Abstract

Dissolved salts are a part of the physiological milieu and can significantly influence the kinetics and thermodynamics of varied biomolecular processes, such as binding and catalysis, so it is important for molecular simulations to reliably describe their effects. The present study uses a simple, non-ionized host-guest model system to study the sensitivity of computed binding enthalpies to the choice of water and salt models. Molecular dynamics simulations of a cucurbit[7]uril host with a neutral guest molecule show striking differences in the salt dependency of the binding enthalpy across four water models, TIP3P, SPC/E, TIP4P-Ew and OPC, with additional sensitivity to the choice of parameters for sodium and chloride. In particular, although all of the models predict that binding will be less exothermic with increasing NaCl concentration, the strength of this effect varies by 7 kcal/mol across models. The differences appear to result primarily from differences in the number of sodium ions displaced from the host on binding the guest, rather than from differences in the enthalpy associated with this displacement; and it is the electrostatic energy that contributes most to the changes in enthalpy with increasing salt concentration. That a high sensitivity of salt effects to the choice of water model is observed for the present host-guest system, despite its being non-ionized, raises issues regarding the selection and adjustment of water models for use with biological macromolecules, especially as these typically possess multiple ionized groups which can interact relatively strongly with ions in solution.
INTRODUCTION
The types and concentrations of dissolved salts can profoundly influence the stability and function of biomolecular systems, including ones involving proteins, nucleic acids, and lipid membranes. Such effects are particularly important for highly charged molecules, such as nucleic acids, and for those where ions are of central functional importance, such as ion channels1–6. Even simple salts can affect the thermodynamics of noncovalent association. For instance, increased concentrations of NaCl cause an apparent conformational shift and enhanced catalytic activity of HIV-1 protease7, and the concentration of NaCl can strongly influence the binding enthalpy of protein-nucleic acid systems8. A more complex array of salt effects emerges when one considers the full palette of dissolved cations and anions that play roles in the physiological milieu and in vitro. Thus, similar cations, such as Na+ and K+, or Mg2+ and Ca2+, can interact very differently with biomolecules in solution9,10, while the well-known Hofmeister series3,11 catalogs the spectrum of effects due to various anions.
Computer simulations of biomolecules must therefore include realistic representation of ions and their interactions, in order to provide meaningful mechanistic explanations of experimental observations and reliable predictions for applications like enzyme engineering and drug design. Accordingly, substantial effort has been devoted to the development of reliable force field parameters, not only for biomolecules12–29 and water30–33, but also for the dissolved salts34,35. In addition, molecular dynamics (MD) simulations of biomolecular systems often only include a few Na+ or Cl− counterions to make the simulation system electrically neutral, rather than modeling a more realistic concentration and composition of the electrolyte. This approach may suffice for some applications, but an assumption that dissolved salts do not affect simulation results is not necessarily justified. Computing salt effects can be difficult, however, due to the challenges of obtaining convincingly converged experimental observables for large, complex biomolecules.
The small size and chemical simplicity of host-guest systems make them appealing models for mechanistic studies of noncovalent binding, via both experiments11,36–48 and computational methods49–54, and for the evaluation of force fields52–55. Host molecules are miniature receptors, whose affinities for the guest molecules they bind span the same range as protein-ligand systems38,39,50,56, and they share many key features of their larger, biomolecular relatives. In particular, salt effects can substantially influence host-guest binding thermodynamics. For example, the association of the non-ionic hosts cucurbit[6]uril (CB6) and cucurbit[7]uril (CB7; Figure 1) with various guests are strongly salt-dependent,35,55,56, while the enthalpy of binding for the octa-acid host41 with adamantane carboxylic acid varies by nearly 10 kcal/mol when 100 mM Na2SO4 in the buffer is replaced by 100 mM NaClO4 40.
Figure 1.
Structures of the host molecule CB7 and the guest molecule B2. Cyan: carbon; blue: nitrogen; red: oxygen; white: hydrogen.
We recently reported that host-guest binding enthalpies can be computed efficiently to high numerical precision with GPU-equipped computers55 and that the computed enthalpies depend strongly on the choice of the water model. In particular, the TIP4P-Ew31 model yielded binding enthalpies systematically more favorable, by about 4 kcal/mol, than those obtained with the TIP3P30 model, across a set of eight CB7-guest systems55. This high degree of sensitivity to details of the water model suggested that the results might also be sensitive to the treatment of salt in the simulations, and, furthermore, that the salt effects might themselves depend on the choice of water model. The present study explores these issues by studying the consequences of salt concentration, water model, and ion model, on the association of the non-ionic CB7 host59 with a non-ionic guest50 (B2 in Figure 1) in aqueous solution. Strong sensitivities are indeed observed, in calculations spanning salt concentrations of 0 to 500 mM, for several three-site (TIP3P30, SPC/E32) and four-site (TIP4P30, TIP4P-Ew31, OPC33) water models. Three models of NaCl are also considered. Structural analysis and energy decompositions of the results provide insight into the mechanisms underlying the observed effects, and suggest possible explanations for the substantial differences among the results from the various water models. The present results have implications for force field development and biomolecular simulations.
METHODS
Microsecond timescale molecular dynamics simulations, with explicit solvent comprising water and dissolved ions, were used to compute the binding enthalpies at 300K for the host CB7 with guest B2 at four different NaCl concentrations, 0, 100, 300, 500 mM. Results were obtained and analyzed for several water models, TIP3P, SPC/E, TIP4P, TIP4P-Ew and OPC, with three sets of NaCl force field parameters. Methodological details follow.
Binding Enthalpy Calculations
The binding enthalpy of the CB7-B2 system was computed as the difference between the mean potential energies associated with four separate simulations. The two initial states are the free host and guest in aqueous solution; the two final states are the complex in a matching solvent, and a solvent-only simulation which balances solvent counts between the initial and final states:
| Eq 1 |
No salt was included in the simulations for 0 mM salt. For the simulations with NaCl at 100 mM, 300 mM and 500 mM, the numbers of Na+ and Cl− ions included in each of the four simulations were 3, 9 and 15, respectively. The mean potential energies in Eq 1 were computed by using the sander. MPI module of Amber 1460 to postprocess all trajectory snapshots from the simulations. Convergence of the total potential energy from a simulation was evaluated by plotting the cumulative average as a function the total simulation time, and by using blocking analysis61 to estimate the standard error of the mean(SEM). All average potential energies in the present calculations have a SEM of 0.11 kcal/mol or less (Table S1 and Figures S1, S2); thus, if the same simulation were repeated multiple times, with different randomly assigned initial velocities, the resulting set of mean potential energies should have a standard deviation within about 0.11 kcal/mol. Because the binding enthalpy in Eq 1 is an additive combination of four mean energies, the variances of the mean energies add, and the standard deviation of the reported binding enthalpy is given by , where σi is the SEM for simulation i.
Calculation of density distributions for ions and water
Spatial density distributions for ions and water were calculated using the grid command in the cpptraj module of Amber 1460. Prior to performing the grid analysis, each frame of the trajectory was aligned to a reference frame via mass-weighted RMS fit of the CB7 atoms. The grid bin size in each dimension (X, Y, and Z) was 0.5 Å. Additionally, we computed the conditional density distribution of the solvent for trajectory frames in which a Na+ ion was bridging two specified CB7 portal oxygens. This was accomplished by performing the grid command on only a subset of trajectory frames for which bridging condition was met. The subset trajectory was generated using cpptraj’s native contacts and filter commands. To analyse the distributions of cations and anions qualitatively, we divided a spherical region centered on CB7 and having a radius of 10 Å into ten shells, each of thickness 1 Å. The average numbers of both Na+ and Cl− ions in the MD simulations of free host and CB7-B2 complex were calculated for each shell, and the standard errors of these means were estimated by blocking analysis55,61.
Molecular Dynamics Simulations
All simulations were set up and run with the Amber suite of programs60, using the GPU-enabled version of pmemd for production. The general AMBER force field (GAFF)15 was used for all bonded and Lennard-Jones parameters of CB7 and the guest molecule, with partial atomic charges generated with the AM1-BCC method62,63 as implemented in the program Antechamber60. In addition, RESP charges64, based on Hartree-Fock calculations with the 6–31G* basis set, were assigned to the host and guest with Antechamber by performing a two-stage fit on an electrostatic potential grid generated with Gaussian 0965. These charges were used for a sample calculation at 0 M NaCl, with the TIP3P water model. Calculations were run with the TIP3P, TIP4P, TIP4P-Ew, SPC/E and OPC water models. For most calculations, parameters for Na+ and Cl− were drawn from the work of Joung and Cheatham34, who provided ionic parameters specific for the TIP3P, TIP4P, TIP4P-Ew and SPC/E water models; note that TIP4P and TIP4P-Ew use the same salt parameters. We considered the ionic model associated with TIP4P-Ew to be the default match to the OPC water model, based on a case study of ion-ion interactions in OPC33. We first used each water model with its matching ion model, then experimented with nonstandard combinations in order to distinguish the effects of water models from the effects of ion models. Calculations at 100 mM NaCl were also carried out with TIP3P and TIP4P-Ew using alternative Na+ and Cl− parameters recently proposed for use with these water models35.
The starting conformations of CB7 is from a crystal structures59, B2 was built using Gaussview 566, and the initial conformation of their complex was obtained from a previous computational study67. The host/guest system was placed in a cubic water box containing 1665 water molecules, along with the appropriate number of Na+ and Cl− ions (see above) under periodic boundary conditions. To remove potential bad contacts, the initial model was subjected to two rounds of energy minimization: first 1000 steps of steepest descent followed by 9000 steps of conjugate gradient with the host and guest atoms restrained with a harmonic potential with a force constant of 100 kcal/mol; and then another 1000 steps of steepest descent and 9000 steps of conjugate gradient without any restraints. A heating phase, at constant volume, lasted 2 ns and raised the temperature from 0 to 300 K. Equilibration and production runs then were carried out at constant temperature (300 K) and pressure (1 bar) with a cubic orthorhombic box having equal side lengths of about 37 Å. The temperature was set using the Langevin thermostat with the collision frequency set to 1.0 ps−1, and pressure was set with the Berendsen barostat with the relaxation time set to 2 ps. The first 10 ns of each production simulation was discarded to allow for additional equilibration. The production simulation times were at least 1 μs for each system, for a total simulation time of over 100 μs across the various force field combinations.
RESULTS
Calculated binding enthalpies as a function of NaCl concentration
Prior isothermal titration calorimetry experiments for binding of the host CB7 to its guest B2 (Figure 1), carried out in the absence of dissolved NaCl, yielded a measured binding enthalpy of −15.8 kcal/mol50. The corresponding results from molecular dynamics calculations overestimate it, for all four water models examined here, TIP3P, TIP4P-Ew, SPC/E and OPC (Table 1, Figure 2). This observation is consistent with previously published results for TIP3P and TIP4P-Ew55. The recently developed OPC model is closest to experiment, at −18.9±0.2 kcal/mol, while the results from TIP3P, SPC/E and TIP4P-Ew are all more negative than −21 kcal/mol. As a check of the AM1-BCC atomic partial charges, we reran the 0mM NaCl concentration with TIP3P water and RESP partial charges. This calculation led to a binding enthalpy of 23.2±0.4 kcal/mol, which overestimates the binding enthalpy even more than the corresponding AM1-BCC result, −21.6±0.2 kcal/mol.
Table 1.
Non-bonded Lennard-Jones (LJ) and Coulombic (Coul) contributions to the computed binding enthalpies (ΔH; in kcal/mol), at a variety of NaCl concentrations (in mM). The bonded contributions, not listed, make up the differences. The experimental binding enthalpy is −15.8 kcal/mol at 0 mM NaCl50. The SEM values are less than 0.2 kcal/mol for all computed terms. The TIP3P, SPC/E and TIP4P-Ew models were used with their corresponding Na+ and Cl− parameters from Joung and Cheatham34, and OPC was used with the parameters tuned for TIP4P-Ew.
| NaCl conc | TIP3P | SPC/E | TIP4P-Ew | OPC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LJ | Coul | ΔH | LJ | Coul | ΔH | LJ | Coul | ΔH | LJ | Coul | ΔH | |
|
|
|
|
|
|
||||||||
| 0 | −13.3 | −7.0 | −21.6 | −10.5 | −10.8 | −22.2 | −9.1 | −12.8 | −23.4 | −7.7 | −9.6 | −18.9 |
| 100 | −12.7 | −5.9 | −19.7 | −10.1 | −4.0 | −15.0 | −9.6 | −4.0 | −14.7 | −10.0 | 0.6 | −10.9 |
| 300 | −12.8 | −4.7 | −18.7 | −10.1 | −3.5 | −14.8 | −10.0 | −2.1 | −13.5 | −10.2 | 1.2 | −10.4 |
| 500 | −12.7 | −4.6 | −18.2 | −10.0 | −2.8 | −13.9 | −10.0 | −1.2 | −13.1 | −10.7 | 2.2 | −10.0 |
Figure 2.
Computed binding enthalpies with four different water models (TIP3P, SPC/E, TIP4P-Ew, OPC) for the CB7-B2 system, in the absence and the presence of various NaCl concentrations (100, 300, 500 mM). The black dot represents the experimental value measured in the absence of salt. All calculations used AM1-BCC partial charges for the host and guest, except one calculation at 0 M NaCl, where RESP charges were used along with the TIP3P water model, as indicated in the legend.
The computed binding enthalpies become less favorable (i.e., more positive) with increasing NaCl concentration (Table 1, Figure 2). The largest changes occur between 0 and 100 mM, with smaller changes for successive increases in the salt concentration. The choice of water molecule strongly impacts the sensitivity of the binding enthalpy to salt concentration. Thus, while the change in binding enthalpy between 0 to 100 mM is only 1.9 kcal/mol for TIP3P, the changes for SPC/E, TIP4P-Ew, and OPC are 7–9 kcal/mol (Table 1, Figure 2). Nonetheless, the overall picture of less exothermic binding with increasing salt concentration is broadly consistent with experimental studies of CB7 with various guest molecules; for example, the enthalpy of association of CB7 with two dipeptides becomes less favorable by about 4 kcal/mol when the NaCl concentration increases from 0 to 100 mM68.
Coulombic and Lennard-Jones contributions to computed binding enthalpies
The computed binding enthalpies may be decomposed into physically interpretable terms, due to the additive form of the force field55. Accordingly, Table 1 lists the electrostatic and van der Waals contributions to the binding enthalpies, as reflected in the Coulombic and Lennard-Jones terms, respectively. (The contributions of bond-stretch, bond-angle and dihedral terms, are much smaller; these are not explicitly listed, but their sum may be inferred from the data in the table.) In the absence of dissolved NaCl, both the Coulombic and Lennard-Jones terms contribute strongly (−7 to −13.3 kcal/mol) to the total binding enthalpy, for all water models.
The Coulombic contribution to the binding enthalpy is strongly sensitive to the salt concentration, becoming much less favorable with increasing salt concentration, for all water models. Indeed, it is the Coulombic term which accounts for the salt sensitivity of the overall binding enthalpy evident in Figure 2. The Coulombic term thus accounts for the strikingly reduced salt-sensitivity of the TIP3P results relative to the other three models: whereas raising the salt concentration from 0 to 500 mM makes the Coulombic term more positive by 8.0 to 11.8 kcal/mol for SPC/E, TIP4P-Ew and OPC, the corresponding change for TIP3P is only 2.4 kcal/mol. The largest Coulombic change due to added salt, 11.8 kcal/mol, is observed for the OPC model, but this large Coulombic change is partly cancelled by the opposite shift in its Lennard-Jones term, noted above.
In contrast, the Lennard-Jones term is relatively insensitive to salt concentration. It changes by at most 1 kcal/mol with increasing NaCl, except in the case of the OPC model, where the Lennard-Jones term becomes more favorable by −3 kcal/mol as the NaCl concentration rises from 0 to 500 mM. Another notable difference among the water models is that the Lennard-Jones term for TIP3P is several kcal/mol more negative than that obtained with the other water models, across all salt concentrations.
Dissecting the roles of the water and ion model as determinants of computed binding enthalpies
Each calculation above uses the Na+ and Cl− force field parameters deemed most appropriate for the water model used, as is standard practice34,35. Thus, the TIP3P and TIP4P-Ew models were paired with their own tailored ion models34, here termed Ion3P and Ion4P, respectively; the SPC/E model also was used with its own tailored salt parameters34; and the OPC model was paired with the Ion4P ion model. The latter selection was motivated by the fact that both OPC and TIP4P-Ew are 4-point water models, and the fact that salt parameters tuned for OPC are not yet available. Because of these pairings, the differences in computed binding enthalpy across water models could result from differences in the water models themselves, differences in the ion parameters, or the combination. In order to dissect these dependencies, we ran new calculations for several water models with the “wrong” ion parameters. To limit the number of calculations, results were obtained only for 100 mM NaCl.
We find that, for every water model tested, the Ion3P model yields binding enthalpies which are less sensitive to NaCl concentration than those from the Ion4P model, by 1.3 to 2.4 kcal/mol (Figure 3, and Table S2). At the same time, the TIP3P water model still gives the least salt-sensitive results, even if TIP3P is made maximally sensitive by using Ion4P while other models are made minimally sensitive by using Ion3P. Therefore, the water model plays a greater role than the ion model in determining the salt-dependence of the binding enthalpies, for this system, at least for the models tested here.
Figure 3.
Binding enthalpies computed with mixed combinations of water models and ion parameters as indicated. The results of TIP3P, TIP4P and TIP4P-Ew were computed with their default parameters.
For completeness, we also computed the binding enthalpies with the TIP4P model at 0 M and 100 mM NaCl, with the Ion4P NaCl model. Perhaps not surprisingly, the results match the corresponding TIP4P-Ew results to within ~1 kcal/mol. It is nonetheless worth noting that TIP4P provides the most problematic overestimate of the experimental binding enthalpy at 0mM NaCl of any water model tried here, at −24.3 kcal/mol.
Recently, four new sets of NaCl parameters have been tuned for use with the TIP3P and the TIP4P-Ew water models35. For each of these water models, one set of ion parameters focuses on reproducing the hydration free energies of the ions, and the other focuses on replicating ion-oxygen distances. Enthalpy calculations using these parameters with their corresponding water models give slightly weaker salt effects than those obtained with the Ion3P and Ion4P models. For TIP3P and 100mM NaCl, the computed enthalpies are −20.6 and −20.2 kcal/mol, respectively, while the results are −15.5 and −15.4 kcal/mol for TIP4P-Ew. These values may be compared with −19.7 and −14.7 Kcal/mol for TIP3P and TIP4P-Ew with the Ion3P and Ion4P parameters, respectively.
Displacement of Na+ ions upon binding and the salt-dependence of binding enthalpy
Experimental studies indicate that dissolved cations compete with guest molecules for binding to cucurbiturils36, and crystal structures of cucurbiturils often reveal cations near the portal carbonyl oxygens36. In order to determine whether the salt-dependence of the binding enthalpies computed here could be accounted for by competitive displacement of Na+ ions from the host by the guest, we analyzed the distributions of Na+ ions around CB7 in the simulations. Direct inspection of the animated simulation trajectories revealed that, for the host in solution, Na+ ions are frequently present near the carbonyl oxygens at the portals of CB7, and occasionally enter its binding cavity. However, they are not bound tightly, but instead exchange frequently with those in solution. In contrast, the Cl− ions barely form any direct interactions with the host molecule. In the presence of bound guest, Na+ ions associate less with the complex, though they still tend to lie closer to it than the Cl− ions.
Visualization of the spatial distribution of Na+ ions in and around the CB7 host (Figure 4a) shows peaks in Na+ density at sites between each pair of neighboring carbonyl oxygens at each portal of the host. The degree of localization appears somewhat less for TIP3P than for TIP4P-Ew; the densities computed with the SCP/E and OPC water models are similar to those from TIP4P-Ew (Figure S3). Unlike the other three water models, TIP3P also yields a peak of Na+ density within the binding cavity, although this is less pronounced than those at the carbonyl oxygens.
Figure 4.
(a) Average number density of Na+ around free CB7, contoured at 1.5 Å−3, from simulations with 100 mM NaCl and with TIP3P (left) and TIP4P-Ew (right) water models (The results for the SPC/E and OPC models, which are similar to those for TIP4P-Ew, shown in Figure S3). (b) Average number densities of Na+ (orange) and TIP4P-Ew water (transparent) around free CB7, both conditioned on the presence of a Na+ ion between the leftmost carbonyl oxygens. Contour isovalues are 1.0 Å−3 for Na+ and 4.5 Å−3 for water. Strong spatial perspective is used in the top panel of (b) so that densities at both portals can be seen.
Despite the symmetric appearance of the Na+ densities in Figure 4a, Na+ ions are not normally present simultaneously at all seven sites at each portal. This point is elucidated in Figure 4b, which shows the distribution of Na+ density conditioned on the presence of a Na+ ion at one of the sites between the carbonyl oxygens, for the TIP4P-Ew model; results are similar for the other water models. The presence of one Na+ ion at the left-most site (large orange contour) essentially deletes any density from the four nearest sites, while allowing some density at the two sites across the portal. However, the distribution of Na+ at the other portal is not perturbed much by the conditioning Na+. Interestingly, there are four clear-cut peaks in the density of water molecules around the conditioning Na+ site. These, combined with the two carbonyl oxygens, make for hexa-coordination of the Na+ ion, in a structure is similar to the coordination of a Na+ ion by the host and water molecules in a crystal structure of CB636. It is worth noting that, when B2 is bound to the CB7 molecule, the localization of Na+ around the host is markedly reduced (Figure S4); the displacement of Na+ by the guest is analyzed below.
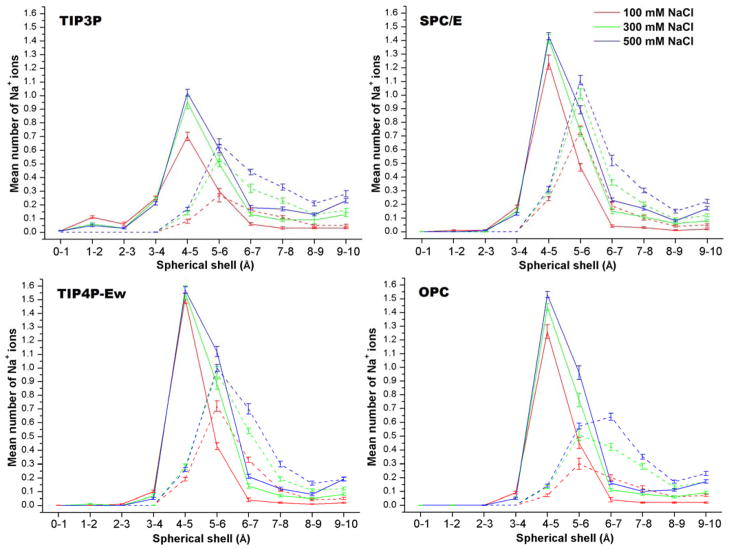
Graphs of Na+ occupancy as a function of distance from the center of CB7 without bound guest show strong peaks in the 4–5 Å range, for all water models (Figure 5, solid lines). This distance corresponds to the sites between the carbonyl oxygens highlighted in Figure 4, and the peak heights range from a low of 0.7 Na+ ions, for TIP3P, to a high of 1.5 Na+, for TIP4P-Ew; the results for SPC/E and OPC are nearer to TIP4P-Ew than to TIP3P. The lower Na+ count for TIP3P than for the other models is consistent with the three-dimensional visualizations in Figure 4. The Na+ occupancy graphs also show that increasing the salt concentration above 100 mM leads to only modest and diminishing increments in Na+ occupancy (Figure 5). The diminution presumably results, at least in part, from growing steric and electrostatic competition among bound Na+ ions.
Figure 5.
Average number of Na+ ions in spherical shells centered in CB7, for free CB7 (solid lines) and on the CB7-B2 complex (dashed lines). Error bars provide the standard errors of the means, computed by blocking analysis (see Methods).
Binding of B2 to CB7 abolishes Na+ occupancy from 0 to 4 Å and markedly lowers the number of Na+ ions in the 4–5 Å region, to 0.24 or less (Figure 5, dashed lines). However, the number of Na+ ions in the shells from 5 to 10 Å is not affected much by binding of B2. This result, combined with visual inspection of the MD trajectories, suggests that the binding of B2 in effect expels most of the Na+ ions previously associated with CB7 into the bulk; i.e. to a distance beyond 10 Å. As a consequence, binding of the neutral B2 guest to CB7 requires overcoming attractive electrostatic interactions between CB7 and Na+ ions localized in and near the CB7 host. In contrast, the dissolved Cl− ions reside in the outermost spherical shells (6–10 Å) whether or not B2 is bound (Figure 6). The displacement by B2 of Na+ ions from favorable sites in and near CB7 is consistent with the decreasingly favorable electrostatic contribution to the binding enthalpy with increasing ionic strength, particularly on going from 0 to 100 mM NaCl (Table 1). Furthermore, the observation that NaCl has less effect on the binding enthalpy in the case of TIP3P, relative to the other water models, is consistent with the observation that CB7 associates with fewer Na+ ions when the TIP3P water model is used.
Figure 6.
Average number of Cl− ions in spherical shells centered on free CB7 (solid lines) and on the CB7-B2 complex (dashed lines). Error bars provide the standard errors of the means, computed by blocking analysis (see Methods).
There is a clear correlation between the number of Na+ ions displaced when B2 binds CB7 and the effect of salt on the binding enthalpy, across all ionic strengths and all water and ion models (Figure 7 and Table S3). The slope of a linear regression fit to these data, about 7 kcal/mol per Na+ ion, represents an estimate of the enthalpy penalty for displacing one Na+ ion. The scatter plot reveals that the reduced salt effect for TIP3P, relative to the other water models, results both from a smaller number of displaced Na+ ions, but also from a reduced enthalpic penalty per Na+ ion, since the TIP3P points lie both to the left of the other points and also below the regression line. One may see that going from the Ion4P to the Ion3P model decreases the number of displaced Na+ ions for the three water models tested; i.e., TIP3P, TIP4P-Ew and OPC. These results are all consistent with the reduced sensitivity of binding enthalpy to ionic strength consistently observed for the TIP3P water model, as well as for the Ion3P model relative to Ion4P (Figure 3). Thus, if fewer Na+ ions are bound to free CB7, then fewer are displaced when B2 binds, and the sensitivity of the binding enthalpy to dissolved NaCl concentration is diminished.
Figure 7.

Scatter plot of the average number of Na+ ions displaced from the 4–5 Å region on binding of B2 to CB7, against the difference in the computed binding enthalpy from that in the absence of dissolved NaCl.
DISCUSSION
Overview
The trends and correlations observed in these calculations of binding enthalpy offer mechanistic insights, and highlight technical issues and opportunities. The results have important implications for the accuracy of binding calculations. First, all the water models lead to overestimates of the experimental binding enthalpy in the absence of salt by −3 to −8 kcal/mol. This strong, consistent bias, as combined with a similar bias observed for CB7 for eight different guest molecules studied with TIP3P and TIP4P-Ew water models55, raises significant concerns regarding the accuracy of the water models and/or the GAFF/AM1-BCC force field parameters used here and in the prior study. It should be emphasized that the tight numerical convergence of these and the prior results leaves the simulation force field as the only credible source of these errors. Given the goal of achieving “chemical accuracy” in drug design calculations—i.e., accuracy to within about 1 kcal/mol—the observation of large errors for these simple host-guest systems highlights the critical need for further improvement in force field accuracy.
On a more encouraging note, the strong sensitivity of the computed binding enthalpies to the concentration of NaCl is in qualitative agreement with experimental data for CB7 binding to other guest molecules68. Analysis of the simulations indicates that this salt-sensitivity results from the displacement by the guest molecule of Na+ ions which were associated with the free host. This mechanism, too, is consistent with prior experimental studies of CB6, which show two sodium ions coordinated to each portal of the cucurbituril without a bound guest36. However, the magnitude of the computed salt effect depends upon the choice of water models and ion parameters. In fact, the difference in the salt effect between models ranges up to 8 kcal/mol. Unfortunately, experimental enthalpies are not available for this host-guest system in salt water, so it is not possible to determine which models are the most reliable in this regard. Nonetheless, it follows from the wide variation among these widely used models that some of them must be significantly inaccurate. As a consequence, further force field work is necessary to ensure reliable modeling of salt effects, not only for host-guest binding, but also, presumably, for biomolecular systems.
Factors influencing the computed binding enthalpies
Because of the complexity and nonlinearity of the systems studied here, it would be difficult to establish a definitive and complete mechanistic explanation for how the variations among the water and salt models considered here lead to the observed variations in salt effects. Nonetheless, some insights may be gained. A central observation in this regard is that the computed decrement in the binding enthalpy due to the dissolved salt is roughly proportional to the number of Na+ ions displaced from CB7 when the neutral guest binds (Figure 8a). Thus, the variation in salt effect across water and ion models largely reflects variations in the affinity of CB7 for Na+, rather than variations in the enthalpy contribution per Na+ ion displaced. The association of Na+ ions with the electronegative carbonyl oxygens of the host is presumably driven largely by electrostatic interactions, which diminish with increasing dielectric constant. Accordingly, it is physically reasonable that the mean number of Na+ ions in the major peak at 4–5 Å (Figure 5) correlates with the reciprocal of the dielectric constant of the water model, as evident in Figure 8. On the other hand, the OPC model would lie noticeably above a regression line based on the other three water models. We speculate that this difference results from the fact that the partial charges of the OPC model are all situated near the center of the water molecule, while the hydrogen charges of the other three models are more peripheral33. The more central placement of OPC’s charges could weaken its electrostatic interactions with polar solutes while maintaining a realistic bulk dielectric constant.
Figure 8.
Scatter plot of the mean number of Na+ ions in a 4–5 Å spherical shell centered on free CB7, versus (a)the reciprocal of the dielectric constant of the water model used in the simulation33; (b) the hydration free energy of Na+ for the TIP3P, SPC/E, and TIP4P-Ew water models34. Data on the y-axis pertain to simulations at 100 mM NaCl, but similar results are obtained at higher concentrations.
Alternatively, one may interpret the present results in terms of the hydration free energies of Na+ ions across the various models: the more favorable the interactions between Na+ and water, the less the tendency of Na+ to bind CB7, since the ion is partly dehydrated upon binding. In fact, the hydration free energies of Na+, using Jensen and Jorgensen’s ion parameters34, correlate well with the mean number of Na+ ions in the 4–5 Å shell of CB7 (Figure 8b) for TIP3P, SPC/E and TIP4P-Ew. (Corresponding hydration data are not yet available for the more recent OPC model.) At the same time, it is not a given that a water model which gives a greater Na+ dehydration energy will lead to greater Na+ binding to CB7, because these water molecules might also stick more tightly to CB7, making them harder to displace when Na+ binds. Similarly, the increased affinity for CB7 of Na+ modeled with Ion4P parameters relative to Ion3P parameters (see above) reflects the balance of the difference in these models’ interactions with water versus CB7.
Conclusions
Water models are typically adjusted based on the properties of pure water30–33, and ion parameters based on hydration free energies, ion-water interactions, and the properties of ion pairs and salt crystals34,35. Here, we find that water and ion models which all perform reasonably well against these reference data yield significant errors relative to experiment and widely different salt effects when used to compute host-guest binding enthalpies. We conclude that calculations of binding thermodynamics and their associated salt effects can surface operational differences among force fields that are not apparent in calculations of other experimental observables. Given the profound biological and medical importance of specific binding in physiological salt solutions, these differences have implications for practical applications like computer-aided drug design. We would therefore argue for beginning to incorporate the broad set of experimental host-guest binding data11,36–48 into the reference dataset used to assess and parameterize force fields.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (NIH) for grant GM61300. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. M.K.G. has an equity interest in, and is a cofounder and scientific advisor of VeraChem LLC.
Footnotes
Notes
The authors declare no competing financial interest.
Convergence and Numerical Error Analysis of Simulations; supplemental tables and figures about binding enthalpies computed with various combinations of water models and ion parameters, mean Na+ density around free CB7 and CB7-B2, mean number of Na+ ions in a 4–5 Å spherical shell centered on CB7. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Melander W, Horváth C. Salt Effect on Hydrophobic Interactions in Precipitation and Chromatography of Proteins: An Interpretation of the Lyotropic Series. Arch Biochem Biophys. 1977;183:200–215. doi: 10.1016/0003-9861(77)90434-9. [DOI] [PubMed] [Google Scholar]
- 2.Kumar CV, Turner RS, Asuncion EH. Groove Binding of a Styrylcyanine Dye to the DNA Double Helix: The Salt Effect. J Photochem Photobiol A Chem. 1993;74:231–238. [Google Scholar]
- 3.Cacace MG, Landau EM, Ramsden JJ. The Hofmeister Series: Salt and Solvent Effects on Interfacial Phenomena. Q Rev Biophys. 1997;30:241–277. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 4.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.Kozlov AG, Lohman TM. Calorimetric Studies of E. Coli SSB Protein-Single-Stranded DNA Interactions. Effects of Monovalent Salts on Binding Enthalpy. J Mol Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien R, DeDecker B, Fleming KG, Sigler PB, Ladbury JE. The Effects of Salt on the TATA Binding Protein-DNA Interaction from a Hyperthermophilic Archaeon. J Mol Biol. 1998;279:117–125. doi: 10.1006/jmbi.1998.1743. [DOI] [PubMed] [Google Scholar]
- 7.Szeltner Z, Polgár L. Conformational Stability and Catalytic Activity of HIV-1 Protease Are Both Enhanced at High Salt Concentration. J Biol Chem. 1996;271:5458–5463. doi: 10.1074/jbc.271.10.5458. [DOI] [PubMed] [Google Scholar]
- 8.Lohman TM, Overman LB, Ferrari ME, Kozlov AG. A Highly Salt-Dependent Enthalpy Change for Escherichia Coli SSB Protein-Nucleic Acid Binding due to Ion-Protein Interactions. Biochemistry. 1996;35:5272–5279. doi: 10.1021/bi9527606. [DOI] [PubMed] [Google Scholar]
- 9.Remko M, Rode BM. Effect of Metal Ions (Li+, Na+, K+, Mg2+, Ca2+, Ni2+, Cu2+, and Zn2+) and Water Coordination on the Structure of Glycine and Zwitterionic Glycine. J Phys Chem A. 2006;110:1960–1967. doi: 10.1021/jp054119b. [DOI] [PubMed] [Google Scholar]
- 10.Remko M, Fitz D, Rode BM. Effect of Metal Ions (Li+, Na+, K+, Mg2+, Ca2+, Ni2+, Cu2+ and Zn2+) and Water Coordination on the Structure and Properties of L-Histidine and Zwitterionic L-Histidine. Amino Acids. 2010;39:1309–1319. doi: 10.1007/s00726-010-0573-8. [DOI] [PubMed] [Google Scholar]
- 11.Gibb CLD, Oertling EE, Velaga S, Gibb BC. Thermodynamic Profiles of Salt Effects on a Host–Guest System: New Insight into the Hofmeister Effect. J Phys Chem B. 2015;119:5624–5638. doi: 10.1021/acs.jpcb.5b01708. [DOI] [PubMed] [Google Scholar]
- 12.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weinerl P. A New Force Field for Molecular Mechanical Simulation of Nucleic Acids and Proteins. J Am Chem Soc. 1984;106:765–784. [Google Scholar]
- 13.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 14.Mackerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and Testing of a General Amber Force Field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 16.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 17.Ponder JW, Case DA. Force Fields for Protein Simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 18.Cheatham TE, Case DA. Twenty-Five Years of Nucleic Acid Simulations. Biopolymers. 2013;99:969–977. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickson CJ, Madej BD, Skjevik ÅA, Betz RM, Teigen K, Gould IR, Walker RC. Lipid14: The Amber Lipid Force Field. J Chem Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, De Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 21.Maisuradze GG, Senet P, Czaplewski C, Liwo A, Scheraga HA. Investigation of Protein Folding by Coarse-Grained Molecular Dynamics with the UNRES Force Field. J Phys Chem A. 2010;114:4471–4485. doi: 10.1021/jp9117776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott WRP, Hünenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Krüger P, van Gunsteren WF. The GROMOS Biomolecular Simulation Program Package. J Phys Chem A. 1999;103:3596–3607. [Google Scholar]
- 23.Jorgensen WL, Tirado-Rives J. The OPLS Potential Functions for Proteins. Energy Minimizations for Crystals of Cyclic Peptides and Crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 24.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 25.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 26.Chapman DE, Steck JK, Nerenberg PS. Optimizing Protein-Protein van Der Waals Interactions for the AMBER ff9x/ff12 Force Field. J Chem Theory Comput. 2014;10:273–281. doi: 10.1021/ct400610x. [DOI] [PubMed] [Google Scholar]
- 27.Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF. A Biomolecular Force Field Based on the Free Enthalpy of Hydration and Solvation: The GROMOS Force-Field Parameter Sets 53A5 and 53A6. J Comput Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 28.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved Side-Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins Struct Funct Bioinforma. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 31.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an Improved Four-Site Water Model for Biomolecular Simulations: TIP4P-Ew. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 32.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 33.Izadi S, Anandakrishnan R, Onufriev AV. Building Water Models_: A Different Approach. J Phys Chem Lett. 2014;5:3863–3871. doi: 10.1021/jz501780a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joung IS, Cheatham TE. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Song LF, Merz KM. Systematic Parameterization of Monovalent Ions Employing the Nonbonded Model. J Chem Theory Comput. 2015;11:1645–1657. doi: 10.1021/ct500918t. [DOI] [PubMed] [Google Scholar]
- 36.Jeon YM, Kim J, Whang D, Kim K. Molecular Container Assembly Capable of Conrolling Binding and Release of Its Guest Molecules: Reversible Encapsulation of Organic Molecules in Sodium Ion Complexed Cucurbituril. J Am Chem Soc. 1996;118:9790–9791. [Google Scholar]
- 37.Rekharsky MV, Mori T, Yang C, Ko YH, Selvapalam N, Kim H, Sobransingh D, Kaifer AE, Liu S, Isaacs L, Chen W, Moghaddam S, Gilson MK, Kim K, Inoue Y. A Synthetic Host-Guest System Achieves Avidin-Biotin Affinity by Overcoming Enthalpy-Entropy Compensation. Proc Natl Acad Sci U.S.A. 2007;104:20737–20742. doi: 10.1073/pnas.0706407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houk KN, Leach AG, Kim SP, Zhang X. Binding Affinities of Host-Guest, Protein-Ligand, and Protein-Transition-State Complexes. Angew Chemie - Int Ed. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Ruspic C, Mukhopadhyay P, Chakrabarti S, Zavalij PY, Isaacs L. The Cucurbit[n]uril Family: Prime Components for Self-Sorting Systems. J Am Chem Soc. 2005;127:15959–15967. doi: 10.1021/ja055013x. [DOI] [PubMed] [Google Scholar]
- 40.Gibb CLD, Gibb BC. Anion Binding to Hydrophobic Concavity Is Central to the Salting-in Effects of Hofmeister Chaotropes. J Am Chem Soc. 2011;133:7344–7347. doi: 10.1021/ja202308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choudhury R, Gupta S, Da Silva JP, Ramamurthy V. Deep-Cavity Cavitand Octa Acid as a Hydrogen Donor: Photofunctionalization with Nitrenes Generated from Azidoadamantanes. J Org Chem. 2013;78:1824–1832. doi: 10.1021/jo301499t. [DOI] [PubMed] [Google Scholar]
- 42.Piñeiro Á, Banquy X, Pérez-Casas S, Tovar E, García A, Villa A, Amigo A, Mark AE, Costas M. On the Characterization of Host-Guest Complexes: Surface Tension Calorimetry and Molecular Dynamics of Cyclodextrins with a Non-Ionic Surfactant. J Phys Chem B. 2007;111:4383–4392. doi: 10.1021/jp0688815. [DOI] [PubMed] [Google Scholar]
- 43.Witlicki EH, Hansen SW, Christensen M, Hansen TS, Nygaard SD, Jeppesen JO, Wong EW, Jensen L, Flood AH. Determination of Binding Strengths of a Host-Guest Complex Using Resonance Raman Scattering. J Phys Chem A. 2009;113:9450–9457. doi: 10.1021/jp905202x. [DOI] [PubMed] [Google Scholar]
- 44.Inoue Y, Hakushi T, Liu Y, Tong LH, Shen BJ, Jin DS. Thermodynamics of Molecular Recognition by Cyclodextrins 1. Calorimetric Titration of Inclusion Complexation of Naphthalenesulfonates with Alpha.-, Beta.-, and Gamma.-Cyclodextrins: Enthalpy-Entropy Compensation. J Am Chem Soc. 1993;115:475–481. [Google Scholar]
- 45.Freeman WA, Mock WL, Shih NY. Cucurbituril. J Am Chem Soc. 1981;17:7367–7368. [Google Scholar]
- 46.Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Cucurbituril Homologues and Derivatives: New Opportunities in Supramolecular Chemistry. Acc Chem Res. 2003;36:621–630. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]
- 47.Kim HJ, Jeon WS, Ko YH, Kim K. Inclusion of Methylviologen in cucurbit[7]uril. Proc Natl Acad Sci U S A. 2002;99:5007–5011. doi: 10.1073/pnas.062656699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeon YJ, Kim SY, Ko YH, Sakamoto S, Yamaguchi K, Kim K. Novel Molecular Drug Carrier: Encapsulation of Oxaliplatin in cucurbit[7]uril and Its Effects on Stability and Reactivity of the Drug. Org Biomol Chem. 2005;3:2122–2125. doi: 10.1039/b504487a. [DOI] [PubMed] [Google Scholar]
- 49.Moghaddam S, Inoue Y, Gilson MK. Host Guest Complexes with Protein Ligand-like Affinities: Computational Analysis and Design. J Am Chem Soc. 2009;131:4012–4021. doi: 10.1021/ja808175m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moghaddam S, Yang C, Rekharsky M, Ko YH, Kim K, Inoue Y, Gilson MK. New Ultrahigh Affinity Host -Guest Complexes of Cucurbit[7]uril with Bicyclo[2.2.2] Octane and Adamantane Guests: Thermodynamic Analysis and Evaluation of M2 Affinity Calculations. J Am Chem Soc. 2011;133:3570–3581. doi: 10.1021/ja109904u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ. The SAMPL2 Blind Prediction Challenge: Introduction and Overview. J Comput Aided Mol Des. 2010;24:259–279. doi: 10.1007/s10822-010-9350-8. [DOI] [PubMed] [Google Scholar]
- 52.Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK. Blind Prediction of Host-Guest Binding Affinities: A New SAMPL3 Challenge. J Comput Aided Mol Des. 2012;26:475–487. doi: 10.1007/s10822-012-9554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muddana HS, Fenley AT, Mobley DL, Gilson MK. The SAMPL4 Host-Guest Blind Prediction Challenge: An Overview. J Comput Aided Mol Des. 2014;28:305–317. doi: 10.1007/s10822-014-9735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickstrom L, He P, Gallicchio E, Levy RM. Large Scale Affinity Calculations of Cyclodextrin Host-Guest Complexes: Understanding the Role of Reorganization in the Molecular Recognition Process. J Chem Theory Comput. 2013;9:3136–3150. doi: 10.1021/ct400003r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fenley AT, Henriksen NM, Muddana HS, Gilson MK. Bridging Calorimetry and Simulation through Precise Calculations of Cucurbituril Guest Binding Enthalpies. J Chem Theory Comput. 2014;10:4069–4078. doi: 10.1021/ct5004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rekharsky MV, Inoue Y. Complexation Thermodynamics of Cyclodextrins. Chem Rev. 1998;98:1875–1918. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- 57.Ong W, Kaifer AE. Salt Effects on the Apparent Stability of the Cucurbit[7]uril-Methyl Viologen Inclusion Complex. J Org Chem. 2004;69:1383–1385. doi: 10.1021/jo035030+. [DOI] [PubMed] [Google Scholar]
- 58.Tang H, Fuentealba D, Ko YH, Selvapalam N, Kim K, Bohne C. Guest Binding Dynamics with cucurbit[7]uril in the Presence of Cations. J Am Chem Soc. 2011;133:20623–20633. doi: 10.1021/ja209266x. [DOI] [PubMed] [Google Scholar]
- 59.Jeon WS, Moon K, Park SH, Chun H, Ko YH, Lee JY, Lee ES, Samal S, Selvapalam N, Rekharsky MV, Sindelar V, Sobransingh D, Inoue Y, Kaifer AE, Kim K. Complexation of Ferrocene Derivatives by the cucurbit[7]uril Host: A Comparative Study of the Cucurbituril and Cyclodextrin Host Families. J Am Chem Soc. 2005;127:12984–12989. doi: 10.1021/ja052912c. [DOI] [PubMed] [Google Scholar]
- 60.Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Luchko T, Luo R, Madej B, Merz KM, DMY, PAK . AMBER 2015. University of California; San Francisco: 2015. [Google Scholar]
- 61.Flyvbjerg H, Petersen HG. Error Estimates on Averages of Correlated Data. J Chem Phys. 1989;91:461. [Google Scholar]
- 62.Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: I. Method. J Comput Chem. 2000;21:132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 63.Jakalian A, Jack DB, Bayly CI. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 64.Bayly CCI, Cieplak P, Cornell WD, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 65.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Had M, DJF . Gaussian 09, Revision D.01. Gaussian, Inc; Wallingford CT: 2009. [Google Scholar]
- 66.Dennington Roy, Keith Todd, JM . GaussView, Version 5. Semichem Inc; Shawnee Mission, KS: 2009. [Google Scholar]
- 67.Muddana HS, Gilson MK. Calculation of Host-Guest Binding Affinities Using a Quantum-Mechanical Energy Model. J Chem Theory Comput. 2012;8:2023–2033. doi: 10.1021/ct3002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rekharsky MV, Yamamura H, Ko YH, Selvapalam N, Kim K, Inoue Y. Sequence Recognition and Self-Sorting of a Dipeptide by cucurbit[6]uril and cucurbit[7]uril. Chem Commun (Camb) 2008;(19):2236–2238. doi: 10.1039/b719902c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.