Abstract
Mountain gorillas are an endangered great ape subspecies and a prominent focus for conservation, yet we know little about their genomic diversity and evolutionary past. We sequenced whole genomes from multiple wild individuals and compared the genomes of all four Gorilla subspecies. We found that the two eastern subspecies have experienced a prolonged population decline over the past 100,000 years, resulting in very low genetic diversity and an increased overall burden of deleterious variation. A further recent decline in the mountain gorilla population has led to extensive inbreeding, such that individuals are typically homozygous at 34% of their sequence, leading to the purging of severely deleterious recessive mutations from the population. We discuss the causes of their decline and the consequences for their future survival.
Mountain gorillas came to the attention of the western world as recently as 1902 (1) and survive today only in two small and critically endangered populations in central Africa (2). Current estimates place their total population at around 800, of which just over half are in the Virunga volcanic mountain range on the borders of Rwanda, Uganda, and the Democratic Republic of Congo (Fig. 1A) (3, 4). Mountain gorillas face a number of threats to their continued existence (5), and their survival became an urgent concern during the 1960s, when a precipitous decline in their numbers appeared to put them at grave risk of extinction (6). Active conservation efforts have since reversed the decline (3), but a loss of genetic diversity associated with small population size may still threaten their long-term viability. Infectious diseases such as Ebola, the cause of substantial recent mortality in the western lowland population (7), represent a particularly serious threat (8).
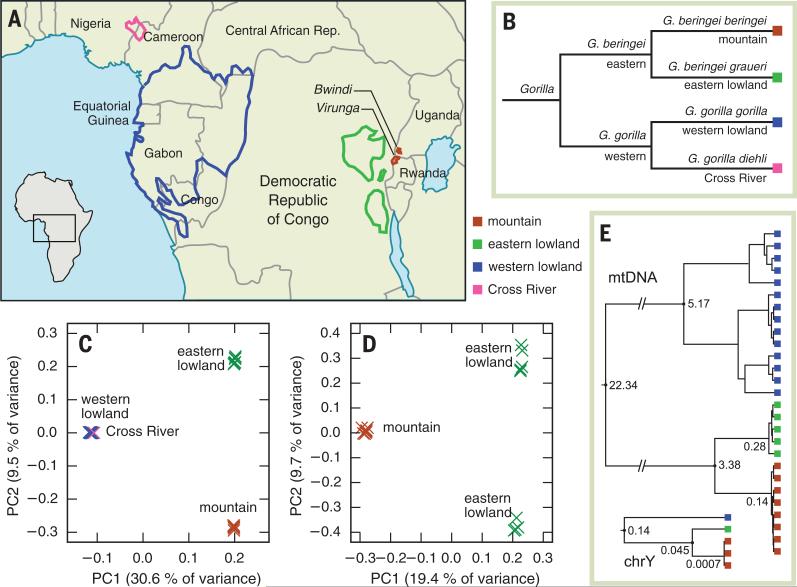
Fig. 1. Geography, taxonomy and genetic structure of gorilla species.
(A) Distribution of gorilla subspecies (2). (B) Gorilla taxonomy. (C) PCA plot of SNP data for all four gorilla subspecies. (D) PCA plot of SNP data from mountain and eastern lowland gorilla samples only. (E) mtDNA and Y-chromosomal phylogenies. Node heights are in units of substitutions per base pair; each tree is drawn to a separate scale.
Despite extensive study of mountain gorillas in the field, few genetic analyses have been carried out, and these have been confined to mitochondrial sequences and a limited number of autosomal loci (9–11). Unlike the other great apes (12), mountain gorillas have not been studied on a genome-wide scale, which is key to understanding their biology, evolution, and relationship to sister taxa (Fig. 1B). It is also important in assessing their current status and in forming strategies for future conservation efforts. Phenotypic indicators of inbreeding such as syndactyly have been reported (13, 14), but the full genetic impact of their decline is unknown. Additionally, the severe population bottleneck experienced by mountain gorillas provides an opportunity to study processes that may have played a recurring role in hominin evolution and extinction.
We performed whole-genome sequencing and analysis for 13 eastern gorillas, comprising seven mountain gorillas from the Virunga volcanoes region and six eastern lowland gorillas. The mountain gorillas originate from both sides of the Virunga massif, including from three groups spanning a densely populated sector on the Rwandan side, and plausibly represent genetic variation and ancestry in the Virunga population at large (15). Sequencing was performed to an average 26×depth (table S1), and combining these data with published gorilla genome sequences (12) yielded a total data set of 44 samples spanning all four gorilla subspecies (table S2).
Sequences were aligned to the gorilla reference genome (16), and several tools were used to call variants across samples (15). Comparing populations, we found that genetic diversity was lower in the eastern species than in the western by a factor of 2 to 3 (table S3), with mean autosomal heterozygosity (frequency of between-chromosomal differences) of 6.5 × 10−4 per base pair in mountain and eastern lowland gorillas, versus 1.9 × 10−3 per base pair in the western lowland population. These values are consistent with the small reported census population sizes of eastern gorillas relative to those in the west (2). A detailed analysis of variation in mitochondrial DNA (mtDNA) showed little diversity in mountain gorillas, with only three haplotypes in total differing by just one to three mutations (15). A similar situation was observed on the Y chromosome, with only two sites differentiating the three male mountain gorillas (15).
A principal components analysis (PCA) of 11,743,407 single-nucleotide polymorphisms (SNPs) across all samples (Fig. 1C) showed a hierarchical structure consistent with the accepted Gorilla taxonomy and the geographical distribution of these populations. In particular, the separation between eastern lowland and mountain gorilla samples confirms them as genetically distinct populations. Another PCA focusing on the eastern species only (Fig. 1D) revealed no substructure within the mountain gorillas sampled here, but did show a separation of eastern lowland gorillas into two subgroups, which may reflect structure within the subspecies as a whole or may only be a feature of the individuals we have sampled. Similar patterns are also found in a genetic structure analysis using the program ADMIXTURE (fig. S3).
Copy number variation (CNV) among gorillas was assessed with a read depth–based approach (17), and the number of fixed loss and gain events between populations reflects the accepted taxonomy (table S5), although eastern lowland and mountain gorillas showed an excess of shared fixed deletions relative to western lowland gorillas, potentially reflecting a small ancestral population prior to their divergence. Diversity patterns at polymorphic CNVs also resembled those seen in SNPs (fig. S8) (15).
Accurate population monitoring and management are of central importance for conservation, and we cataloged 25,628 autosomal and 89 mtDNA ancestry-informative SNP markers (AIMs) uniquely identifying eastern lowland and mountain gorillas (as sampled here), as well as 1127 lineage-specific CNV events (data files S1 and S2) (15).
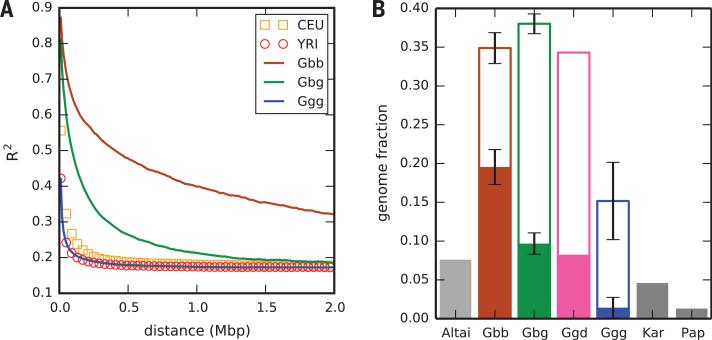
We found that genome-wide linkage disequilibrium (LD) varies markedly between gorilla populations (Fig. 2A). Both eastern subspecies—particularly mountain gorillas—show much more extensive LD than western lowland gorillas, in whom the decay of LD with genomic distance is similar to that in African humans (15). These patterns reflect differing demographic histories and suggest a recent population bottleneck in the two eastern subspecies.
Fig. 2. Linkage disequilibrium and homozygosity in gorillas.
(A) LD decay (15) in gorilla and human populations. Human samples are Utah residents with European ancestry (CEU) or Yoruba in Ibadan, Nigeria (YRI). (B) Mean per-sample genome fractions found in homozygous tracts. Open bars show total fractions for mountain (Gbb), eastern lowland (Gbg), Cross River (Ggd), and western lowland (Ggg) gorillas; solid bars show fractions in tracts of length 2.5 to 10 Mb (gorillas) or 2.5 to 10 cM in an Altai Neandertal and two human individuals [Karitiana (Kar) and Papuan (Pap)] (19). Error bars are ±1 SD.
An analysis of chromosomal sequence sharing within individuals (genomic tracts of homozygosity) provided insight into recent ancestry and a measure of parental relatedness. Within mountain and eastern lowland gorilla individuals, chromosomes are typically homozygous over one-third of their length (on average 34.5% and 38.4%, respectively) (Fig. 2B), much higher than in western lowland individuals (13.8%) and exceeding even the most inbred human populations (18). We observed longer tracts in the eastern species, particularly in mountain gorillas, and a clear distinction in tract length distribution between eastern lowland and mountain gorillas (fig. S15) (15). Very long tracts (2.5 to 10 Mb) are particularly indicative of recent inbreeding, and homozygosity on this scale in mountain gorillas exceeds not only that in other gorilla populations but also that observed in the Altai Neandertal (19), consistent with parental relatedness equivalent to that between two half-siblings (19). These data suggest that mountain gorillas may have experienced several recent generations of close inbreeding. We also examined haplotype matching between individuals, finding subclustering among eastern lowland gorillas consistent with that seen in Fig. 1D; individuals in one cluster displayed a 10% higher mean level of sequence sharing than those in the other (fig. S16).
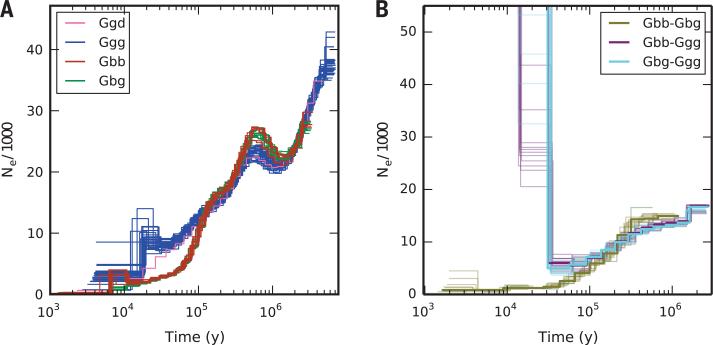
Whole-genome data also enabled us to explore the longer-term demographic history of these populations. Effective population size (Ne) in each of the four subspecies, as inferred by the PSMC algorithm (20), has varied over time (Fig. 3A), and we note the decline of Ne particularly in the eastern species within the past 100,000 years. The most recent values of inferred Ne are just 273 ± 54 in mountain gorillas and 290 ± 18 in eastern lowland gorillas. Ne, which reflects genetic diversity, is usually lower than census population size; however, in 1981 the estimated census size in the Virunga region was as low as 254 (3).
Fig. 3. Ancestral effective population size and gene flow between gorilla populations.
(A) Inferred effective population size (Ne) history for each of the samples studied. (B) Cross-population Ne history, based on paired male X-chromosomal sequences. Both plots are scaled using a generation time of 19.3 years and an autosomal mutation rate of 1.25 × 10−8 per base pair per generation.
As judged from the date when inferred Ne began to differ, the divergence of eastern and western gorillas began at least 150,000 years ago, but a more direct analysis using male X-chromosomal sequences suggests that they exchanged genetic material until around 20,000 years ago (Fig. 3B) (15). Given that this also coincides with a notable decline in western lowland gorilla Ne (Fig. 3A), it may be that environmental changes during the Last Glacial Maximum (26,000 to 19,000 years ago), when dry savannah replaced tropical forest over much of the Congo basin (21), triggered a collapse in the western population and complete separation of the two species. Indeed, changes in forestation across the center of the continent over the past 150,000 years may have had a substantial influence on speciation and diversity in African apes more generally, including ancient humans (12).
We found no evidence for more recent east-west genetic contact, and D-statistic analysis provided no strong support for differences in east-west gene flow between subspecies pairs in either species (table S17).
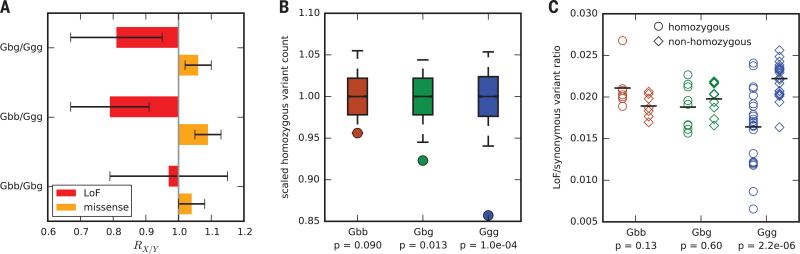
These evolutionary differences between subspecies allowed us to investigate the relationship between demography and selection at protein-coding loci. To facilitate this, we classified coding sequence variants into three groups: synonymous, missense, and loss-of-function (LoF) (15). Accounting for potential reference bias, we found a relatively larger number of derived missense alleles in eastern gorillas, particularly in mountain gorillas, relative to western lowland gorillas (Fig. 4A). This is consistent with long-term differences in Ne (Fig. 3A), because selection in a larger population is more effective at removing moderately deleterious variants. In contrast, we found that mountain gorillas have relatively fewer LoF variants than western lowland gorillas. This is again consistent with low Ne and inbreeding, wherein alleles are more frequently exposed in a homozygous state, and because LoF variants are more likely to be strongly deleterious, they are less likely to persist in the population even if recessive (22). Further evidence for the purging of severely deleterious mutations from mountain and eastern lowland gorillas comes from the lack of any deficit of homozygous LoF genotypes in mountain gorillas (Fig. 4B) and the observation that LoF variants in both subspecies occur at the same rate in homozygous and heterozygous tracts (Fig. 4C) (15).
Fig. 4. The genetic burden of missense mutations and purging of LoF mutations in eastern gorillas.
(A) Relative number of derived alleles at LoF (red) and missense (orange) sites that are frequent in one population and not another (15). Error bars represent ±2 SD. (B) Circles indicate the scaled number of LoF variant sites in each population where at least one sample is homozygous for the derived allele. Boxplots show distributions of the same statistic for matched samples of synonymous sites (15); whiskers show 5th and 95th percentiles; P values are the proportion of each sample distribution smaller than the corresponding LoF count. (C) Circles show the rate of LoF variants relative to synonymous variants in homozygous tracts for each sample; diamonds show the same ratio in nonhomozygous regions. Horizontal bars indicate population means; P values for each subspecies correspond to a Kolmogorov-Smirnov test for difference in distribution between homozygous and nonhomozygous regions.
Finally, we looked at variation in genes using function inferred from homology, focusing on possible selection and adaptation in mountain gorillas. Such adaptation might be expected from the fact that mountain gorillas range over high altitudes (1500 to 4000 m), with consequences for diet, morphology, and physiology (23). However, we found no significant enrichment in any functional category of genes, although there are interesting examples related to nervous system morphology, immunoglobulin quantity, and red blood cell morphology (15). Mountain gorillas carry a significant excess of variants in genes associated with blood coagulation in humans (fig. S21), perhaps linked to high-altitude living (15). We also identified variants associated with cardiomyopathy, including in one deceased individual (Kaboko) in whom post mortem analysis revealed evidence of muscular hypertrophy (15). Cardiovascular disease has been identified as a notable cause of death in captive western lowland gorillas (24).
Concern about the survival of mountain gorillas is focused largely on the threat from human encroachment on their habitat (2, 5), and their severe recent population decline may be responsible for the high level of inbreeding that we observe. An increased burden of deleterious mutation and low genetic diversity—including at the major histocompatibility locus, of central importance to the immune system (fig. S22)—have likely reduced their resilience to environmental change and pathogen evolution. However, the origins of this condition extend far into their history, because both eastern subspecies have experienced a long decline over tens of millennia. Indeed, the demographic histories of mountain and eastern lowland gorillas (Fig. 3A) bear unhappy resemblance to similar histories inferred from Neandertals before their disappearance (19).
Nonetheless, the same evidence suggests that such a fate is not inevitable: These subspecies have survived for thousands of generations at low population levels and may have developed physiological and behavioral strategies to mitigate inbreeding, such as natal dispersal and gene flow between isolated populations (25). The purging of strongly deleterious mutations may also be an important factor. Our findings reinforce the imperative to sustain conservation efforts that have helped to keep mountain gorillas from the brink of extinction, and provide a genomic resource for future conservation and research.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Institut Congolais pour la Conservation de la Nature and the Rwandan Development Board for the use of samples; J. Ramer, D. Zimmerman, J. F. Kinani, J. Bosco Noheri, M. Bahizi, J. P. Lukasa, J. Iyanya, M. Kabuyaya, E. Kambale, and J. Sohl of Gorilla Doctors for sample collection and processing; and M. Pollard, P. Hallast, M. Jobling, and L. Lowenstine for assistance. Supported by Royal Society grant RG130105 (A.S.), Wellcome Trust grants 098051 (Q.A., Y.C., V.N., L.P., M.A.Q., M.S., C.T.-S., Y.X., B.Y.) and 099769/Z/12/Z (V.N.), NIH grant HG002385 (E.E.E.), a European Research Council Starting Grant (260372), and Ministerio de Ciencia e Innovacion grant BFU2011-28549 (T.M.-B.). E.E.E. is an investigator of the Howard Hughes Medical Institute. Accession numbers for sequence data are given in table S1. Gorilla blood samples sequenced in this study were collected and transferred in compliance with CITES legislation and subject to a material transfer agreement between the Mountain Gorilla Veterinary Project and the Office Rwandais du Tourisme et des Parcs Nationaux. D.N.C. acknowledges support from BIOBASE GmbH through a license agreement with Cardiff University; E.E.E. is a member of the external scientific advisory board for DNAnexus Inc.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/348/6231/242/suppl/DC1
Materials and Methods
Figs. S1 to S22
Tables S1 to S18
Data files S1 and S2
References (26–62)
REFERENCES AND NOTES
- 1.Schaller GB. The Year of the Gorilla. Univ. of Chicago Press; Chicago: 2010. p. 7. [Google Scholar]
- 2.Robbins M, et al. Gorilla beringei ssp. beringei (listing in IUCN Red List of Threatened Species. 2008 www.iucnredlist.org/details/39999/0.
- 3.Gray M, et al. Biol. Conserv. 2013;158:230–238. [Google Scholar]
- 4.Guschanski K, et al. Biol. Conserv. 2009;142:290–300. [Google Scholar]
- 5.Harcourt AH. Biol. Conserv. 1996;75:165–176. [Google Scholar]
- 6.Weber AW, Vedder A. Biol. Conserv. 1983;26:341–366. [Google Scholar]
- 7.Le Gouar PJ, et al. PLOS ONE. 2009;4:e8375. doi: 10.1371/journal.pone.0008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.IUCN Ebola outbreak highlights critical links between biodiversity loss and human health, says IUCN’s Wildlife Health Specialist Group. 2014 www.iucn.org/?18439.
- 9.Garner KJ, Ryder OA. Mol. Phylogenet. Evol. 1996;6:39–48. doi: 10.1006/mpev.1996.0056. [DOI] [PubMed] [Google Scholar]
- 10.Jensen-Seaman MI, Kidd KK. Mol. Ecol. 2001;10:2241–2247. doi: 10.1046/j.0962-1083.2001.01365.x. [DOI] [PubMed] [Google Scholar]
- 11.Roy J, et al. Biol. Lett. 2014;10:2014.0811. [Google Scholar]
- 12.Prado-Martinez J, et al. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fossey D. Gorillas in the Mist. Harcourt Brace; New York: 1983. p. 72. [Google Scholar]
- 14.Routh A, Sleeman J. Proceedings of the British Veterinary Zoological Society. Howletts and Port Lympne Wild Animal Parks; Kent: Jun, 1997. pp. 14–15.pp. 22–25. [Google Scholar]
- 15.See supplementary materials on Science Online.
- 16.Scally A, et al. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alkan C, et al. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pemberton TJ, et al. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prüfer K, et al. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anhuf D, et al. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2006;239:510–527. [Google Scholar]
- 22.Glémin S. Evolution. 2003;57:2678–2687. doi: 10.1111/j.0014-3820.2003.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 23.Watts DP. Am. J. Primatol. 1984;7:323–356. doi: 10.1002/ajp.1350070403. [DOI] [PubMed] [Google Scholar]
- 24.McManamon R, Lowenstine L. Fowler's Zoo and Wild Animal Medicine. Vol. 7. Elsevier; St. Louis: 2012. pp. 408–415. [Google Scholar]
- 25.Pusey A, Wolf M. Trends Ecol. Evol. 1996;11:201–206. doi: 10.1016/0169-5347(96)10028-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.