Abstract
Objectives
Mutator strains play an important role in the emergence of antibiotic-resistant bacteria. Campylobacter jejuni is a leading cause of foodborne illnesses worldwide and is increasingly resistant to clinically important antibiotics. The objective of this study was to identify the genetic basis that contributes to a mutator phenotype in Campylobacter and determine the role of this phenotype in the development of antibiotic resistance.
Methods
A C. jejuni isolate (named CMT) showing a mutator phenotype was subjected to WGS analysis. Comparative genomics, site-specific reversion and mutation, and gene knockout were conducted to prove the mutator effect was caused by a single nucleotide change in the mutY gene of C. jejuni.
Results
The C. jejuni CMT isolate showed ∼100-fold higher mutation frequency to ciprofloxacin than the WT strain. Under selection by ciprofloxacin, fluoroquinolone-resistant mutants emerged readily from the CMT isolate. WGS identified a single nucleotide change (G595→T) in the mutY gene of the CMT isolate. Further experiments using defined mutant constructs proved its specific role in elevating mutation frequencies. The mutY point mutation also led to an ∼700-fold increase in the emergence of ampicillin-resistant mutants, indicating its broader impact on antibiotic resistance. Structural modelling suggested the G595→T mutation probably affects the catalytic domain of MutY and consequently abolishes the anti-mutator function of this DNA repair protein.
Conclusions
The G595→T mutation in mutY abolishes its anti-mutator function and confers a mutator phenotype in Campylobacter, promoting the emergence of antibiotic-resistant Campylobacter.
Introduction
Campylobacter jejuni, a Gram-negative microaerophilic bacterium, is one of the most prevalent bacterial foodborne pathogens causing gastroenteritis in humans.1 Infection with C. jejuni is also associated with the development of a paralysing neuropathy, the Guillain–Barré syndrome.2 The main sources of human Campylobacter infections are poultry meat, water and milk contaminated by Campylobacter.3,4
Fluoroquinolone (FQ) antimicrobials are often prescribed for clinical treatment of diarrhoea caused by enteric bacterial pathogens, including Campylobacter.5,6 However, Campylobacter resistance to FQ antimicrobials is increasing,7–9 and some countries have even reported 98%–100% FQ resistance (FQR) rates among Campylobacter isolates originating from broiler chickens.10,11 In a recent report from the US CDC,12 drug-resistant Campylobacter is listed as a serious threat to public health.
The main targets of FQs in bacteria are DNA gyrase (GyrA) and/or topoisomerase IV.13 However, Campylobacter species lack topoisomerase IV, and point mutations in the quinolone resistance-determining region (QRDR) of the gyrA gene confer resistance to FQ antimicrobials. For example, the Thr-86→Ile amino acid modification in GyrA is frequently associated with high-level resistance to FQs,14,15 while other amino acid substitutions in the QRDR region are linked to low to intermediate levels of resistance.16 In addition to the point mutations, the chromosomally encoded and constitutively expressed multidrug efflux pump CmeABC is also a key player in FQR. This was shown by gene-specific inactivation of the cmeABC operon, which led to significant reductions in FQ MICs even in mutant strains that harboured resistance-conferring mutations in GyrA.17–21 Plasmid-mediated FQR genes, such as qnr, aac(6′)-Ibcr and qepA, which have been discovered in other Gram-negative bacteria, have not been reported in Campylobacter.22 Thus, GyrA mutations and CmeABC are synergistically responsible for conferring FQR in Campylobacter.
In addition to contributing to FQR, point mutations mediate resistance to other antimicrobials in Campylobacter, such as macrolides and β-lactams. Macrolide resistance is mainly mediated by a single nucleotide transition (A→G) or transversion (A→C) at position 2058 or 2059 (Escherichia coli coordinates) of 23S RNA.8,23 Recently, a G→T transversion in the promoter region of blaOXA-61, which encodes the sole β-lactamase gene in C. jejuni, has been linked to high-level ampicillin resistance (AmpR) by restoring the expression of blaOXA-61.24
Bacterial strains exhibiting elevated mutation frequencies are mutators, and the mutator phenotype facilitates bacterial adaptation to environmental changes.25 Mutators also play an important role in the emergence of antibiotic resistance in the clinical setting.26 Mutators have also been reported among natural isolates of different bacterial species, including E. coli, Salmonella enterica,27 Helicobacter pylori28 and Streptococcus pneumoniae.29 The majority of naturally occurring mutators are related to defects in the methyl-directed mismatch repair (MMR) system and the nucleotide excision repair (NER) system, such as mutations in mutS, mutH, mutL and uvrD.26 In E. coli, the MMR system, including MutS, repairs replication errors that arise from misincorporations (mismatches) and strand slippage (frameshift errors). In addition, the MMR system inhibits recombination between homologous sequences.30 Epsilonproteobacteria, including Campylobacter and Helicobacter, do not have most of the MMR genes, except for a mutS homologue in both species. However, inactivation of mutS in H. pylori did not confer a mutator phenotype and there is no evidence that MutS mutations are present in naturally occurring mutators of H. pylori.28 Gaasbeek et al.31 characterized several putative DNA repair genes in Campylobacter and found that none of the investigated genes (including recA, uvrA, uvrC, mutS and ung) altered the spontaneous FQR mutation frequency. However, mutation of ung was found to be associated with increased mutation frequency in H. pylori.32 A previous study documented high variability (up to 700-fold) in FQR mutation frequencies among C. jejuni and Campylobacter coli strains,16 suggesting mutator strains may exist among Campylobacter isolates. However, the genetic basis conferring the mutator status in Campylobacter is still unknown.
In this study, we identified a variant (named CMT in this study) of C. jejuni NCTC 11168,33 which demonstrated a spontaneous FQR mutation frequency >100-fold higher than that of 11168 and other C. jejuni isolates. This finding prompted us to examine the mutator phenotype by utilizing WGS, comparative genomics analysis and other genetic methods. A single nucleotide change (G595→T transversion) was identified in the putative base excision repair (BER) gene mutY and was found to be responsible for the elevated frequency of mutation to FQR resistance in Campylobacter. Additionally, the G595→T mutation in mutY also significantly increases the emergence of spontaneous AmpR mutants in C. jejuni. To our knowledge, this is the first identified genetic factor conferring the naturally occurring mutator phenotype in Campylobacter, promoting the emergence of antibiotic resistance mutants.
Materials and methods
Bacterial strains and growth conditions
The CMT isolate is a natural variant of C. jejuni 11168 identified in our laboratory. All of the C. jejuni strains used in this study are listed in Table 1. C. jejuni was cultured using Mueller–Hinton (MH) broth or agar (Difco) at 42°C under microaerobic conditions (5% O2, 10% CO2 and 85% N2). E. coli was grown on LB agar or in LB broth at 37°C for 24 h under aerobic conditions. When needed, culture media were supplemented with ciprofloxacin (1 mg/L), ampicillin (100 mg/L), kanamycin (30 mg/L) or chloramphenicol (10 mg/L).
Table 1.
C. jejuni strains used in this study
| Bacterial strain | Genotype or phenotype | Source |
|---|---|---|
| NCTC 11168 | C. jejuni isolate | 33 |
| W7 | C. jejuni isolate | 42 |
| IA3902 | C. jejuni isolate | 41 |
| CMT | a naturally occurring variant of C. jejuni NCTC 11168 | this study |
| CMTW199G::Km | CMT derivative; cj1621::KmR, 199W→G reversion in MutY | this study |
| CMT::Km | CMT derivative; cj1621::KmR | this study |
| 11168G199W::Km | NCTC 11168 derivative; cj1621::KmR, 199G→W mutation in MutY | this study |
| 11168::Km | NCTC 11168 derivative; cj1621::KmR | this study |
| 11168 mutY::cat | NCTC 11168 derivative; mutY::cat insertional mutation | this study |
| CMT mutY::cat | CMT derivative; mutY::cat insertional mutation | this study |
Ciprofloxacin susceptibility testing
The susceptibilities of C. jejuni strains to ciprofloxacin (Fisher Bioreagents) were determined by a standard microtitre broth dilution method with an inoculum of 106 cfu/mL as described previously.19 MICs were determined as the lowest concentration of ciprofloxacin showing complete inhibition of bacterial growth after 24 or 48 h of incubation at 42°C.
C. jejuni survival assay under ciprofloxacin treatment in vitro
In order to test the ability of C. jejuni cells to generate FQR mutants under FQ selective pressure, different strains were tested for their survival under treatment with a lethal concentration of ciprofloxacin. The assay was carried out in 96-well plates. Overnight cultures on MH agar plates were resuspended in fresh MH broth and adjusted to an inoculum of 106 cfu/mL. Ciprofloxacin was added at a final concentration of 1 mg/L (∼10–20 × MIC). Aliquots of 200 μL inoculum (∼105 cfu) with ciprofloxacin were added to each well. The plates were then incubated at 42°C for 48 h in the incubator under microaerobic conditions. Triplicate samples were taken at 0, 6, 12, 24, 36 and 48 h for viable counts by dropping 10 μL of different dilutions onto non-selective MH agar plates. Then, 50 colonies (if any) were randomly picked from the counting plates at each timepoint, and streaked onto MH plates containing 1 mg/L ciprofloxacin and checked for growth after overnight incubation to confirm the susceptibility of C. jejuni cells in the culture to ciprofloxacin at designated timepoints. To ensure that the inoculum was free of pre-existing resistant mutants, three 200 μL precultures were plated on selection plates. The test results were used only if no FQR mutants were present in these 200 μL precultures.
Assay for spontaneous FQR and AmpR mutation frequencies
To determine spontaneous mutation frequencies for FQR or AmpR in Campylobacter, the methods of Bjorkholm et al.28 and Hanninen et al.16 with minor modifications were applied. Briefly, for each C. jejuni isolate 30 μL (∼105 cfu) overnight cultures were distributed into 20 tubes containing 3 mL of MH broth. Cultures were shaken for 24 h at 42°C. After incubation, colony counts (cfu) of evolved mutants in each tube were determined by spreading 1.5 mL on MH agar plates containing 1 mg/L ciprofloxacin or 100 mg/L ampicillin (∼10 × MIC). The number of total viable bacteria was determined from three tubes by dropping 10 μL of 10−4, 10−5 and 10−6 dilutions onto non-selective MH agar plates. After 2 days of incubation in a microaerobic atmosphere at 42°C, colonies were counted. The frequency of resistant mutants was expressed as the mean number of resistant colonies divided by the mean of the total number of viable cells. The mutation frequencies were calculated from the mean of 20 cultures for the respective isolate. To ensure that the 30 μL of bacteria used to inoculate the 3 mL sample cultures was free of pre-existing resistant mutants, three 30 μL precultures were plated on selection plates. The sample cultures were used to measure the mutation frequency only if no mutants were present in these 30 μL precultures. Nine or 10 colonies growing on selective MH plates from the spontaneous FQR or AmpR mutation frequency test were randomly picked to sequence the QRDR in the gyrA gene and the promoter region of the blaOXA-61 gene, respectively (primers are listed in Table 2). Meanwhile, these colonies were sub-cultured and tested for their susceptibility to ciprofloxacin and ampicillin, respectively, as described above.
Table 2.
PCR primers used in this study
| Primer | Sequence | PCR products |
|---|---|---|
| mutY-FFm | CAAAGCCCaTATATCAAGCAAA | 712 bp fragment containing mutY(T595) for MutY mutation |
| mutY-FF | CAAAGCCCcTATATCAAGCAAA | 712 bp fragment containing mutY(G595) for MutY reversion |
| mutY-FR | CATTCTATAGATATATTGATAAGCGAAAAACCTACACCTAAACCTT | with mutY-FFm/mutY-FF for amplifying the 712 bp fragment for MutY mutation/reversion |
| aphA3-F | CGCTTATCAATATATCTATAGAATG | aph gene |
| aphA3-R | GATAATGCTAAGACAATCACTAAA | |
| mutY-RF | TTTAGTGATTGTCTTAGCATTATCTTTGTTGCAGGAGTAGTTTTTA | 896 bp fragment for MutY mutation/reversion |
| mutY-RR | TTTTAAGTCCCTTGTGTCTACC | |
| mutY-5F | TCGGGTTTTAGCGTATTGCT | mutY-5′ end |
| mutY-5R | CGGGGTACCAGCCCCAAACTTATCCACG | |
| cat-F | CGGTGGTACCTGGAGCGGACAACGAGTAAA | cat gene |
| cat-R | CGCGGATCCTCAGTGCGACAAACTGGGATT | |
| mutY-3F | CGCGGATCCATTTGCGATACAGAAAAGCCAA | mutY-3′ end |
| mutY-3R | GCTGTTTTTGGAGGATCTGC | |
| GyrAF1 | CAACTGGTTCTAGCCTTTTG | gyrA QRDR region |
| GyrAR1 | AATTTCACTCATAGCCTCACG | |
| p0299-F | TCTCATTTTGCATACCTCAA | blaOXA-61 promoter region |
| p0299-R | CTCCATAGCCCTTGAAAAGT |
Lowercase letters represent the T595 or G595 nucleotide in the mutY gene.
The nucleotides in bold represent sequences from the aphA3 gene designed for overlap PCR.
The underlined nucleotides represent the restriction sites added to the primers (KpnI and BamHI).
WGS of C. jejuni CMT
Briefly, genomic DNA of C. jejuni NCTC 11168 and its CMT variant were prepared from bacteria grown on MH agar medium using a Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturer's instructions. An Illumina Genome Analyzer was used to sequence the genome of the CMT isolate according to the manufacturer's instructions, with read lengths of 150 bp. The generated reads were assembled de novo into contigs using the Velvet (v 1.2.10) and VelvetOptimiser (v 2.2.5).34
Comparative genomics analysis
The contigs were aligned against the reference genome C. jejuni NCTC 1116833 by using Mauve (v 2.3.1).35,36
Site-specific reversion (W199→G)/mutation (G199→W) of MutY in C. jejuni isolates
In order to investigate the role of the single nucleotide change (G→T) of mutY in promoting the emergence of FQR mutants in C. jejuni, this mutation in the CMT isolate was reverted to the WT sequence using a method previously reported, with some modifications.20,37 Cj1621 and mutY (cj1620c) are tandemly positioned on the chromosome of C. jejuni, but transcribed in opposite directions. Cj1621 encodes a possible periplasmic protein with unknown function. A KmR marker was inserted in the cj1621 gene upstream of mutY to facilitate the reversion of the specific mutation in mutY by homologous recombination. Briefly, a 896 bp fragment containing 702 bp of the cj1621 encoding sequence with 194 bp immediately downstream of the gene, and another 712 bp fragment containing the 5′ region of cj1621, the intergenic region and the 5′ mutY sequence up to the mutation site, were amplified by PCR using C. jejuni 11168 DNA as the template and primers listed in Table 2. These two PCR fragments were then linked with the KmR marker by overlap PCR using the listed primers. The overlap PCR product was purified and used to naturally transform C. jejuni CMT. Transformants were screened on MH agar plates containing 30 mg/L kanamycin and confirmed by chromosomal DNA amplification of the gene flanking the insertion site. This resulted in CMTW199G::Km with reversion (W199→G) in MutY through homologous recombination, which was confirmed by DNA sequencing. Another construct, CMT::Km, containing only a KmR cassette in cj1621 of the CMT isolate (without the reversion) served as a control to demonstrate that neither the presence nor the location of the KmR cassette in cj1621 had any effect on the emergence of FQR mutants.
A similar protocol was utilized to construct 11168G199W::Km, with a point mutation (G199→W) in MutY of C. jejuni 11168 in order to confirm its mutator effect in C. jejuni cells. Another construct, 11168::Km, containing only a KmR cassette in cj1621 of the 11168 isolate, served as a control as described above.
Construction of an insertional MutY mutant in C. jejuni isolates
Briefly, primers mutY-5F and mutY-5R were used to amplify the 5′ part of mutY and its upstream region (mutY-5′ fragment), while primers mutY-3F and mutY-3R were used to amplify the 3′ part of mutY and its downstream region (mutY-3′ fragment). Primer pair cat-F/cat-R was used to amplify the cat gene from the pRY112 plasmid, encoding chloramphenicol resistance, using Phusion High-Fidelity DNA Polymerase (New England Biolabs, USA). After KpnI and BamHI digestion, the mutY-5′, cat and mutY-3′ PCR fragments were ligated by T4 DNA ligase (New England Biolabs, USA) and amplified utilizing mutY-5F and mutY-3R primers, resulting in the construction of a mutY-5′-cat-mutY-3′ PCR product. The purified mutY-5′-cat-mutY-3′ product was then electroporated into C. jejuni competent cells. Transformants were selected on MH agar plates containing 10 mg/L chloramphenicol. The insertion of the resistance marker into the mutY gene was confirmed by PCR using primers mutY-5F and mutY-3R.
Structural modelling of C. jejuni MutY
The crystal structure of MutY in epsilonproteobacteria is still unknown. To shed light on how the G199→W mutation might affect the function of MutY in C. jejuni, the MutY structure was modelled according to the crystallized MutY structures in other bacterial species. The modelling was conducted and completed using the SWISS-MODEL web site (http://swissmodel.expasy.org/). Briefly, the amino acid sequence of C. jejuni MutY was searched with BLAST38 against the primary amino acid sequences deposited in the SWISS-MODEL template library (SMTL, last update 3 December 2014; last included PDB release 28 November 2014). For each identified template, the template's quality was predicted from features of the target template alignment. The templates with the highest quality were then selected for model building. Structural models were built using the target template alignment with Promod-II39 and MODELLER.40 The modelled structure of MutY of C. jejuni was then animated and visualized by the PyMOL Molecular Graphics System, Version 1.7.4 (Schrödinger, LLC).
Results
Abnormal ciprofloxacin susceptibility pattern of C. jejuni isolate CMT
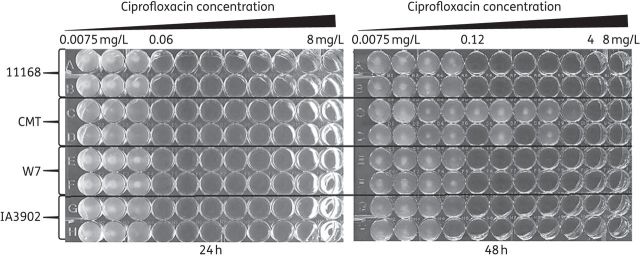
During regular MIC testing, it was observed that the MIC of ciprofloxacin in isolate CMT varied with the duration of incubation. At 24 h of incubation, the CMT isolate showed an MIC of 0.06 mg/L, similar to that of other three C. jejuni strains, NCTC 11168, IA390241 and W742 (Figure 1). However, after 48 h of incubation growth was observed in wells with ciprofloxacin concentrations as high as 2 mg/L for the CMT isolate (Figure 1). This abnormal pattern did not occur with other tested strains. This observation was reproducible in multiple experiments. The wells that did not show visible growth until after 48 h of incubation were passaged onto MH plates with 1 mg/L ciprofloxacin. All these passaged cultures from CMT grew on the selective MH plates after overnight incubation, while the passaged cultures from 11168 did not show any growth. These observations suggested that FQR mutants might have developed from the CMT isolate during the MIC test.
Figure 1.
Ciprofloxacin susceptibility test for C. jejuni 11168, CMT, W7 and IA3902 utilizing a broth microdilution method in a 96-well plate. The concentration range of ciprofloxacin used for this assay was from 0.0075 to 8 mg/L. Each of the isolates was assayed in duplicate. Pictures were taken of the same plate at 24 and 48 h of incubation.
CMT rapidly accumulates FQR mutants under ciprofloxacin treatment in vitro
To further evaluate the ability of isolate CMT to generate FQR mutants under ciprofloxacin treatment, C. jejuni CMT and 11168 were compared for growth in MH broth with addition of 1 mg/L ciprofloxacin (Figure 2a). The cfu number of the initial inoculum in each well was adjusted to that normally used for the broth microdilution test (∼105 cfu/well). The bacterial counts for both CMT and 11168 started to decrease steadily after the addition of ciprofloxacin. Interestingly, CMT experienced greater growth reductions than 11168 during the first 12 h of treatment. However, a drastic increase in cfu/mL was observed with the CMT cultures after 24 h of incubation. Six of the nine cultures of CMT yielded >106 cfu/mL bacteria after 24 h of treatment (Figure 2a). In contrast, eight of the nine cultures of 11168 did not show bacterial growth. For each strain, up to 50 colonies from each timepoint were randomly picked from the enumerating plates (no antibiotics) and streaked onto MH plates with 1 mg/L ciprofloxacin. None of the colonies from 11168 collected at 0–36 h timepoints grew on the selective MH plates, but all 50 colonies from the single 11168 culture showing growth at 48 h grew on the ciprofloxacin plate. For the CMT cultures, no colonies from the 0 h cultures grew on the selective MH plate. One in 50 colonies from 6 h cultures grew on the MH plate with ciprofloxacin. Notably, all of the colonies from the CMT cultures at each of the 12, 24, 36 and 48 h timepoints grew on ciprofloxacin plates, which was distinct from that of 11168 cultures. These results suggest that the deferred growth in the CMT cultures after 24 h of incubation was not due to the efficacy loss of ciprofloxacin in the medium, but was mainly attributable to the growth of FQR mutants in these cultures. It is interesting to note that the percentage of FQR mutants increased rapidly in the CMT cultures during the first 12 h of incubation, which subsequently allowed further accumulation of FQR mutants.
Figure 2.
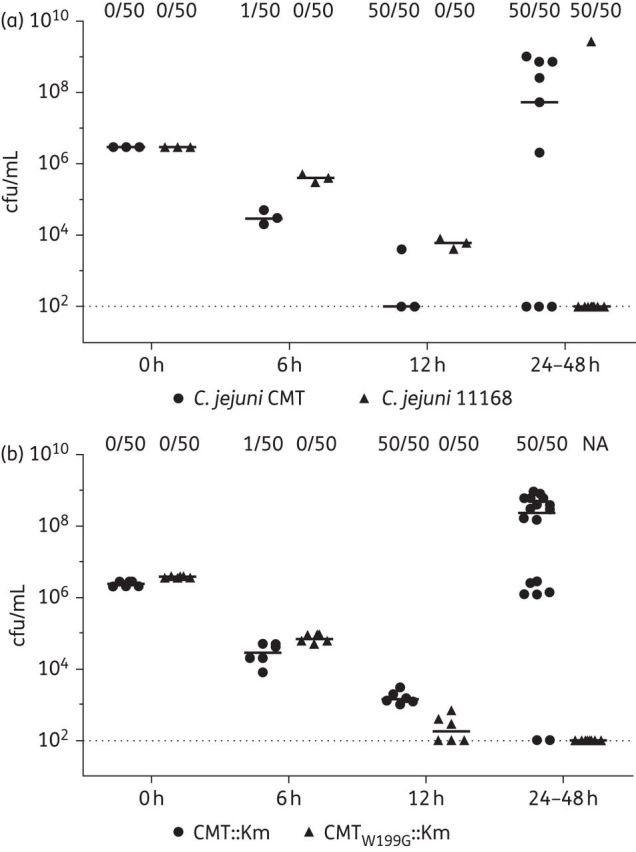
Growth of C. jejuni under ciprofloxacin (1 mg/L) treatment. (a) Comparison of C. jejuni CMT (filled circles) with NCTC 11168 (filled triangles). (b) Comparison of C. jejuni CMTW199G::Km (filled circles) with CMT::Km (filled triangles). For both panels, at each timepoint the number of FQR mutants among 50 randomly picked colonies from antibiotic-free enumerating plates is shown at the top. The broken line in both panels represents the detection limit of the plating method. NA, no colonies available.
CMT has notably elevated spontaneous FQR mutation frequency
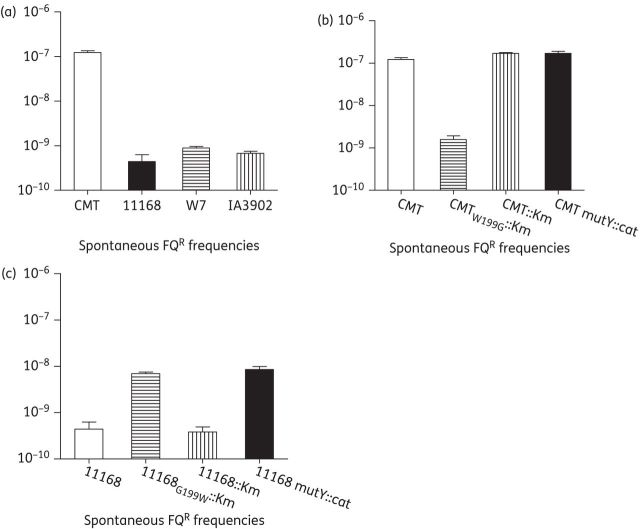
CMT and the other three C. jejuni isolates, 11168, IA3902 and W7, were tested for spontaneous FQR mutation frequency in MH broth under conventional conditions without adding any antibiotics. This assay was performed in triplicate. C. jejuni 11168, IA3902 and W7 had similar spontaneous FQR mutation frequencies (10−9). The average FQR mutation frequency for the CMT isolate was 1.2 × 10−7, ∼2 logs higher than that of the other three C. jejuni isolates (Figure 3a).
Figure 3.
Spontaneous FQR mutation frequencies of various C. jejuni isolates. (a) Elevated mutation frequency in C. jejuni CMT compared with strains 11168, W7 and IA3902. (b) Reduced mutation frequency in the revertant of MutY of the CMT isolate. (c) Mutation of mutY increases spontaneous FQR frequencies in C. jejuni 11168. Each bar represents the mean value of triplicate experiments.
Several FQR colonies growing on selective plates were picked for strains 11168 and CMT and subsequently sequenced for the QRDR region in the gyrA gene and tested for susceptibility to ciprofloxacin (Table 3). For strain 11168, characteristic mutations had occurred either as a single substitution consisting of C257→T or G268→A transition, which leads to a Thr-86→Ile or Asp-90→Asn amino acid change in FQR mutants, with MIC values of 8–16 mg/L. However, all colonies sequenced for the CMT strain carried a C257→A or G268→T transversion in the QRDR region, which resulted in a Thr-86→Lys or Asp-90→Tyr amino acid change. Antibiotic susceptibility tests showed that all FQR mutants carrying Thr-86→Lys or Asp-90→Tyr showed an MIC of 4 mg/L for ciprofloxacin. This result revealed that FQR mutants generated from the CMT strain had distinct gyrA mutation patterns compared with that of 11168 or other FQR mutant C. jejuni isolates that were also sequenced for QRDR regions in the gyrA gene.16 Together, spontaneous FQR mutation tests and the ciprofloxacin treatment assay indicate that that the CMT strain generates FQR mutants much more frequently than does the 11168 isolate.
Table 3.
Differences in the GyrA mutations of FQR mutants from different strains
| C. jejuni isolate | No. of total isolates (colonies) | No. of isolates with different mutations in GyrA |
MIC of ciprofloxacin (mg/L) | |||
|---|---|---|---|---|---|---|
| Asp-90→Asn (G268→A) | Thr-86→Ile (C257→T) | Asp-90→Tyr (G268→T) | Thr-86→Lys (C257→A) | |||
| CMT | 9 | 0 | 0 | 8 | 1 | 4 |
| CMTW199G::Km | 10 | 9 | 1 | 0 | 0 | 8–16 |
| NCTC 11168 | 10 | 6 | 4 | 0 | 0 | 8–16 |
Genome sequencing and comparative genomics analysis
To fully investigate the genetic basis of the high-frequency emergence of FQR mutants in the CMT isolate, this isolate was subjected to WGS analysis. De novo assembled contigs were aligned against the NCTC 11168 genome. Few artificial gaps were observed due to the presence of repetitive sequences in the genome, which makes the software unable to align some tiny contigs appropriately (data not shown). Excluding the gaps, multiple single nucleotide changes, frame shifts (due to single nucleotide deletion or insertion) and previously reported homopolymeric nucleotide stretches were observed in the CMT genome compared with the genome of 11168. As shown in Table S1 (available as Supplementary data at JAC Online), most of the mutations occurred in genes not related to DNA repair and thus were not expected to affect spontaneous mutation. However, there was a G→T transversion in cj1620, which leads to a G199→W amino acid change in the coding sequence. cj1620 is an orthologue of DNA repair gene mutY, encoding a putative adenine glycosylase that has the ability to remove adenines from adenine/7,8-dihydro-8-oxoguanine (8-oxoG) mismatches in E. coli, which is specific for the repair of the G→T or C→A transversion.43 In C. jejuni 11168, the mutY gene encodes a protein of 339 amino acids. At the amino acid level, the MutY products of C. jejuni 11168 and Escherichia coli K-12 MG1655 share 35% identity. The G199 is well conserved in MutY proteins among the 10 bacteria species that were analysed in this study (Figure S1), suggesting that it might be important for the function of MutY. Thus we hypothesized that the single point mutation in mutY was related to the increased spontaneous mutation frequencies in the CMT isolate.
Reversion of the G199→W mutation in MutY significantly decreases the spontaneous FQR mutation frequency of the CMT isolate
To define the role of the G→T transversion in mutY in affecting the spontaneous mutation rate in C. jejuni, the G199→W mutation in the CMT isolate was reverted by homologous recombination, creating construct CMTW199G::Km, in which the reversion was accompanied by insertion of a kanamycin resistance cassette in the adjacent gene cj1621. Another construct, CMT::Km, which only contained the kanamycin resistance cassette in cj1621, was also made as a control. As shown in Figure 3(b), the CMT::Km isolate did not show any change in spontaneous FQR mutation frequency (∼1.7 × 10−7) compared with that of the CMT isolate (∼1.2 × 10−7), while CMTW199G::Km showed a drastic decrease in spontaneous FQR mutation frequency (∼1.6 × 10−9). Sequence analysis of the QRDR region in the gyrA gene of FQR mutants from the CMTW199G::Km isolate revealed that the mutation patterns had changed to C257→T or G268→A transition in FQR mutants, instead of the C257→A or G268→T transversion in FQR mutants from the CMT isolate (Table 3). Together, these results indicated that the G199→W mutation was responsible for the increased spontaneous FQR mutation frequency in the CMT isolate, and reversion of this mutation recovered the function of MutY in repairing G:C→T:A transversion in C. jejuni cells, substantially decreasing the emergence of FQR mutants in C. jejuni.
The CMT::Km and CMTW199G::Km constructs were further compared for growth in MH broth in the presence of 1 mg/L ciprofloxacin. This assay was conducted twice under the same conditions; the results of the two independent experiments were combined and are shown in Figure 2(b). Similar to the results for CMT and 11168 isolates, the cfu/mL of both CMT::Km and CMTW199G::Km decreased steadily in the first 12 h of incubation. As expected, significant increases in cfu/mL were observed in the CMT::Km cultures after 24 h of incubation. Sixteen of the 18 cultures of CMT::Km yielded >106 cfu/mL bacteria after 24 h of incubation. In contrast, none of the 18 cultures of CMTW199G::Km showed bacterial growth after 24 h, which indicated that the W199→G reversion in mutY prevented the emergence of FQR mutants in the CMT isolate under the experimental conditions used in this study. For each isolate, up to 50 colonies were randomly picked for each strain and at each timepoint from the non-antibiotic plates, and streaked onto MH plates with 1 mg/L ciprofloxacin. None of the colonies from CMTW199G::Km cultures at any timepoints grew on the selective MH plates. For CMT::Km, no colonies from the 0 h cultures grew on the selective MH plate, while one of the 50 colonies from 6 h cultures grew on the ciprofloxacin-containing plates. Similar to the results of CMT cultures, all 50 colonies from CMT::Km cultures at each of the 12, 24, 36 and 48 h timepoints grew on the selective MH plates. These results from the selective plates confirmed that the regrowth after 12 h of treatment was due to emergence of FQR mutants and that the W199→G reversion in MutY prevented the emergence of the mutants.
The G199→W mutation in MutY abolishes the anti-mutator function of MutY in Campylobacter
To verify the mutator effect of the G199→W mutation in MutY, the point mutation was introduced into C. jejuni 11168 by homologous recombination. A MutY insertional mutant of 11168 was also constructed to confirm the anti-mutator role of MutY in Campylobacter. As shown in Figure 3(c), the G199→W mutation in MutY of 11168 significantly increased the spontaneous FQR mutation frequency compared with that of the 11168 isolate. The same increased frequency was also observed in the mutY insertional mutant. As a control, the KmR insertion in the cj1621 gene alone did not change the mutation frequency of 11168. These results indicated that the G199→W mutation had the same effect on mutation frequencies as the mutY insertional mutation, suggesting that this mutation significantly compromised the anti-mutator effect of MutY in C. jejuni. Additionally, an isogenic MutY insertional mutant was also made in strain CMT and compared with the CMT isolate for spontaneous FQR mutation frequencies. No difference in the FQR mutation frequency was observed between CMT and its isogenic mutY insertional mutant (Figure 3b). Together, these results indicate that the G199→W mutation abolishes the anti-mutator function of MutY, which leads to the elevated frequency of emergence of FQR mutants in C. jejuni.
The G199→W mutation in MutY also significantly increases the spontaneous AmpR mutation frequency in C. jejuni
To investigate whether the G199→W mutation in MutY has a general effect on the emergence of antimicrobial-resistant mutants in C. jejuni, the CMT::Km and CMTW199G::Km constructs were compared for the spontaneous AmpR mutation frequency. As shown in Table 4, there was a ∼700-fold difference in mutation frequency between the CMT::Km isolate and the CMTW199G::Km revertant, in which the G199→W mutation was reverted, Sequencing of the spontaneous AmpR mutants randomly picked from both the CMT::Km and the CMTW199G::Km background revealed the same G→T transversion in the −10 region of the blaOXA-61 promoter. This G→T transversion was previously shown to restore the TATA box, leading to expression of β-lactamase blaOXA-61 and consequently high-level resistance to ampicillin in C. jejuni.24 These results indicate that the G199W mutation in MutY also results in a significant increase in the emergence of AmpR mutants, due to the defect of the MutY mutant in repairing G:C→T:A transversions in the C. jejuni genome.
Table 4.
Spontaneous AmpR mutation frequencies and the point mutation in the blaOXA-61 promoter region in ampicillin-resistant C. jejuni mutants
| C. jejuni isolate | Spontaneous AmpR mutation frequencies | No. in (total no.) picked ampicillin-resistant colonies with the G→T mutation in blaOXA-61 promoter region | Ampicillin MIC (mg/L) before mutation | Ampicillin MIC (mg/L) after G→T mutation |
|---|---|---|---|---|
| CMT::Km | 3.31E−07 | 7 (7) | 16 | 256 |
| CMTW199G::Km | 4.26E−10 | 8 (8) | 16 | 256 |
The G199→W mutation probably affects the catalytic domain of MutY in C. jejuni
Structural modelling of C. jejuni using the published MutY structure from Geobacillus stearothermophilus44 suggests that G199 is located in helix 10 (Figure S2) and is close to the catalytic domain (the six-helix barrel module and the [4Fe–4S] cluster), which is vital for MutY catalytic enzyme activity. In the MutY of the CMT isolate a hydrophobic amino acid, tryptophan (W), with an extra indole ring replaced the simple amino acid glycine (Figure S2). This modification may affect the function of the catalytic domain and thus interfere with the removal of adenine by the MutY catalytic domain.
Discussion
Antibiotic resistance in C. jejuni is an increasing problem worldwide and is considered a serious threat to public health in the USA.12 Both acquisition of antimicrobial resistance genes and mutations contribute to the prevalence of antibiotic-resistant Campylobacter. Multiple antimicrobial resistance genes have been found in Campylobacter, such as the tet(O) gene for tetracycline resistance, blaOXA-61 for β-lactam resistance, erm(B) for macrolide resistance and several aminoglycoside resistance genes.14,45,46 However, mutation is the most common mechanism of resistance in Campylobacter for several classes of clinically important antimicrobial agents, such as FQs45,47 and macrolides.15 Except for Mfd, which was previously reported to promote mutation in Campylobacter,48 little is known about genetic factors that affect spontaneous mutation frequencies in this organism. In this study, the single G595→T transversion in mutY of C. jejuni was found to promote spontaneous FQR and AmpR mutation frequencies. This mutation abolishes the anti-mutator function of MutY and leads to significantly increased emergence of FQR and AmpR mutants in Campylobacter. To our knowledge, genetic factors contributing to the naturally occurring mutator Campylobacter have not been reported and the findings in this study represent a previously undescribed genetic mechanism that is involved in hypermutation for antibiotic resistance in this important foodborne pathogen.
The G595→T mutation in mutY was initially identified by WGS analysis. Although the mutator isolate CMT shows significantly elevated spontaneous mutation frequencies compared with the WT 11168 isolate, there were few ambiguous sites in the alignment of the genome sequences of CMT and 11168 genomes (Table S1). This is due to the fact that the assembled whole-genome sequence was actually a consensus sequence from multiple reads in a mixed population. In the mutator CMT strain, although the mutation frequency is elevated, the vast majority of DNA alleles are still WT. Without a selection step to enrich the mutations, these mutations are only a tiny fraction and massively outnumbered by the WT sequences in the population, Thus, the high mutation frequency in CMT cannot be revealed by WGS alone.
To prove that the G199W change in MutY was linked to elevated mutation frequencies, the mutation was reverted at its original location utilizing homologous recombination. Significant decreases in spontaneous FQR and AmpR mutation frequencies were observed in the resulting revertants (Figure 3 and Table 4). Additionally, the same strategy was utilized to make the site-specific G199→W change in MutY of C. jejuni 11168, which showed an effect on the spontaneous FQR mutation frequency similar to that of the MutY insertional mutant. Furthermore, insertional inactivation of the mutY gene in the CMT isolate did not further increase its mutation frequency, suggesting that G199W alone is sufficient to abolish the anti-mutator function of MutY. Together, these results establish that the naturally occurring mutation in mutY confers a mutator phenotype in Campylobacter.
The MutY protein in C. jejuni is specific for the repair of the G→T or C→A transversion, which is similar to that observed in E. coli.43 The G199→W mutation resulted in loss of the repair function and consequently an elevated mutation rate with the G→T or C→A transversion. This was shown by the findings that all of the FQR mutants of the CMT isolate carried a C257→A or G268→T transversion in the QRDR region of GyrA (Table 3) and also all of the AmpR mutants of the CMT isolate carried a G→T transversion in the blaOXA-61 promoter region (Table 4). We also tested the mutation rate for macrolides, and no apparent difference was observed in the frequency of erythromycin resistance between WT 11168 and the CMT isolates (data not shown). This can be explained by the fact that macrolide resistance is mediated by an A→G transition or A→C transversion in the ribosomal RNA of C. jejuni,8,23 and MutY is not expected to repair these types of mutations in C. jejuni. Thus, loss of MutY function selectively affects antibiotic resistance mechanisms that are mediated by G:C→T:A transversions in target genes.
In this study, all of the FQR mutants from the CMT isolate showed intermediate-level FQR (MIC = 4 mg/L). When the concentration of ciprofloxacin was increased to 4 mg/L in the mutant-enumerating plates, no obvious difference was observed in the spontaneous FQR mutation frequency between NCTC 11168 and the CMT isolates (data not shown), consistent with the MIC results and suggesting that 4 mg/L prevented the emergence of the FQR mutants carrying a C257→A or G268→T transversion. Blondeau et al.49 introduced the concept of mutant prevention concentration (MPC), which represents a threshold above which the selective proliferation of resistant mutants is expected to occur only rarely. In this study, increasing the ciprofloxacin concentration from 1 to 4 mg/L significantly reduced the spontaneous FQR mutation frequency of the CMT isolate. Thus, understanding MPC and managing the doses of FQs may significantly reduce the development of resistance in Campylobacter.
The crystal structures of MutY from E. coli and another thermophilic bacterium (Geo), Bacillus stearothermophilus, were resolved,44,50 revealing the basis for recognizing lesions in the A·oxoG pair and for catalysing removal of the adenine base. However, the function of G199 in MutY was not defined in these crystallization studies. To shed light on how the G199→W mutation might affect the function of MutY, we conducted modelling of the C. jejuni MutY structure (Figure S2), which suggested that G199 is located next to the MutY catalytic domain and that the G199→W mutation may interfere with the function of this vital domain. However, this finding from structural modelling remains to be confirmed experimentally in future studies.
The growth rates of CMT, its revertant and C. jejuni 11168 appeared to be similar, as measured by their growth curves in MH broth and the colony sizes on MH plates (data not shown), which suggests that there is no obvious growth defect related to the G199W mutation in MutY of Campylobacter. However, it is still unknown whether there is a long-term fitness cost of the G→T transversion in mutY for Campylobacter. Although many mutations are deleterious to organisms, there are multiple reports of naturally occurring pathogenic organisms that exhibit permanent mutator phenotypes due to defects in the DNA repair system.28,29,51–53 Thus, bacterial organisms have great potential to adapt to stress by fixing novel mutations that increase fitness.54 There are only limited numbers of C. jejuni MutY sequences available in the NCBI database. However, examination of the 35 published C. jejuni genomes indeed revealed a number of nucleotide sequence polymorphisms of MutY (Figure S3), some of which are also located near the functional domains of the MutY protein. Whether these mutations in MutY also contribute to a mutator phenotype in Campylobacter remain to be determined.
To our best knowledge, this is the first identification of a genetic factor conferring a natural mutator phenotype in Campylobacter. This mutator phenotype is due to a single amino acid change (G199W) in MutY, which results in loss of the repairing function. Antibiotic resistance associated with a G→T or C→A transversion is influenced by the G199W mutation in MutY. Thus, the mutator phenotype conferred by the mutY mutation may help Campylobacter adapt to antibiotic stresses. Mutators are a risk factor for clinical treatment of bacterial infections as they tend to promote the selection of mutants with resistance to antibiotics.26 Thus, identification of mutator strains and understanding their associated mechanisms may help to guide the clinical use of antibiotics and treatment regimens, reducing the development of antibiotic resistance.
Funding
This work was supported by grants R21AI098742 and R56AI118283 from the National Institute of Allergy and Infectious Diseases.
Transparency declarations
None to declare.
Disclaimer
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding agency.
Supplementary data
References
- 1.Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect 2004; 10: 868–76. [DOI] [PubMed] [Google Scholar]
- 2.Yuki N, Koga M. Bacterial infections in Guillain-Barre and Fisher syndromes. Curr Opin Neurol 2006; 19: 451–7. [DOI] [PubMed] [Google Scholar]
- 3.Tauxe RV. Emerging foodborne pathogens. Int J Food Microbiol 2002; 78: 31–41. [DOI] [PubMed] [Google Scholar]
- 4.Kassenborg HD, Smith KE, Vugia DJ, et al. Fluoroquinolone-resistant Campylobacter infections: eating poultry outside of the home and foreign travel are risk factors. Clin Infect Dis 2004; 38 Suppl 3: S279–84. [DOI] [PubMed] [Google Scholar]
- 5.Takkinen J, Ammon A, Robstad O, et al. European survey on Campylobacter surveillance and diagnosis 2001. Euro Surveill 2003; 8: 207–13. [DOI] [PubMed] [Google Scholar]
- 6.Oldfield EC, 3rd, Wallace MR. The role of antibiotics in the treatment of infectious diarrhea. Gastroenterol Clin North Am 2001; 30: 817–36. [DOI] [PubMed] [Google Scholar]
- 7.Gupta A, Nelson JM, Barrett TJ, et al. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg Infect Dis 2004; 10: 1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engberg J, Aarestrup FM, Taylor DE, et al. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis 2001; 7: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White DG, Zhao S, Simjee S, et al. Antimicrobial resistance of foodborne pathogens. Microbes Infect 2002; 4: 405–12. [DOI] [PubMed] [Google Scholar]
- 10.Maćkiw E, Korsak D, Rzewuska K, et al. Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from food in Poland. Food Control 2012; 23: 297–301. [Google Scholar]
- 11.Chen X, Naren GW, Wu CM, et al. Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet Microbiol 2010; 144: 133–9. [DOI] [PubMed] [Google Scholar]
- 12.CDC. Antibiotic Resistance Threats in the United States, 2013 2014. http://www.cdc.gov/drugresistance/threat-report-2013/.
- 13.Hooper DC. Emerging mechanisms of fluoroquinolone resistance. Emerg Infect Dis 2001; 7: 337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfredson DA, Korolik V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett 2007; 277: 123–32. [DOI] [PubMed] [Google Scholar]
- 15.Payot S, Bolla JM, Corcoran D, et al. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect 2006; 8: 1967–71. [DOI] [PubMed] [Google Scholar]
- 16.Hanninen ML, Hannula M. Spontaneous mutation frequency and emergence of ciprofloxacin resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 2007; 60: 1251–7. [DOI] [PubMed] [Google Scholar]
- 17.Yan M, Sahin O, Lin J, et al. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother 2006; 58: 1154–9. [DOI] [PubMed] [Google Scholar]
- 18.Luo N, Sahin O, Lin J, et al. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother 2003; 47: 390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 2002; 46: 2124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge B, McDermott PF, White DG, et al. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 2005; 49: 3347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cagliero C, Maurel MC, Cloeckaert A, et al. Regulation of the expression of the CmeABC efflux pump in Campylobacter jejuni: identification of a point mutation abolishing the binding of the CmeR repressor in an in vitro-selected multidrug-resistant mutant. FEMS Microbiol Lett 2007; 267: 89–94. [DOI] [PubMed] [Google Scholar]
- 22.Chatzipanagiotou S, Ioannidou V, Ioannidis A, et al. Absence of the plasmid-mediated quinolone resistance qnrA gene among Campylobacter jejuni clinical isolates from Greece. Int J Antimicrob Agents 2005; 26: 261–2. [DOI] [PubMed] [Google Scholar]
- 23.Gibreel A, Taylor DE. Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 2006; 58: 243–55. [DOI] [PubMed] [Google Scholar]
- 24.Zeng X, Brown S, Gillespie B, et al. A single nucleotide in the promoter region modulates the expression of the β-lactamase OXA-61 in Campylobacter jejuni. J Antimicrob Chemother 2014; 69: 1215–23. [DOI] [PubMed] [Google Scholar]
- 25.Taddei F, Radman M, Maynard-Smith J, et al. Role of mutator alleles in adaptive evolution. Nature 1997; 387: 700–2. [DOI] [PubMed] [Google Scholar]
- 26.Chopra I, O'Neill AJ, Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat 2003; 6: 137–45. [DOI] [PubMed] [Google Scholar]
- 27.LeClerc JE, Li B, Payne WL, et al. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 1996; 274: 1208–11. [DOI] [PubMed] [Google Scholar]
- 28.Bjorkholm B, Sjolund M, Falk PG, et al. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci USA 2001; 98: 14607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morosini MI, Baquero MR, Sanchez-Romero JM, et al. Frequency of mutation to rifampin resistance in Streptococcus pneumoniae clinical strains: hexA and hexB polymorphisms do not account for hypermutation. Antimicrob Agents Chemother 2003; 47: 1464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet 1991; 25: 229–53. [DOI] [PubMed] [Google Scholar]
- 31.Gaasbeek EJ, van der Wal FJ, van Putten JP, et al. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J Bacteriol 2009; 191: 3785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Kang J, Blaser MJ. Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J Bacteriol 2006; 188: 6224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000; 403: 665–8. [DOI] [PubMed] [Google Scholar]
- 34.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008; 18: 821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS one 2010; 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darling AC, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14: 1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han J, Wang Y, Sahin O, et al. A fluoroquinolone resistance associated mutation in gyrA affects DNA supercoiling in Campylobacter jejuni. Front Cell Infect Microbiol 2012; 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18: 2714–23. [DOI] [PubMed] [Google Scholar]
- 40.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993; 234: 779–815. [DOI] [PubMed] [Google Scholar]
- 41.Wu Z, Sahin O, Shen Z, et al. Multi-omics approaches to deciphering a hypervirulent strain of Campylobacter jejuni. Genome Biol Evol 2013; 5: 2217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plummer P, Sahin O, Burrough E, et al. Critical role of LuxS in the virulence of Campylobacter jejuni in a guinea pig model of abortion. Infect Immun 2012; 80: 585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol 1992; 174: 6321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan Y, Manuel RC, Arvai AS, et al. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol 1998; 5: 1058–64. [DOI] [PubMed] [Google Scholar]
- 45.Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int 2013; 2013: 340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin S, Wang Y, Zhang Q, et al. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother 2014; 69: 964–8. [DOI] [PubMed] [Google Scholar]
- 47.Smith JL, Fratamico PM. Fluoroquinolone resistance in Campylobacter. J Food Prot 2010; 73: 1141–52. [DOI] [PubMed] [Google Scholar]
- 48.Han J, Sahin O, Barton YW, et al. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog 2008; 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blondeau JM, Zhao X, Hansen G, et al. Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 2001; 45: 433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fromme JC, Banerjee A, Huang SJ, et al. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 2004; 427: 652–6. [DOI] [PubMed] [Google Scholar]
- 51.Denamur E, Bonacorsi S, Giraud A, et al. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J Bacteriol 2002; 184: 605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliver A, Canton R, Campo P, et al. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000; 288: 1251–4. [DOI] [PubMed] [Google Scholar]
- 53.Richardson AR, Yu Z, Popovic T, et al. Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc Natl Acad Sci USA 2002; 99: 6103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacLean RC, Torres-Barcelo C, Moxon R. Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat Rev Genet 2013; 14: 221–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.