thank the reviewers for their constructive comments and suggestions in improving the DSigDB web resource.
Funding
This work was partly supported by the >Cancer League of Colorado, the >National Institutes of Health (P30CA046934, P50CA058187) and the David F. and Margaret T. Grohne Family Foundation.
Conflict of Interest: none declared.
References
- Choi J., et al. (2012) BOSS: context-enhanced search for biomedical objects. BMC Med. Inform. Decis Mak., 12 (Suppl. 1), S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldren C.D., et al. (2006) Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol. Cancer Res., 4, 521–528. [DOI] [PubMed] [Google Scholar]
- Culhane A., et al. (2009) GeneSigDB—a curated database of gene expression signatures. Nucleic Acids Res., 38, D716–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., et al. (2013) A CTD-Pfizer collaboration: manual curation of 88,000 scientific articles text mined for drug-disease and drug-phenotype interactions. Database, 2013, bat080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., et al. (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science, 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Liberzon A., et al. (2011) Molecular signature database (MSigDB) 3.0. Bioinformatics, 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., et al. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet., 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Qin C., et al. (2014) TTD: Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res., 42, D1118–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam A., et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.