Inflammation is an essential defense strategy mounted by the innate immune system to eradicate infections and repair tissue damage. The inflammasome is an intracellular signaling complex involved in inflammation initiation and perpetuation. These multimeric protein assemblies promote the activation of proteases and the maturation of proinflammatory cytokines, as well as a form of cell death (pyroptosis) that incites further inflammation (1). Excessive inflammasome activation has been implicated in chronic inflammatory disorders such as diabetes, atherosclerosis, multiple sclerosis, and Alzheimer’s disease (2). However, inflammasomes also play protective roles in response to microbial pathogens. On pages 404 and 399 of this issue, Zhang et al. (3) and Hu et al. (4) report the structural basis for activation of an inflammasome implicated in both infectious disease response and autoinflammatory disorders (see the first figure).
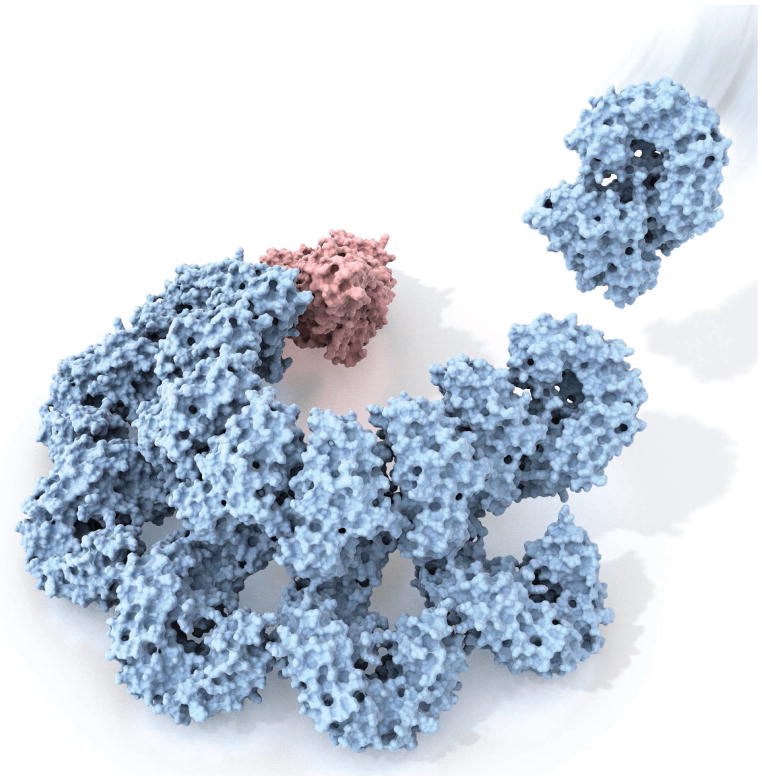
Figure 1. Getting ready for defense.
Reconstruction of the assembling inflammasome complex reported by Hu et al. and Zhang et al. Red, NAIP2 protein; cyan, NLRC4 proteins.
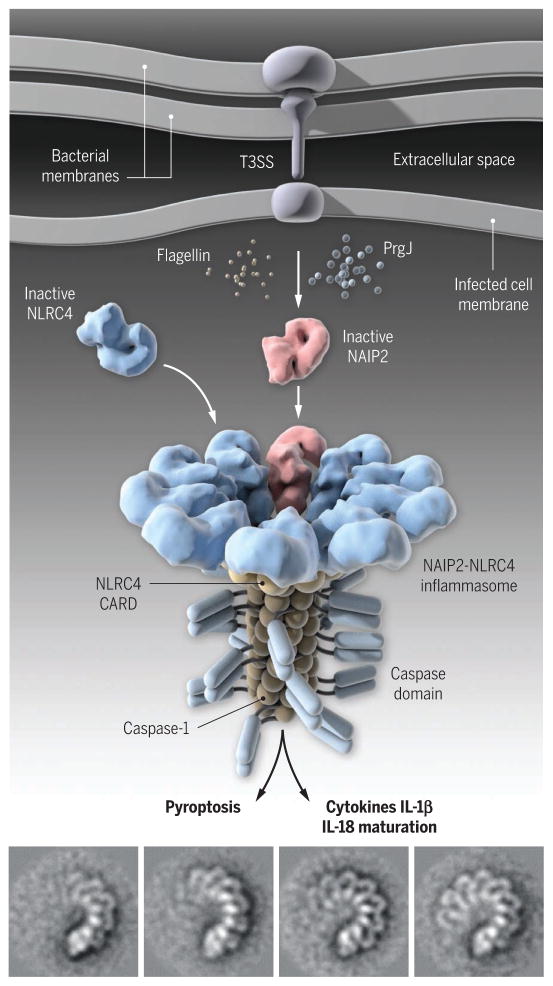
Several inflammasomes have been shown to be activated by both microbial and host stimuli. Among these inflammasomes, NLRC4 [where NLR denotes NOD-like receptor and C denotes caspase activation and recruitment domain (CARD)] is a unique inflammasome that uses host NAIP proteins (NLR family apoptosis inhibitory proteins) to recognize two bacterial proteins, flagellin and PrgJ, which are essential for bacterial movement and pathogenesis (see the second figure) (5, 6). Previous work suggested that the flagellin-activated NLRC4 inflammasome adopts disk-like structures containing 11 or 12 protomers (7), but the details of the inflammasome assembly are lacking.
Figure 2. Add just one NAIP.
Bacterial infections activate the NAIP2-NLRC4 inflammasomes, which promote the activation of caspase-1 and maturation of proinflammatory cytokines interleukin-1β (IL-1β) and IL-18, as well as pyroptotic cell death. Hu et al. and Zhang et al. show that a single ligand-bound NAIP2 protein triggers the progressive assembly of the NAIP2-NLRC4 inflammasomes through large conformational changes in NLRC4 and perhaps NAIP2. Similar mechanisms could govern the assembly of the NLRP3 inflammasome. (Bottom) Cryo-EM images of the assembling inflammasome.
Using cryo–electron microscopy (cryo-EM) methods, Hu et al. and Zhang et al. now show that just one PrgJ-bound NAIP2 molecule is sufficient to trigger a dramatic conformational change in NLRC4; 9 to 11 activated NLRC4 molecules then oligomerize around one NAIP2 molecule to form a 10- to 12-spoke wheel-like structure (see the second figure). Nano-gold labeling demonstrated that there was only one NAIP2 and one PrgJ molecule in the inflammasome wheel (see the second figure). Unlike the assembly of the Apaf-1 (apoptotic protease activating factor–1) apoptosome, which requires ligand binding by each of its seven protomers (8), a single ligand-bound NAIP2 molecule triggers the assembly of an entire NAIP2-NLRC4 inflammasome complex. This complex then oligomerizes the caspase-1 protein and promotes its activation (see the second figure). Comparison of the cryo-EM structures and a previous crystal structure of NLRC4 in an autoinhibited state reveals striking conformational changes in NLRC4. In particular, NLRC4’s C-terminal half pivots on a helix within the NOD (nucleotide-binding oligomerization) domain, leading to a ~90° rotation. This large structural reorganization is necessary to expose two oligomerization surfaces that facilitate the protein’s progressive oligomerization. Mutations near the NLRC4 hinge region cause severe autoinflammatory diseases typified by recurrent fever and pyroptotic cell death (9–11). The two new structural studies (3, 4) reveal the molecular mechanisms for this autoinflammatory pathology involving NLRC4.
Notwithstanding the remarkable new findings, many mysteries remain regarding inflammasome activation. A key question is the mode of ligand binding. Tenthorey et al. have shown that the NAIP NOD domains confer the capacity to recognize flagellin or T3SS proteins (12). The resolution of the current cryo-EM structures was too low to identify the ligand-bound NAIP2 molecule within the wheel-like structures. Therefore, the structural basis for ligand recognition by the NOD modules remains to be deciphered. Because the NAIP2 NOD module also recruits NLRC4, it is possible that the ligand may engage both NAIPs and NLRC4 in the fully assembled inflammasomes. Therefore, NLRC4 may function as a co-receptor instead of an adapter.
Another unsolved mystery is the role of adenosine diphosphate (ADP) or adenosine tri-phosphate (ATP). The assembly of the apoptosome requires the exchange of ADP for ATP (8). Halff et al. have shown that ATP binding by NAIP5 was not essential for inflammasome assembly, but the role of ATP for NLRC4 function was not clear (7). It remains to be shown whether exchange of ADP for ATP is necessary for inflammasome assembly or disassembly or whether, as suggested by Zhang et al., release of ADP suffices.
The results presented by Hu et al. and Zhang et al. may also help in understanding the NLRP3 inflammasome, which is perhaps the most intensively studied because it is activated by various microbial and environmental stimuli. Because of their similar domain architecture, the NLRP3 inflammasome may adopt quaternary structures resembling the NAIP2-NLRC4 inflammasome. Many chemically distinct stimuli for the NLRP3 inflammasome may converge on as yet unidentified host protein(s), which then trigger the assembly of the NLRP3 inflammasome in a fashion similar to how NAIP2 accomplishes the NLRC4 oligomerization.
Acknowledgments
In memory of William E. Paul, a mentor, colleague and friend to T.S.X.
REFERENCES AND NOTES
- 1.Schroder K, Tschopp J. Cell. 2010;140:821. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Guo H, Callaway JB, Ting JPY. Nat Med. 2015;21:677. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, et al. Science. 2015;350:404. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Z, et al. Science. 2015;350:399. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 5.Kofoed EM, Vance RE. Nature. 2011;477:592. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Nature. 2011;477:596. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 7.Halff EF, et al. J Biol Chem. 2012;287:38460. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai J, Shi Y. Natl Sci Rev. 2014;1:101. [Google Scholar]
- 9.Canna SW, et al. Nat Genet. 2014;46:1140. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romberg N, et al. Nat Genet. 2014;46:1135. doi: 10.1038/ng.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura A, Sasaki Y, Abe T, Kano H, Yasutomo K. J Exp Med. 2014;211:2385. doi: 10.1084/jem.20141091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Mol Cell. 2014;54:17. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]