Abstract
Background
Colorectal cancer (CRC) is the second most common type of cancer in the Western world. The treatment of this disease has evolved greatly, particularly for patients with metastatic disease. The advent of combination chemotherapy plus targeted agents has led to more curative resections and improved survival rates in these patients. A deeper understanding of the mechanisms of tumorigenesis has facilitated tumor characterization, prognosis and patient stratification, bringing us one step closer towards personalized medicine.
Summary
There are two main pathways of CRC development: (1) chromosomal instability, also known as the classical adenoma-carcinoma sequence, and (2) microsatellite instability, caused by a defective mismatch repair (dMMR) system. Analysis of these pathways has uncovered key prognostic and predictive biomarkers to guide patient selection and treatment strategy. This review summarizes the current treatment regimens and recent advances in the personalized therapy of CRC.
Key Message
Understanding of the mechanisms of CRC pathogenesis has led to new developments in tumor characterization, patient stratification, prognosis and treatment, bringing us closer to personalized therapy.
Practical Implications
In the adjuvant setting, the treatment decision is driven by clinical and histopathological factors. dMMR status is one of the most robust positive prognostic factors in resected colon cancer. More and more guidelines recommend refraining from adjuvant chemotherapy in patients with dMMR. In the metastatic setting, the introduction of effective compounds, including agents that target the epidermal growth factor receptor and vascular endothelial growth factor pathways, has significantly improved survival. The presence of wild-type KRAS and NRAS (all RAS) is a positive predictive factor for epidermal growth factor receptor antibody treatment. Therefore, analysis of all RAS status is recommended for all patients with metastatic disease prior to the initiation of first-line chemotherapy.
Key Words: Adjuvant therapy, Colorectal cancer, Microsatellite instability, Palliative therapy, Personalized therapy
Introduction
Colorectal cancer (CRC) is the second most common cancer type in the Western world, accounting for approximately 450,000 new cases in Europe each year. More than 200,000 patients die of the disease each year, which makes CRC still the second leading cause of cancer death in the Western world [1]. Over the past decade the treatment of CRC has changed markedly, in particular in metastatic disease, mostly through the introduction of combination chemotherapy with targeted agents, leading to more curative resections and also prolonging survival in patients with unresectable disease.
In the past years, a better understanding of the pathogenesis and progression of cancer has led to the identification of distinct cancer subtypes and an increasing number of treatment targets. Thereby, patients can now be better categorized into specific prognostic and predictive groups. Moreover and importantly, more effective drugs could be developed. This improvement in treatment options has been markedly noticed in various cancer types, such as breast cancer as well as non-small-cell lung cancer, where a number of new targeted agents have been recently approved for systemic treatment. In this short review, standard treatments and recent advances in the personalized therapy of CRC will be briefly summarized, focusing on prognostic (independent of treatment) and predictive (treatment effect) biomarkers and approved targeted therapies in the adjuvant as well as the palliative treatment setting.
The Pathogenesis of CRC
CRC develops along distinct pathways involving various genetic and epigenetic alterations [2]. Two major pathways of CRC development are presently known. One, called the classical adenoma-carcinoma sequence, is through chromosomal instability (CIN), and one through microsatellite instability (MSI), which is caused by a defective mismatch repair (dMMR) gene system following the so-called serrated pathway [3]. Beyond the division into these two major pathways, colon cancers are further grouped into five subtypes through their genetic and epigenetic alterations and prognosis (table 1) [3,4]. Important molecular criteria for this classification are chromosomal stability (CIN), CpG island methylator phenotype (CIMP) status, microsatellite instability (MSI, MSI-H, MSI-L, MSS), called dMMR status, as well as alterations (mutations and methylation) in key genes such as APC, KRAS, MLH1, MGMT and BRAF. Most recently the different molecular subgroups of colon cancer have been linked to prognosis and survival in stage III cancer and in a population-based registry [5,6].
Table 1.
Classification of colon cancer subtypes based on genetic and epigenetic alterations (according to [3, 4])
| Type 1 | Type 2 | Type 3 | Type 4 | Type 5 (Lynch syndrome) | |
| MSI status | MSI-H | MSS/MSI-L | MSS/MSI-L | MSS/MSI-L | MSI-H |
| CIMP | + | + | - | - | - |
| Mutations | |||||
| BRAF | mutant | mutant | wild-type | wild-type | wild-type |
| KRAS | wild-type | wild-type | mutant | wild-type | wild-type |
| 5-year survival | |||||
| n | 100 | 55 | 353 | 631 | 50 |
| Overall | 80.5% | 46.2% | 67.8% | 78.0% | 84.1% |
| Disease-specific | 89.5% | 49.2% | 72.4% | 82.5% | 93.1% |
The root of dMMR is either a germline mutation in one of the mismatch repair proteins MLH1, MSH2, MSH6 or PMS2 as in the hereditary Lynch syndrome. Alternatively, a mismatch repair defect is induced by hypermethylation of the promotor region and thus epigenetic inactivation of the MLH1 gene. Hypermethylation of promotor regions of cancer genes generally occurs associated with the CpG island methylator phenotype high [7,8].
In sporadic cancer with MLH1 inactivation one often finds mutations in the BRAF gene at V600E, whereas BRAF mutations are never found in Lynch syndrome. BRAF is a component of the raf kinase family and like KRAS and NRAS a regulator of the epidermal growth factor receptor (EGFR)/MAP kinase/ERK signaling pathway. Between 5 and 10% of colon cancers are mutant for BRAF. BRAF mutations are known to occur early in tumor development. There is a high concordance between primary tumor and metastasis regarding BRAF mutations. BRAF mutations are associated with right-sided tumors, high-grade histology, older age and female sex and more often occur in tumors developing along the so-called serrated pathway of CRC [3]. KRAS exon 2 mutations appear in approximately 40% of CRCs early in tumor development. BRAF and KRAS exon 2 mutations are virtually mutually exclusive.
Adjuvant Therapy in Colon Cancer: Clinical and Molecular Features for Treatment Decision
Through the introduction of screening colonoscopies e.g. in Germany, more and more cancers of the colorectum are detected at earlier stages of cancer development [9]. Since survival in colon cancer is largely dependent on stage, disease-related mortality should thus be gradually declining. Patients with UICC stage I cancers show excellent 5-year overall survival (OS) of >90%. Similarly, patients with UICC stage II cancers without known specific clinical risk factors also survive disease-free in >80% of cases [10]. Such clinical risk factors comprise T4 category, tumor perforation, surgery with complete bowel obstruction and too few lymph nodes examined (understaging, <12). Some also consider G3 grading and vascular (V1) or lymphatic invasion (L1) clinical or histopathological risk factors [11].
After curative resection of colon cancer, adjuvant chemotherapy should be considered mainly depending on stage. While adjuvant therapy is recommended in all patients with stage III disease, adjuvant therapy in stage II disease is more complex. Most guidelines recommend adjuvant chemotherapy with fluoropyrimidines in stage II disease if the above-mentioned clinical and histopathological risk factors are present, leading to a survival benefit of approximately 8-10%. In patients without those risk factors, OS is only improved by 3.6% under 6 months of 5-FU therapy [12]. Thus, large efforts are made to further characterize these approximately 5% of patients who benefit from adjuvant treatment in stage II low-risk cancer. Unfortunately, no predictive biomarkers have so far been identified for this patient group. In contrast, a number of prognostic factors have been found which help to further subdivide this population and guide treatment decisions. These factors comprise single genetic and epigenetic markers, combination of markers as well as gene signatures.
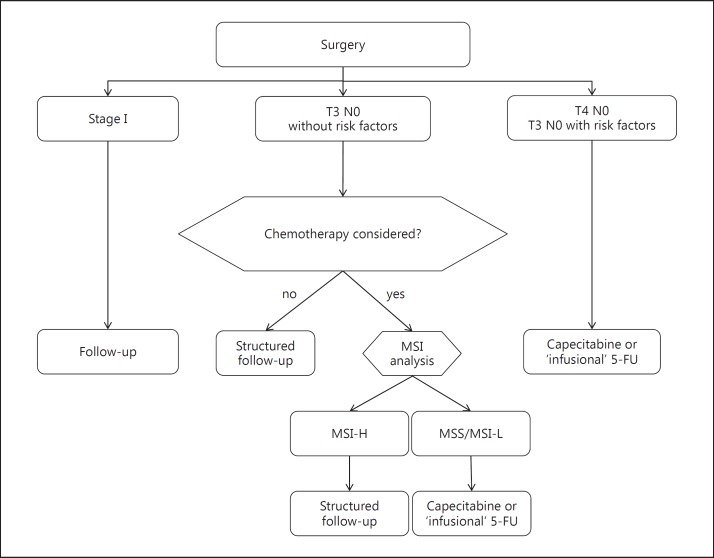
MSI or dMMR are the most clinically relevant molecular markers in stage II colon cancer at present. A large number of studies has identified dMMR as a strong and robust positive prognostic marker, in particular in stage II cancers with hazard ratios (HRs) for survival ranging from 0.3 to 0.46 [13,14,15]. The incidence of dMMR is stage-dependent, with approximately 20% MSI high in stage II, 12% in stage III and <4% in stage IV [16]. Patients with stage II MSI high tumors without clinical risk factors survive 5 years in >90% [17]. While dMMR as a prognostic marker is firmly established, there is no evidence that dMMR is also predictive. In fact, the only prospective randomized study analyzing dMMR as a predictive marker found no difference in chemosensitivity between MSS and MSI high cancers [17]. Moreover, some studies indicate that adjuvant fluoropyrimidine chemotherapy in dMMR cancers is harmful, lowering survival in these patients [18]. Interestingly, in a recent retrospective analysis of the NSABP C-08 trial, patients with dMMR cancers seemed to benefit from the addition of the anti-vascular endothelial growth factor (anti-VEGF) monoclonal antibody bevacizumab while the study was negative for the entire study population [19]. In stage III patients the addition of oxaliplatin is beneficial for both dMMR and proficient mismatch repair cancers [20,21,22]. In conclusion, guidelines more and more integrate the analysis of MSI into the management of resected colon cancer (fig. 1). Thus, in patients with low-risk stage II cancer where adjuvant 5-FU chemotherapy is considered, mismatch repair status should be analyzed. In MSI high cancers with OS >90% the absolute benefit to 5-FU is low. Therefore, follow-up only should be recommended.
Fig. 1.
Treatment algorithm of early colon cancer based on clinical and molecular markers (according to [47]).
Beyond single markers, some have found that the combination of genetic and epigenetic markers seems to improve the prediction of survival in patients with resected colon cancer [e.g. [8]]. There in particular, presence or absence of BRAF mutations separate survival in patients with microsatellite-stable cancers, with BRAF-mutant MSS (proficient mismatch repair) patients having the worst prognosis [23,24]. Most recently, analysis of mismatch repair in combination with mutation detection of KRAS and BRAF and hypermethylation of MLH1 (methylator phenotype) in patients with stage III colon cancer under adjuvant FOLFOX therapy identified significant differences in survival [5]. Thus, the prognosis in this patient population can be better predicted using the combination of these markers. Here again, the analysis did not identify markers indicating benefit from adjuvant therapy (predictive marker).
Lastly, a number of prognostic gene signatures have been identified, such as Oncotype DX or ColoPrint signature, both of which are under intense evaluation. The prognostic score using Oncotype DX can be retrieved from paraffin-embedded tissue [25] while the test ColoPrint uses fresh frozen tissue [26]. The signatures seem especially useful in stage II patients who do not carry a dominant prognostic marker such as T4 or dMMR [25,27]. In addition, Oncotype DX seems useful in stage IIIA and B cancers because it may predict benefit from oxaliplatin in a specific subgroup [28,29]. So far, none of the signatures have been introduced into the standard management of patients with colon cancer.
Systemic Treatment in Advanced Disease: Patient Groups, Targeted Therapy and Relevant Biomarkers
In the past decades survival in patients with metastatic CRC (mCRC) was significantly improved through the introduction of effective systemic therapy and an increase in surgical interventions [e.g. [30]]. Most practice guidelines presently recommend to divide stage IV patients – apart from cases where metastases are primarily resectable – into three clinical groups according to the extent and the dynamics of their disease and the resulting treatment goal [11,31]. Group 1 represents patients with hepatic and/or pulmonary potentially resectable metastases and group 2 patients with non-resectable disease with a high tumor burden, rapid disease progression or tumor-related symptoms. In both groups intensive systemic therapy should be offered if patient comorbidities and age allow such treatment. Especially in potentially resectable metastases, intensive combination therapy may lead to shrinking of lesions and eventually to R0 resectability of metastases. Thereby, more and more patients are being referred to secondary surgery with increase in survival if complete resection is achieved. Group 3 patients comprise patients with never-resectable disease, but lack of symptoms and less aggressive cancers. There, less intensive therapy may be applied [31]. As part of the improvement in systemic therapy in mCRC, monoclonal antibodies directed against the EGFR (cetuximab and panitumumab) or directed against the VEGF (bevacizumab) have been introduced.
Role of the Predictive Markers KRAS and NRAS in Anti-EGFR Antibody Combinations
The antibody cetuximab was tested in the first-line treatment of mCRC in combination with the FOLFIRI regimen within the CRYSTAL study, leading to a small but significant improvement in progression-free survival (PFS) [32]. Through a number of studies the mutational status of codon 12/13 in the KRAS gene was identified as a negative predictive marker for anti-EGFR antibody treatment in mCRC [33,34]. Only patients with a wild-type status in KRAS showed benefit from these antibodies, while in patients with mutant KRAS anti-EGFR treatment was detrimental, especially when combined with FOLFOX. When evaluating the KRAS wild-type exon 2/3 subgroup within the CRYSTAL study, one found a median OS benefit of 3.5 months reaching an OS of 23.5 months (HR = 0.79) [35]. A further retrospective analysis of the PRIME study identified additional exons 3 and 4 in the KRAS gene as well as exons 2, 3 and 4 of the NRAS gene as strong predictive markers for anti-EGFR therapy, in this case panitumumab [36]. The combined analysis of both KRAS and NRAS is called all RAS analysis. While approximately 40% of mCRC patients carry a mutation in KRAS exon 2, the ‘new’ RAS mutations occur in a further 10-12%. In the all RAS wild-type population of the PRIME study, median OS in the panitumumab FOLFOX arm was 26 months compared to 20.2 months in the FOLFOX alone group (HR = 0.78). Importantly, survival in patients with any RAS mutation under anti-EGFR and FOLFOX combinations is shorter than with chemotherapy alone [36]. Therefore, the determination of the all RAS status should be obligatory before initiating systemic treatment with anti-EGFR antibodies in patients with mCRC. In fact, in Europe cetuximab and panitumumab are approved in patients with all RAS wild-type status only. Trials testing anti-EGFR antibodies in the first-line treatment of patients with mCRC are shown in table 2.
Table 2.
| Treatment | PFS, months | OS, months | ORR, % | |
|---|---|---|---|---|
| KRAS wiid-type | ||||
| CRYSTAL (n = 666) [35] | FOLFIRI ± cetuximab | 9.9 vs. 8.4 | 23.5 vs. 20.0 | 57 vs. 40 |
| HR = 0.696 | HR = 0.796 | p (CMH test) < 0.001 | ||
| p (log-rank test) = 0.0012 | p (log-rank test) = 0.0093 | |||
| PRIME (n = 656) [48] | FOLFOX ± panitumumab | 10.0 vs. 8.6 | 23.9 vs. 19.7 | 55 vs. 48 |
| HR = 0.80 | HR = 0.83 | p (stratified log-rank test) = 0.068 | ||
| p (stratified log-rank test) = 0.02 | p (stratified log-rank test) = 0.072 | |||
| OPUS (n = 179) [49] | FOLFOX ± cetuximab | 8.3 vs. 7.2 | 22.8 vs. 18.5 | 57 vs. 34 |
| HR = 0.567 | HR = 0.855 | p (stratified CMH test) = 0.0027 | ||
| p (stratified log-rank test) = 0.0064 | p (stratified log-rank test) = 0.39 | |||
| COIN (n = 729) [50] | XELOX/FOLFOX± cetuximab | 8.6 vs. 8.6 | 17.9 vs. 17.0 | 64 vs. 57 |
| HR = 0.96 | HR = 1.04 | p (log-rank test) = 0.049 | ||
| p (log-rank test) = 0.60 | p (log-rank test) = 0.67 | |||
| NORDIC (n = 126) [51] | FLOX ± cetuximab | 8.7 vs. 7.9 | 22.0 vs. 20.1 | 47 vs. 46 |
| HR = 1.07 | HR = 1.14 | p = 0.89 | ||
| p (log-rank test) = 0.66 | p (log-rank test) = 0.48 | |||
| all RAS wild-type | ||||
| OPUS (n = 78) [52] | FOLFOX ± cetuximab | 12.0 vs. 5.8 | 19.8 vs. 17.8 | 58 vs. 29 |
| HR = 0.53 | HR = 0.94 | p (CMH test) = 0.008 | ||
| p (log-rank test) = 0.062 | p (log-rank test) = 0.80 | |||
| CRYSTAL (n = 367) [53] | FOLFIRI ± cetuximab | 11.4 vs. 8.4 | 28.4 vs. 20.2 | 66 vs. 39 |
| HR = 0.56 | HR = 0.69 | p < 0.000 | ||
| p = 0.0002 | p = 0.0024 | |||
| PRIME (n = 512) [36] | FOLFOX ± panitumumab | 10.1 vs. 7.9 | 26.0 vs. 20.2 | n.a. |
| HR = 0.72 | HR = 0.78 | |||
| p = 0.004 | p = 0.04 | |||
CMH = Cochran-Mantel-Haenszel; n.a. = not assessed.
BRAF, on the other hand, is not a predictive marker for anti-EGFR antibody treatment [36], but a known strong negative prognostic factor in mCRC. Patients with mutant BRAF present with the shortest survival, with a median of <20 months.
The anti-VEGF antibody bevacizumab has been shown to improve survival in combination with chemotherapy compared to chemotherapy alone, while overall response rates (ORRs) are less increased [37,38]. The efficacy of bevacizumab is independent of the mutational status in RAS, so bevacizumab is the preferred antibody combination when RAS mutations are present. Interestingly, for patients where the detrimental BRAF mutation is detected and who are in good condition, the combination of irinotecan, oxaliplatin, 5-FU (FOLFOXIRI) and bevacizumab is considered the most effective combination [39]. In the recent TRIBE study, patients were randomized to either FOLFIRI plus bevacizumab or FOLFOXIRI plus bevacizumab. In the FOLFOXIRI arm, median OS exceeded 30 months in the entire study population (HR = 0.79), with BRAF mutant patients displaying an impressive HR of 0.55. We therefore recommend testing for BRAF in very fit patients with all RAS wild-type mCRC to identify the prognostically unfavorable BRAF V600E mutation.
Head-to-Head Comparison of Bevacizumab and Anti-EGFR Therapy in mCRC
So far two phase III and one phase II trial have directly compared bevacizumab and an anti-EGFR antibody in mCRC patients initially selected for KRAS exon 2 wild-type (table 3). The German FIRE-3 study compared FOLFIRI-cetuximab with FOLFIRI-bevacizumab in almost 600 patients with mCRC [40]. The US American CALGB/SWOG 80405 study compared cetuximab or bevacizumab with either FOLFOX or FOLFIRI chemotherapy backbone in almost 1,200 patients [41]. The primary endpoint of the FIRE-3 study was the ORR, with OS being a secondary endpoint. The ORR was 62% in the cetuximab arm and 58% in the bevacizumab arm (investigator assessment), which was not significantly different. Also, PFS was not different in the two treatment arms (10.0 vs. 10.2 months). In contrast, OS was increased by 3.7 months to 28.7 months in the cetuximab arm compared to the bevacizumab arm (HR = 0.77; p = 0.017). At ESMO 2014 the extended RAS data were presented [42]. 475 samples were analyzed for all RAS (more than 80% of samples), with 75 patients displaying a new RAS mutation (15.8%). Interestingly, in the subpopulation with all RAS wild-type, median OS was 33.1 months in the cetuximab arm versus 25 months in the bevacizumab arm (HR = 0.69). A central review of CT scans revealed a significant increase in overall response in the KRAS exon 2 as well as the all RAS wild-type population. How do we explain the large increase in OS in the cetuximab arm without PFS being different? Cetuximab may cause a deeper response with more shrinkage of lesions than bevacizumab, potentially leading to a longer time interval before lethal tumor load occurs. Anti-EGFR treatment also augments early tumor shrinkage in patients with mCRC, which may be used as another on-treatment marker in patients with wild-type tumors.
Table 3.
Randomized trials comparing anti-EGFR and anti-VEGF antibody treatment in mCRC [40 – 43, 54, 55]
| Treatment | Gene status | PFS, months | OS, months | ORR, % | |
|---|---|---|---|---|---|
| FIRE-3 (n= 178) [54] | FOLFIRI + cetuximab | KRAS mutant | 7.5 vs. 10.1 | 20.3 vs. 20.6 | 38 vs. 51 |
| vs. | p = 0.085 | HR = 1.09 | p = 0.097 | ||
| FOLFIRI + bevacizumab | p = 0.60 | ||||
| FIRE-3 (n= 592) [40] | FOLFIRI + cetuximab | KRAS wild-type | 10.0 vs. 10.3 | 28.7 vs. 25.0 | 62 vs. 58 |
| vs. | p = 0.55 | HR = 0.77 | p = 0.183 | ||
| FOLFIRI + bevacizumab | p = 0.017 | ||||
| FIRE-3 (n = 400) [42] | FOLFIRI + cetuximab | all RAS wild-type | 10.3 vs. 10.2 | 33.1 vs. 25.0 | 66 vs. 58 |
| vs. | p = 0.77 | HR = 0.697 | p = 0.92 | ||
| FOLFIRI + bevacizumab | p = 0.0059 | ||||
| CALGB (n = 1,137) [41] | FOLFOX/FOLFIRI + cetuximab | KRAS wild-type | 10.4 vs. 10.8 | 29.9 vs. 29.0 | 66 vs. 57 |
| vs. | codons 12/13 | HR = 1.04 | HR = 0.925 | p = 0.02 | |
| FOLFOX/FOLFIRI + bevacizumab | p = 0.55 | p = 0.34 | |||
| CALGB (n = 526) [43] | FOLFOX/FOLFIRI + cetuximab | all RAS wild-type | 11.4 vs. 11.3 | 32.0 vs. 31.2 | 69 vs. 54 |
| vs. | HR = 1.1 | HR = 0.9 | p < 0.01 | ||
| FOLFOX/FOLFIRI + bevacizumab | p = 0.31 | p = 0.40 | |||
| PEAK [55] | FOLFOX + panitumumab | KRAS wild-type | 10.9 vs. 10.1 | 34.2 vs. 24.3 | 58 vs. 54 |
| vs. | exon 2 | HR = 0.87 | HR = 0.62 | p = n.s. | |
| FOLFOX + bevacizumab | p = 0.353 | p = 0.009 | |||
| all RAS wild-type | HR = 0.65 | 41.3 vs. 28.9 | |||
| p = 0.029 | HR = 0.63 | ||||
| p = 0.058 | |||||
n.s. = Not significant.
The US American CALGB/SWOG 80405 study used median OS as the primary endpoint [41,43]. Most patients received FOLFOX as chemotherapy backbone (74%), 26% received FOLFIRI. Within the entire study population (n = 1,137), OS was the same in the cetuximab and the bevacizumab arm (29.9 vs. 19.0 months, HR = 0.92; p = 0.34). Furthermore, under both chemotherapy regimens separately no difference in survival was found (HR = 0.9 for FOLFOX, HR = 1.2 for FOLFIRI) [41]. At ESMO 2014 the all RAS population was presented although the group could retrieve <60% of tissue samples for analysis only (n = 670). 256 patients in the bevacizumab arm and 270 patients in the cetuximab arm were all RAS wild-type. Objective response rates (investigator-assessed) were 69% under cetuximab and 54% under bevacizumab. OS under FOLFOX as chemotherapy or FOLFIRI was not significantly different between the arms, with high OS times of 29-35.2 months [43]. The reasons for the differences in results of these two large phase III studies are not completely clear. Data for the CALGB trial are still considered preliminary because information about second-line therapy and dosing is missing.
At present, we would recommend to test the biomarker all RAS (KRAS exon 2, 3 and 4 and NRAS exon 2, 3 and 4) in all patients with mCRC before initiation of first-line chemotherapy. In all RAS wild-type patients, one should consider offering anti-EGFR antibody treatment if the patient qualifies for combination chemotherapy. As shown in the FIRE-3 study, OS is increased and also early responses and especially the deepness of response are significantly improved under cetuximab combinations compared to combination with bevacizumab.
Future Developments
A number of studies have recently tried to further characterize patients with CRC, aiming at a further individualization of treatment and the identification of new treatment targets.
In a recent comprehensive analysis of more than 1,200 patients, Sadanandam et al. [44] used unsupervised clustering of gene expression profiles to group patients into distinct cancer subtypes. They there found six specific subgroups of cancer types, which interestingly responded differently to cetuximab and/or chemotherapy with irinotecan. The specific subtypes shared properties of normal epithelial cells of the non-transformed colon crypt with more or less stem cell properties. These distinct subtypes may help to further individualize therapy for patients with CRC in the adjuvant or metastatic setting.
In another comprehensive characterization of human colon and rectal cancer published in Nature 2012 [45], whole-genome sequencing of 276 patients with CRC was performed by exome sequencing, DNA copy number analysis, promotor methylation and messenger and micro RNA expression. 16% of cancers were hypermutated, 75% of them were MSI-H. Expected mutations found were APC, TP53, SMAD4, PIK3CA and KRAS. Some unexpected mutations found were ARID1A, SOX9, FAM123B and ERBB2 [45]. Thereby, new targets may be identified for treatment.
At present, a number of trials have been initiated also testing combinations of targeted drugs, for example for the unfavorable BRAF mutation [46]. There, BRAF inhibitors are combined with anti-EGFR antibodies and MEK or Pi3K inhibitors. First activity has been demonstrated.
Importantly, with further subdivision of patient populations into smaller and smaller groups according to the mutational status of certain genes, we need to combine efforts to screen a large number of patients for all sorts of mutations simultaneously. This allows us to identify suitable patients for our clinical trials more quickly and efficiently. To this end, screening platform studies such as the EORTC platform SPECTAcolor (PI: G. Folprecht, Dresden) have been initiated and should be supported.
Disclosure Statement
A. Reinacher-Schick: honoraria from Amgen, Celgene, Roche, Pfizer and Sanofi-Aventis; member of advisory boards of Amgen, Celgene, Roche, Pfizer and Merck; studies and research projects supported by Roche and Sanofi-Aventis.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 4.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 5.Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, Bot BM, Tejpar S, Delorenzi M, Goldberg RM, Mahoney M, Sargent DJ, Alberts SR. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty C, Buchanan DD, Potter JD, Newcomb PA. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87. doi: 10.1053/j.gastro.2014.09.038. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinicrope FA, Sargent DJ. Molecular pathways: microsatellite instability in colorectal cancer: prognostic, predictive, and therapeutic implications. Clin Cancer Res. 2012;18:1506–1512. doi: 10.1158/1078-0432.CCR-11-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Jeffrey A, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pox CP, Altenhofen L, Brenner H, Theilmeier A, Von Stillfried D, Schmiegel W. Efficacy of a nationwide screening colonoscopy program for colorectal cancer. Gastroenterology. 2012;142:1460–1467. doi: 10.1053/j.gastro.2012.03.022. e2. [DOI] [PubMed] [Google Scholar]
- 10.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 11.Pox C, Aretz S, Bischoff SC, Graeven U, Hass M, Heußner P, Hohenberger W, Holstege A, Hübner J, Kolligs F, Kreis M, Lux P, Ockenga J, Porschen R, Post S, Rahner N, Reinacher-Schick A, Riemann JF, Sauer R, Sieg A, Scheppach W, Schmitt W, Schmoll HJ, Schulmann K, Tannapfel A, Schmiegel W, Leitlinienprogramm Onkologie der AWMF; Deutschen Krebsgesellschaft e. V; Deutschen Krebshilfe e. V. S3-Leitlinie kolorektales Karzinom Version 1.0 – Juni 2013 AWMF Registernummer: 021/007OL. Z Gastroenterol. 2013;51:753–854. doi: 10.1055/s-0033-1350264. [DOI] [PubMed] [Google Scholar]
- 12.Gray R, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 13.Sargent DJ, Marsoni S, Thibodeau SN, Labianca R, Hamilton SR, Torri V, Monges G, Ribic C, Grothey A, Gallinger S. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): a pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol. 2008;26(suppl):4008. [Google Scholar]
- 14.Ribic C, Sargent D, Moore M, Thibodeau SN, French A, Goldberg R, Hamilton S, Laurent-Puig P, Gryfe R, Shepherd L, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth AD, Delorenzi M, Tejpar S, Yan P, Klingbiel D, Fiocca R, d'Ario G, Cisar L, Labianca R, Cunningham D, Nordlinger B, Bosman F, Van Cutsem E. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst. 2012;104:1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- 16.Klingbiel D, Saridaki Z, Roth AD, Bosman FT, Delorenzi M, Tejpar S. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann Oncol. 2015;26:126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 17.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, Gray R, Quirke P. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 18.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pogue-Geile K, Yothers G, Taniyama Y, Tanaka N, Gavin P, Colangelo L, Blackmon N, Lipchik C, Kim SR, Sharif S, Allegra C, Petrelli N, O'Connell MJ, Wolmark N, Paik S. Defective mismatch repair and benefit from bevacizumab for colon cancer: findings from NSABP C-08. J Natl Cancer Inst. 2013;105:989–992. doi: 10.1093/jnci/djt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flejou JF, André T, Chibaudel B, Scriva A, Hickish T, Tabernero J, Van Laethem JL, Banzi M, Maartense E, Shani A, Carlsson G, Scheithauer W, Papamichael D, Moehler M, Landolfi S, Demetter P, Duval A, Lee M, Colote S, De Gramont A. Effect of adding oxaliplatin to adjuvant 5-fluorouracil/leucovorin (5-FU/LV) in patients with defective mismatch repair (dMMR) colon cancer stage II and III included in the MOSAIC study. J Clin Oncol. 2013;31(suppl) abstr 3524. [Google Scholar]
- 21.Sargent DJ, Shi Q, Yothers G, Tejpar S, Bertagnolli MM, Thibodeau SN, Andre T, Labianca R, Gallinger S, Hamilton SR, Monges G, Pogue-Geile KL, Paik S, Klingbiel D, Roth A, Pavey ES, Kim GP, Sinicrope FA. Prognostic impact of deficient mismatch repair (dMMR) in 7,803 stage II/III colon cancer (CC) patients (pts): a pooled individual pt data analysis of 17 adjuvant trials in the ACCENT database. J Clin Oncol. 2014;32(suppl) abstr 3507. [Google Scholar]
- 22.Tougeron D, Sickersen G, Lecomte T, Mouillet G, Trouilloud I, Coriat R, Aparicio T, Des Guetz G, Lecaille C, Artru P, Cauchin E, Sefrioui D, Boussaha T, Ferru A, Taïeb J, Michel P, Karayan-Tapon L, Vernerey D, Bonnetain F, Zaanan A. Impact of adjuvant chemotherapy with 5-FU or FOLFOX in colon cancers with microsatellite instability: an AGEO multicenter study. J Clin Oncol. 2014;32(suppl) abstr 3508. [Google Scholar]
- 23.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, Fuchs CS, Ogino S. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin PG, Colangelo LH, Fumagalli D, Tanaka N, Remillard MY, Yothers G, Kim C, Taniyama Y, Kim S II, Choi HJ, Blackmon NL, Lipchik C, Petrelli NJ, O'Connell MJ, Wolmark N, Paik S, Pogue-Geile KL. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18:6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, Watson D, Baehner FL, Shak S, Baker J, Cowens JW, Wolmark N. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopetz S, Tabernero J, Rosenberg R, Jiang ZQ, Moreno V, Bachleitner-Hofmann T, Lanza G, Stork-Sloots L, Maru D, Simon I, Capellà G, Salazar R. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist. 2015;20:127–133. doi: 10.1634/theoncologist.2014-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr D, Gray R, Quirke P, Watson D, Yothers G, Lavery IC, Lee M, O'Connell MJ, Shak S, Wolmark N, Genomic Health and QUASAR Colon Teams A quantitative multigene RT-PCR assay for prediction of recurrence in stage II colon cancer: selection of the genes in four large studies and results of the independent, prospectively designed QUASAR validation study. J Clin Oncol. 2009;27(suppl) abstr 4000. [Google Scholar]
- 28.O'Connell M, Lee M, Lopatin M, Yothers G, Kim CL, Millward C, Paik S, Sharif S, Shak S, Wolmark N. Validation of the 12-gene colon cancer recurrence score (RS) in NSABP C07 as a predictor of recurrence in stage II and III colon cancer patients treated with 5-FU/LV (FU) and 5-FU/LV+oxaliplatin (FU+Ox) J Clin Oncol. 2012;30(suppl) doi: 10.1200/JCO.2012.47.3116. abstr 3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yothers G, O'Connell MJ, Lee M, Lopatin M, Clark-Langone KM, Millward C, Paik S, Sharif S, Shak S, Wolmark N. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31:4512–4519. doi: 10.1200/JCO.2012.47.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM, McWilliams RR. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, ESMO Guidelines Working Group Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 2):iii1–iii9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 32.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chien CRC, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 33.Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 34.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 35.Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 36.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 37.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 38.Saltz LB, Clarke S, Díaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, Couture F, Sirzén F, Cassidy J. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol. 2008;26:2013–2019. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- 39.Falcone A, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, Trenta P, Tomasello G, Ronzoni M, Ciuffreda L, Zaniboni A, Tonini G, Buonadonna A, Valsuani C, Chiara S, Carlomagno C, Boni C, Marcucci L, Boni L, Loupakis F. FOLFOXIRI/bevacizumab (bev) versus FOLFIRI/bev as first-line treatment in unresectable metastatic colorectal cancer (mCRC) patients (pts): results of the phase III TRIBE trial by GONO group. J Clin Oncol. 2013;31(suppl) abstr 3505. [Google Scholar]
- 40.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Müller S, Link H, Niederle N, Rost A, Höffkes HG, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 41.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Mahoney MR, O'Neil BH, Shaw JE, Polite BN, Hochster HS, Atkins JN, Goldberg RM, Mayer RJ, Schilsky RL, Bertagnolli MM, Blanke CD. CALGB/SWOG 80405: phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC) J Clin Oncol. 2014;32(suppl) abstr LBA3. [Google Scholar]
- 42.Stintzing S, Modest DP, Fischer von Weikersthal L, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran S, Heintges T, Lerchenmueller C, Kahl C, Seipelt G, Kullmann F, Scheithauer W, Held S, Giessen C, Moehler M, Jagenburg A, Jung A, Kirchner T, Heinemann V. Independent radiological evaluation of objective response, early tumor shrinkage, and depth of response in FIRE-3 (AIO KRK-0306) in the final RAS evaluable population. Ann Oncol. 2014;25(suppl 2):ii117. [Google Scholar]
- 43.Lenz H, Niedzwiecki D, Innocenti F, Blanke C, Mahony MR, O'Neil BH, Shaw JE, Polite B, Hochster H, Atkins J, Goldberg R, Mayer R, Schilsky RL, Bertagnolli M, Venook A. CALGB/SWOG 80405: phase III trial of irinotecan/ 5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (MFOLFOX6) with bevacizumab (bv) or cetuximab (cet) for patients (pts) with expanded RAS analyses untreated metastatic adenocarcinoma of the colon or rectum (MCRC) Ann Oncol. 2014;25(suppl 4) [Google Scholar]
- 44.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, Lhermitte B, Olshen AB, Wiedenmann B, Cantley LC, Gray JW, Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kucherlapati R. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong DS, Morris VK, Fu S, Overman MJ, Piha-Paul SA, Kee BK, Zinner R, Fogelman DR, Mistry R, Shureiqi I, Meric-Bernstam F, Kopetz S. Phase 1B study of vemurafenib in combination with irinotecan and cetuximab in patients with BRAF-mutated advanced cancers and metastatic colorectal cancer. J Clin Oncol. 2014;32(suppl) abstr 3516. [Google Scholar]
- 47.Al-Batran SE, Hofheinz RD. Therapiealgorithmen Onkologie 2014. ed 2. Regensburg: RS Media GmbH; 2014. p. 324. [Google Scholar]
- 48.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 49.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 50.Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL, Kay E, Mitchell JK, Madi A, Jasani B, James MD, Bridgewater J, Kennedy MJ, Claes B, Lambrechts D, Kaplan R, Cheadle JP. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tveit KM, Guren T, Glimelius B, Pfeiffer P, Sorbye H, Pyrhonen S, Sigurdsson F, Kure E, Ikdahl T, Skovlund E, Fokstuen T, Hansen F, Hofsli E, Birkemeyer E, Johnsson A, Starkhammar H, Yilmaz MK, Keldsen N, Erdal AB, Dajani O, Dahl O, Christoffersen T. Phase III trial of cetuximab with continuous or intermittent fluorouracil, leucovorin, and oxaliplatin (Nordic FLOX) versus FLOX alone in first-line treatment of metastatic colorectal cancer: the NORDIC-VII study. J Clin Oncol. 2012;30:1755–1762. doi: 10.1200/JCO.2011.38.0915. [DOI] [PubMed] [Google Scholar]
- 52.Bokemeyer C, Kohne CH, Ciardiello F, Lenz HJ, Heinemann V, Klinkhardt U, Beier F, Duecker K, Tejpar S. Treatment outcome according to tumor RAS mutation status in OPUS study patients with metastatic colorectal cancer (mCRC) randomized to FOLFOX4 with/without cetuximab. J Clin Oncol. 2014;32(suppl) abstr 3505. [Google Scholar]
- 53.Ciardiello F, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, Beier F, Stroh C, Van Cutsem E. Treatment outcome according to tumor RAS mutation status in CRYSTAL study patients with metastatic colorectal cancer (mCRC) randomized to FOLFIRI with/without cetuximab. J Clin Oncol. 2014;32(suppl) abstr 3506. [Google Scholar]
- 54.Stintzing S, Modest DP, Fischer von Weikersthal L, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran S, Heintges T, Lerchenmueller C, Kahl C, Seipelt G, Kullmann F, Scheithauer W, Held S, Giessen C, Moehler M, Jagenburg A, Jung A, Kirchner T, Heinemann V. Analysis of KRAS/NRAS and BRAF mutations in FIRE-3: a randomized phase III study of FOLFIRI plus cetuximab or bevacizumab as first-line treatment for wild-type (WT) KRAS (exon 2) metastatic colorectal cancer (mCRC) patients. Eur J Cancer. 2013;49(suppl 3) [Google Scholar]
- 55.Schwartzberg LS, Rivera F, Karthaus M, Fasola G, Canon JL, Hecht JR, Yu H, Oliner KS, Go WY. PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240–2247. doi: 10.1200/JCO.2013.53.2473. [DOI] [PubMed] [Google Scholar]