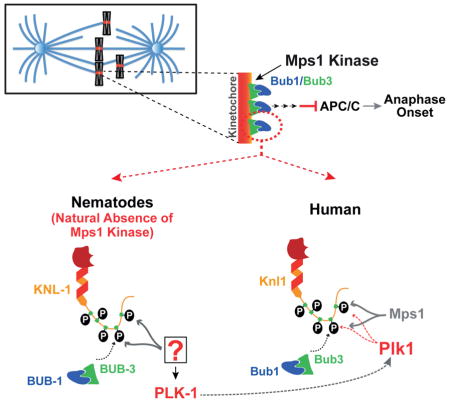
SUMMARY
The spindle checkpoint safeguards against chromosome loss during cell division by preventing anaphase onset until all chromosomes are attached to spindle microtubules. Checkpoint signal is generated at kinetochores, the primary attachment site on chromosomes for spindle microtubules. Mps1 kinase initiates checkpoint signaling by phosphorylating the kinetochore-localized scaffold protein Knl1 to create phospho-docking sites for Bub1/Bub3. Mps1 is widely conserved but is surprisingly absent from the nematode lineage. Here, we show that PLK-1, which targets a similar substrate motif as Mps1, functionally substitutes for Mps1 in C. elegans by phosphorylating KNL-1 to direct BUB-1/BUB-3 kinetochore recruitment. This finding led us to re-examine checkpoint initiation in human cells, where we found that Plk1 co-inhibition significantly reduced Knl1 phosphorylation and Bub1 kinetochore recruitment relative to Mps1 inhibition alone. Thus, the finding that PLK-1 functionally substitutes for Mps1 in checkpoint initiation in C. elegans uncovered a role for Plk1 in species that have Mps1.
Keywords: kinetochore, checkpoint, centromere, mitosis, chromosome segregation, KMN network, KNL-1, BUB-1, Knl1, Bub1, Bub3, CASC5, Plk1, Mps1
Graphical Abstract
Introduction
The spindle checkpoint ensures fidelity in chromosome segregation by monitoring the interaction between chromosomes and microtubules (Lara-Gonzalez et al., 2012; Musacchio and Salmon, 2007). Spindle checkpoint proteins enrich at kinetochores, the microtubule attachment sites on chromosomes, where they generate a diffusible inhibitor of anaphase onset. Following microtubule attachment, spindle checkpoint proteins are removed from kinetochores and the checkpoint is silenced, leading to sister chromatid separation, anaphase chromosome segregation, cytokinesis, and mitotic exit.
At the kinetochore, the protein Knl1 recruits the Bub1/Bub3 complex to activate the checkpoint, recruits the PP1 phosphatase that participates in checkpoint silencing, and interacts with microtubules (Ghongane et al., 2014; Caldas and DeLuca, 2014). The KNL-1 N-terminus contains several “MELT” repeats comprised of repetitions of the M-[E/D]-[L/I]-[T/S] amino-acids sequence (Cheeseman et al., 2004; Desai et al., 2003; Vleugel et al., 2012). In yeast and in human cells, Bub1/Bub3-binding to Knl1 is dependent on Knl1 phosphorylation of MELT repeats, and adjacent motifs, by Mps1 kinase (Krenn et al., 2014; London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Zhang et al., 2014). However, in human cells, whether Mps1 fully accounts for Bub1/Bub3 localization is not clear (Espert et al., 2014; Hewitt et al., 2010; Maciejowski et al., 2010; Santaguida et al., 2010).
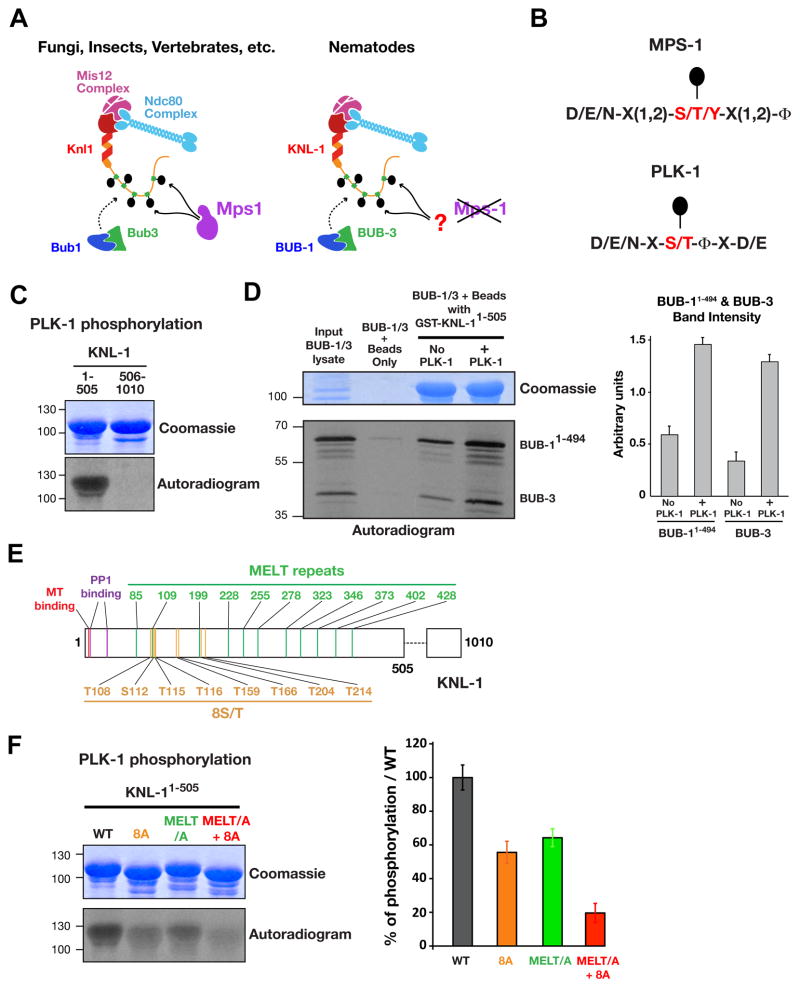
Consistent with the central importance of Mps1 in the spindle checkpoint, Mps1 kinases are widely conserved in fungi, metazoans, and plants. Surprisingly, in the nematode lineage that includes the well-studied model organism C. elegans, Mps1 is absent (Fig. 1A). This singular absence is intriguing since all other spindle checkpoint components are present and C. elegans embryonic cells and adult germline cells mount a checkpoint response at unattached kinetochores (Espeut et al., 2012; Essex et al., 2009; Kitagawa and Rose, 1999). This evolutionary ‘knockout’ suggests that BUB-1 anchorage on KNL-1 is either not regulated by phosphorylation in nematodes or that a kinase other than Mps1 is phosphorylating KNL-1 to direct BUB-1/BUB-3 recruitment. The second possibility appeared likely given the presence of ‘MELT’ motifs in the C. elegans KNL-1 N-terminus (Cheeseman et al., 2004; Desai et al., 2003). Among the potential kinases that could replace Mps1 in C. elegans, Polo-like kinase 1 (PLK-1) was a good candidate as Mps1 and Plk1 have related phosphorylation consensus motifs (Fig. 1B; Dou et al., 2011), which include the Knl1 ‘MELT’ repeats, and they both localize to kinetochores.
Figure 1. The N-terminal half of KNL-1 is a robust Plk1 substrate in vitro.
(A) Schematic of BUB-1/BUB-3 kinetochore recruitment. Mps1 is absent in the nematode lineage.
(B) Mps1 and Plk1 phosphorylation consensus sequences (Dou et al., 2011).
(C) Plk1 phosphorylation of GST-KNL-11–505 and GST-KNL-1506–1010.
(D) Binding of in vitro translated BUB-11–494 and BUB-3 to GST-KNL-11–505 with or without PLK-1 phosphorylation. Quantification (right) is from 3 independent experiments; error bars are the 95% confidence interval.
(E) Schematic of KNL-1 features, highlighting MELT repeats (green) and other PLK-1 target sites (orange) mapped in Fig. S1.
(F) Analysis of KNL-11–505 WT, MELT/A, 8A, MELT/A+8A phosphorylation by PLK-1 performed as in Fig. 1c. Quantification, relative to WT, from 3 independent experiments is shown on the right. Error bars are the 95% confidence interval. See also Figure S1 and S2.
Here we show that in C. elegans, PLK-1 substitutes for Mps1 by phosphorylating the KNL-1 N-terminus to direct recruitment of BUB-1/BUB-3 to the kinetochore. This result led us to analyse Bub1 recruitment in human cells, where a substantial pool of Bub1 was present at kinetochores independently of Mps1 activity and that this pool depended on Plk1 activity. Thus, analysis of how a fundamental cellular pathway— the spindle checkpoint—functions in the natural absence of its central regulator Mps1 revealed a mechanism that is also operating in species that contain Mps1.
Results
The KNL-1 N-terminus is a robust Plk1 substrate in vitro
A straightforward initial test of the hypothesis that PLK-1 functionally substitutes for Mps1 in directing recruitment of BUB-1/BUB-3 to the C. elegans kinetochore would be to inhibit PLK-1 and monitor BUB-1/BUB-3 recruitment. However, depletion of PLK-1 causes a potent meiosis I arrest in C. elegans (Chase et al., 2000; not shown), preventing the generation of mitotic embryos in which BUB-1 kinetochore localization can be monitored. Therefore, we focused on analyzing KNL-1 phosphorylation by PLK-1 and on determining the role of this phosphorylation in BUB-1/BUB-3 recruitment and checkpoint signaling.
We purified C. elegans PLK-1 from insect cells and analyzed phosphorylation of recombinant N-terminal (KNL-11–505) and C-terminal (KNL-1506–1010) KNL-1 fragments, as well as the model Plk1 substrate α–casein (Fig. 1C, S1A). The N-terminal half of KNL-1, which contains 9 M-[E/D]-[L/I]-[T/S] (Cheeseman et al., 2004; Desai et al., 2003; Vleugel et al., 2012) and two related motifs (M199DLD and M473SID), was robustly phosphorylated by PLK-1; in contrast, the C-terminal half was not phosphorylated (Fig 1C). The phospho-signal observed on KNL-11–505, was 7-fold higher than for a similar concentration of casein, a model substrate of Polo kinases (Fig S1A); this could be due to multiplicity of target sites on the KNL-1 N-terminus and/or substrate preference relative to casein.
Next, we assessed the effect of KNL-1 phosphorylation by PLK-1 on interaction with BUB-1 and BUB-3 by incubating beads coated with GST-tagged KNL-11–505 in a reticulocyte lysate expressing BUB-11–494 and BUB-3. Phosphorylation by PLK-1 increased association of BUB-1 and BUB-3 with KNL-11–505 by 2.4 and 3.8 fold respectively (Fig. 1D). Thus, phosphorylation of KNL-1 by PLK-1 promotes interaction of the KNL-1 N-terminus with BUB-1 and BUB-3.
To assess the contribution of the MELT repeats to the phosphorylation of the KNL-1 N-terminus, we compared PLK-1 kinase activity on WT KNL-11–505 to a mutant with the 11 MELT repeats mutated to AEAA (Fig. 1E,F, S1B). Mutation of the MELT repeats reduced KNL-11–505 phosphorylation to ~60 % of WT KNL-11–505 (Fig. 1F) indicating that additional sites are targeted by PLK-1. To identify these other sites, we analysed phosphorylation of recombinant fragments followed by targeted amino acid mutations (Fig. S1C–G). Using this approach, we identified 8 sites (T108, S112, T115, T116, T159, T166, S204, S214) phosphorylated by PLK-1, whose mutation to alanine (8A) decreased phosphorylation of KNL-11–505 by ~50% (Fig. 1F). Combining mutation of the MELT repeats and of the 8 additional sites (MELT/A+8A), additively reduced PLK-1 phosphorylation to ~20% of control (Fig. 1F). Thus, biochemical analysis defined a set of residues whose mutation should enable testing the functional significance of PLK-1 phosphorylation of KNL-1 in vivo.
KNL-1 Mutants Compromised for PLK-1 Phosphorylation Retain Functional Properties Associated with the KNL-1 N-terminus
As the mutations introduced to reduce PLK-1 phosphorylation alter a significant number of residues in KNL-1 (e.g. 41 out of 1010 in MELT+8A), we were concerned about interpreting such mutants in vivo. The KNL-1 N-terminus has a PP1 docking site, a microtubule-binding activity, and behaves as an oligomer on gel filtration (Cheeseman et al., 2006; Espeut et al., 2012; Kern et al., 2014). We therefore tested all three properties for the MELT/A, 8A and MELT/A+8A mutations and found that none of them was affected by mutations in PLK-1 phosphorylation sites (Fig. S2A–C). Thus, any effect of these mutations in vivo is unlikely to be due to a non-specific disruption of the N-terminal half of KNL-1.
A KNL-1 Mutant Compromised for PLK-1 Phosphorylation Significantly Reduces BUB-1 Kinetochore Recruitment
We next generated strains expressing single copy RNAi-resistant versions of MELT/A, 8A and MELT/A+8A mutant forms of KNL-1 in vivo. These transgenes were based on a prior RNAi-resistant knl-1::mCh transgene that was functionally validated (Espeut et al., 2012).
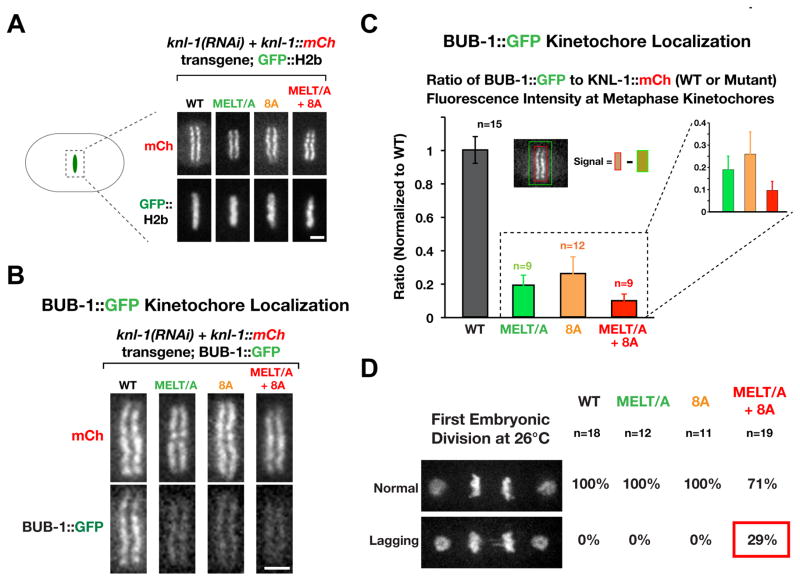
The three KNL-1 mutants generated—MELT/A, 8A and MELT/A+8A—all localized to kinetochores at levels similar to WT KNL-1 (Fig. 2A). To monitor BUB-1 kinetochore localization in these mutants, we introduced a bub-1::gfp transgene into the different knl-1::mCh transgene containing strains, depleted endogenous KNL-1, and measured BUB-1::GFP levels relative to KNL-1::mCherry on kinetochores of aligned chromosomes (Fig. 2B,C). This analysis revealed that the 8A and MELT/A mutants recruited less BUB-1 at kinetochores compared to WT KNL-1 (Fig. 2B,C). Notably, in the MELT/A+8A mutant, significantly less BUB-1 was recruited to kinetochores, compared to MELT/A or 8A alone (Fig. 2B,C). Thus, mutations that compromise PLK-1 phosphorylation of the KNL-1 N-terminus in vitro significantly perturb BUB-1 kinetochore recruitment in vivo, with the MELT/A+8A mutant nearly abolishing BUB-1 localization.
Figure 2. Mutations in KNL-1 that disrupt Plk1 phosphorylation perturb BUB-1 kinetochore recruitment and chromosome segregation.
(A) Images of the metaphase plate in one-cell embryos depleted of endogenous KNL-1 that express indicated RNAi-resistant KNL-1::mCherry variants and GFP::H2b. Scale bar, 2 μm.
(B) Analysis of BUB-1::GFP kinetochore targeting in the indicated KNL-1 variants. Endogenous KNL-1 was depleted in each condition. Scale bar, 2 μm.
(C) Quantification of BUB-1::GFP kinetochore localization in the indicated strains. Graph plots the ratio of BUB-1::GFP (green) to KNL-1::mCh (red) measured at kinetochores of aligned chromosomes. The measured ratios were normalized relative to WT KNL-1. n refers to the number of embryos analyzed. Error bars are the 95% confidence interval.
(D) Images represent normal segregation (top) or segregation with lagging chromatin (bottom) in one-cell embryos at 26°C. The frequency of each for the indicated KNL-1 variants is shown on the right. Scale bar, 5 μm.
To compare the ability of WT and mutant KNL-1 to support chromosome segregation, we crossed in GFP fusions with histone H2b and γ-tubulin and depleted endogenous KNL-1 (Fig. 2D). The MELT/A+8A mutant, but not MELT/A or 8A, showed defects in chromosome segregation at 26°C, with ~30% of first embryonic division embryos exhibiting lagging chromatin in anaphase (Fig. 2D). Moreover, embryonic viability dropped to ~40% at 26°C for the MELT/A+8A transgene in the background of the knl-1(ok3457) deletion mutant (Fig. S2D). Thus, the MELT/A+8A mutant that compromises PLK-1 phosphorylation to the greatest degree greatly reduces BUB-1 kinetochore localization and compromises chromosome segregation in vivo. The phenotypic difference between the MELT/A or 8A mutants and the MELT/A+8A mutant is reminiscent of prior work in human cells showing that ~10% of Bub1 is sufficient to sustain the checkpoint (Johnson et al., 2004; Meraldi and Sorger, 2005).
Overall, these data suggest that PLK-1 phosphorylation of the KNL-1 N-terminus directs BUB-1 kinetochore recruitment in C. elegans.
KNL-1 Phosphorylation by Plk1 is Required for Spindle Checkpoint Activation
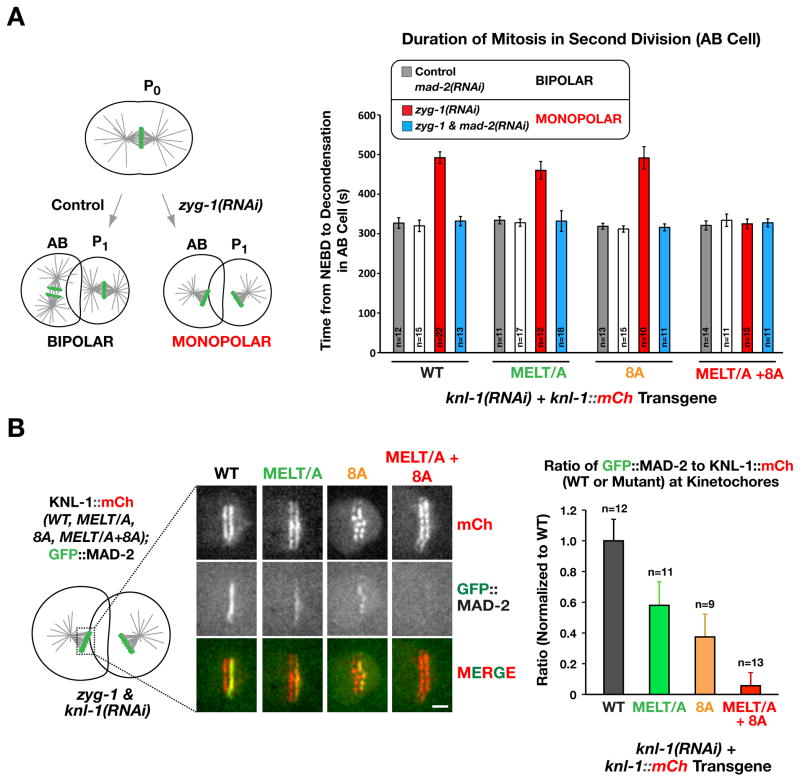
Complete loss of BUB-1 kinetochore localization is expected to abolish spindle checkpoint signalling. In order to monitor checkpoint signaling, we depleted the kinase ZYG-1 to inhibit centriole duplication and generate monopolar spindles in the second embryonic division, and measured the time between nuclear envelope breakdown (NEBD) and chromosome decondensation (Fig. 3A). WT KNL-1::mCherry supported normal checkpoint signaling, as evidenced by the MAD-2-dependent lengthened NEBD-to-decondensation interval in the presence of monopolar spindles, compared to control embryos with bipolar spindles (Fig. 3A; (Espeut et al., 2012; Essex et al., 2009). In cells expressing either the MELT/A or the 8A mutant, the MAD-2-dependent delay induced by monopolar spindles was similar to that observed with WT KNL-1 (Fig. 3A). In contrast, in the MELT/A+8A mutant the monopolar spindle-induced checkpoint delay was abolished (Fig. 3A).
Figure 3. The MELT/A+8A mutant of KNL-1 is checkpoint signaling-defective.
(A) (left) Schematic of the experimental approach used to compare mitotic duration in the AB cell in 2 states: bipolar or monopolar. Monopolar second division cells are generated by depletion of ZYG-1, the kinase required for centriole duplication. (right) NEBD – chromosome decondensation interval measured for the indicated conditions. Error bars are the 95% confidence interval.
(B) (left) GFP::MAD-2 localization at unattached kinetochores on monopolar spindles generated by inhibiting centriole duplication. Scale bar, 5 μm. (right) GFP::MAD-2 intensity on kinetochores normalized relative to the WT KNL-1 transgene control. Error bars are the 95% confidence interval. See also Figure S2.
A marker of spindle checkpoint activation is kinetochore localization of MAD-2. Imaging of GFP::MAD-2 on chromosomes following monopolar spindle formation revealed that the MELT/A or 8A mutants decreased GFP::MAD-2 kinetochore localization (Fig. 3B), consistent with reduced amount of kinetochore-localized BUB-1 (Fig. 2B,C; note that the BUB-1 and MAD-2 measurements are not directly comparable as the BUB-1 was measured on bipolar metaphase kinetochores whereas MAD-2 was measured on monopolar unattached kinetochores, where checkpoint proteins are known to amplify). More importantly, the MELT/A+8A mutation eliminated detectable GFP::MAD-2 kinetochore localization (Fig. 3B), consistent with the abrogation of checkpoint signaling. Thus, the MELT/A+8A mutant of KNL-1, which greatly reduces Plk1 phosphorylation in vitro and BUB-1 kinetochore localization in vivo, lacks a functional spindle checkpoint.
Plk1 contributes to Knl1 phosphorylation and Bub1 kinetochore localization in Mps1-inhibited human cells
Our finding that PLK-1 substitutes for Mps1 in controlling BUB-1 kinetochore recruitment in C. elegans led us to consider the possibility that Plk1 may also contribute to Bub1 targeting in species that contain Mps1. To test this possibility, we re-examined Bub1 kinetochore recruitment in HeLa cells. In published work, the effect of Mps1 on Bub1 kinetochore localization has been variable—several publications indicate a partial effect on Bub1 localization (~65–75% relative to control) (Hewitt et al., 2010; Santaguida et al., 2010) while others suggest near-complete elimination (Espert et al., 2014; Maciejowski et al., 2010).
To inhibit Mps1, we employed three structurally distinct small molecule inhibitors (Table S1): AZ3146 (Hewitt et al., 2010), NMS-P715 (Colombo et al., 2010) and MPI-0479605 (Tardif et al., 2011). When directly compared in radiometric biochemical activity assays, AZ3146, NMS-P715 and MPI-0479605 potently inhibited Mps1 with IC50s in the nanomolar range and, importantly for the analysis here, only significantly affected Plk1 activity at micromolar concentrations (Table S1).
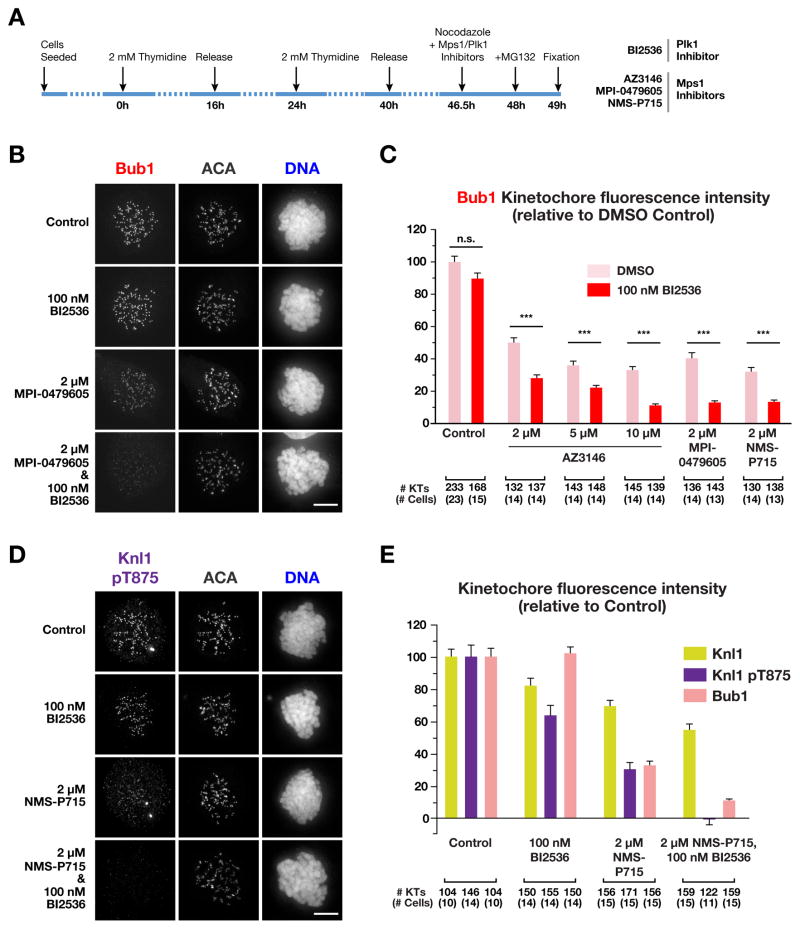
To monitor Bub1 kinetochore recruitment, we released HeLa cells from a double thymidine arrest and treated with each Mps1 inhibitor as well as nocodazole prior to mitotic entry (Fig. 4A). All 3 Mps1 inhibitors, at effective doses, greatly reduced Mad1 kinetochore localization (Fig. S3A,B; (Liu et al., 2003; Martin-Lluesma et al., 2002). In contrast, Mps1 inhibitors only partially affected Bub1 recruitment, with a significant residual pool of Bub1 (~35–50% of control) at kinetochores (Fig. 4B,C; S3A,B). Increasing dosage of AZ3146 did not affect this residual Bub1 pool while eliminating Mad1 localization (Fig. 4C; S3A,B). Thus, Mps1 inhibition on its own does not prevent Bub1 kinetochore recruitment.
Figure 4. Effect of Plk1 inhibition on Knl1 phosphorylation and Bub1 localization in Mps1-inhibited human cells.
(A) Schematic of the protocol used to treat cells with inhibitors prior to mitotic entry under microtubule depolymerization conditions. HeLa cells were used for this analysis and, after fixation, stained for Bub1, Mad1 or Knl1, and centromeres (ACA).
(B) Representative immunofluorescence images of mitotic HeLa cells treated with the Plk1 inhibitor BI2536, the Mps1 inhibitor MPI-0479605, or both, and stained for Bub1. Scale bar, 5 μm.
(C) Quantification of Bub1 kinetochore intensities for the indicated conditions. Values are normalized to controls. Error bars represent the 95% confidence interval for all kinetochores measured. ***: p < 0.0001
(D) Representative immunofluorescence images of mitotic HeLa cells stained for Knl1 pT875 under the indicated conditions. Scale bar, 5 μm.
(E) Quantification of Knl1, Knl1 pT875 and Bub1 kinetochore intensities for the indicated conditions. Values are normalized to controls. Error bars represent the 95% confidence interval for all kinetochores measured. See also Figure S3 and S4.
We next combined the specific Plk1 inhibitor BI2536 (Lenart et al., 2007) with each of the three Mps1 inhibitors. On its own, BI2536 did not significantly affect Bub1 kinetochore recruitment, even at high concentrations (Fig. 4B,C, S3C and not shown). However, in all three of the combination treatments, BI2536 reduced Bub1 kinetochore recruitment to ~10–15% of controls (Fig. 4B, C, S3C).
One explanation for the above result is that the double inhibitor treatments prevent kinetochore recruitment of Knl1. To test this, we co-stained treated cells for Knl1 and Bub1 and measured the levels of both proteins on individual kinetochores. Single Plk1 or Mps1 inhibition (with NMS-P715) reduced Knl1 kinetochore levels to ~70%–80% of controls (Fig. 4E, S4A,). In the double Plk1/Mps1-inhibited cells, there was a modest additive defect in Knl1 recruitment (Fig. 4E, S4A). However, this partial reduction in Knl1 recruitment is insufficient to explain the near-absence of Bub1 recruitment observed in the double Plk1 and Mps1 inhibition (Fig. S4B).
Next, we analysed phosphorylation of the Knl1 MELT repeats, which form docking sites for Bub3/Bub1, using a phospho-specific antibody directed against Thr875 (Yamagishi et al., 2012); this repeat has been shown to be functional for Bub1/Bub3 kinetochore recruitment (Vleugel et al., 2015). Inhibition of Mps1 with NMS-P715 reduced Knl1 pT875 kinetochore staining (Fig. 4D,E), as well as total Knl1 phosphorylation (Fig. S4C); however, as with Bub1, a substantial proportion (~30%) of pT875 staining remained. A partial effect on pT785 staining was observed with Plk1 inhibition alone (Fig. 4D,E), although this is likely due to reduced Knl1 kinetochore levels (Fig. 4D,E, S3A). Strikingly, the double Plk1 and Mps1 inhibition abolished pT875 staining (Fig. 4D,E), analogous to what was observed for Bub1. We conclude that Plk1 activity accounts for the significant residual pool of phospho-MELT staining and kinetochore-localized Bub1 that is observed in Mps1-inhibited human cells.
Conclusion
Here, we investigated how the initiating event of spindle checkpoint signaling—phosphorylation-dependent recruitment of the Bub1/Bub3 complex to the Knl1 kinetochore scaffold—occurs in the absence of Mps1 kinases in the nematode lineage.
We found that PLK-1, which is also kinetochore-localized and targets a similar substrate motif as Mps1, functionally substitutes for Mps1 by phosphorylating the KNL-1 N-terminus and creating recruitment sites for the BUB-1/BUB-3 complex. This result prompted us to re-examine Bub1 kinetochore localization in human cells, where we observed a contribution of Plk1 activity to Bub1 recruitment that was revealed after Mps1 inhibition. While it is possible that the additive effect of Plk1 and Mps1 co-inhibition is indirect, our analysis of Knl1 phosphorylation suggests a direct role. Moreover, an independent study examining Plk1 and Mps1 cooperation in human cells (Von Schubert et al., XXX) also supports a direct role for Plk1 in Knl1 phosphorylation and provides evidence against the observed synergy being due to perturbation of Plk1’s role in haspin activation and Aurora B localization (Ghenoiu et al 2013, Zhou et al 2014). The lack of a significant effect of Plk1 inhibition on its own suggests that the primary kinase targeting Knl1 in human cells is indeed Mps1. However, quantitative phosphoproteomics has shown that Plk1 phosphorylates Knl1 in human cells on at least three sites (Santamaria et al., 2011), two of which are in close vicinity to a MELT motif or a TxxF/Y motif—also shown to be important for Bub1 recruitment (Vleugel et al., 2013). This proteomic data suggests that Plk1 may contribute to recruitment of a pool of Bub1 by phosphorylating Knl1 even in the presence of Mps1 activity, which is consistent with work described in the related, independent study (Von Shubert et al., XXX).
Nematodes are holocentric, with diffuse kinetochores extending along the length of each chromatid in mitosis (Maddox et al., 2004). Mps1 loss in this lineage may have occurred to dampen checkpoint signaling in the context of holocentric chromosome architecture. However, contrary to this notion, recently analyzed holocentric insect species all contain Mps1 family kinases (personal communication; Drinnenberg et al., 2014). Thus, the reason as to why Mps1 is lost in the nematode lineage remains mysterious. Nevertheless, the natural absence of Mps1 provided an opportunity to study how the spindle checkpoint initiates without kinetochore-targeted Mps1 kinase activity. The answer, which is compensation by a similar substrate motif-targeting mitotic kinase, has in turn revealed a potential new contributing mechanism in species that contain Mps1.
Experimental Procedures
Imaging and quantification in C. elegans embryos
Chromosome segregation and checkpoint signaling was followed in embryos expressing GFP::H2b/GFP:γ-tubulin using a Zeiss AxioimagerZ1 microscope equipped with a Coolsnap HQ2 camera at 20°C. Five z-sections (100 ms exposure) were acquired at 2 μm steps at 10s (segregation) or 20s (checkpoint) intervals using a 100×, 1.3 NA Olympus U-Planapo objective with 2×2 binning and a 480×480 pixel area.
For BUB-1::GFP and GFP::MAD-2 localization, embryos were filmed with a a Yokogawa CSU-X1 spinning disk confocal head mounted on an inverted microscope (Ti-Eclipse; Nikon, Tokyo, Japan) equipped with a 100x, 1.45 NA Plan Apochromat lens (Nikon), a solid-state laser combiner (ALC) and an iXon Ultra EM-CCD (Andor Technology, Belfast, Ireland). Acquisition parameters, shutters, and focus were controlled by iQ 3 software. 5 x 2 μm GFP/mCherry z-series with no binning were collected every 20 s at 20°C. Exposures were 200 ms for GFP and 600 ms for mCherry.
Kinase assays
KNL-1 fragments at a concentration of 5.6 μM were incubated for 10 min at room temperature in the presence of 25 nM Plk1 (WT or KD), 100 μM ATP and 0.1 μCi/μl of [γ-32P] ATP. Reactions were analyzed by SDS-PAGE and autoradiography.
Human cell experiments
HeLa cells growing on poly-L-lysine–coated coverslips were synchronized with a double thymidine (2 mM) block, released into the different drug combinations (Fig. 4A), fixed in 1% formaldehyde and stained with the indicated antibodies. Cells were imaged using a Deltavision microscope and kinetochore intensities quantified as described (Hoffman et al., 2001). See supplemental information for more details.
Supplementary Material
Highlights.
Nematodes lack Mps1 kinase despite having a functional spindle checkpoint
PLK-1 substitutes for Mps1 in controlling spindle checkpoint initiation in C. elegans
PLK-1 phosphorylation of KNL-1 directs BUB-1/BUB-3 recruitment in absence of Mps1
Plk1 contributes to Knl1 phosphorylation and Bub1 targeting in human cells with Mps1
Acknowledgments
We thank the Montpellier RIO imaging facility of Campus CNRS route de Mende for their help with imaging, Stephen Taylor, Iain Cheeseman and Yoshinori Watanabe for antibodies, and Federica Bertaso for comments on the manuscript. This work was supported by two ANR grants from the French Research Ministry to A.A., a postdoctorate fellowship from the ARC (French Cancer Research Agency) to J.E., and a grant from the NIH to A.D. (GM074215). P.L.G. is a Pew Latin American Fellow. Salary support for A.A. is provided by INSERM (French Medical Health and Research Institute). A.D. and A.K.S. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Author Contributions
J.E., P.L.G., M.S. carried out experiments; A.K.S. provided chemical inhibitors and determined their specificity; A.D. and A.A. supervised the project; J.E., A.A., A.D. and P.L.G. wrote the manuscript, with input from all authors.
Competing financial interests statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Caldas GV, DeLuca JG. KNL1: bringing order to the kinetochore. Chromosoma. 2014;123:169–181. doi: 10.1007/s00412-013-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, Golden A. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis. 2000;26:26–41. doi: 10.1002/(sici)1526-968x(200001)26:1<26::aid-gene6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R, Caldarelli M, Mennecozzi M, Giorgini ML, Sola F, Cappella P, Perrera C, Depaolini SR, Rusconi L, Cucchi U, et al. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res. 2010;70:10255–10264. doi: 10.1158/0008-5472.CAN-10-2101. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, von Schubert C, Korner R, Santamaria A, Elowe S, Nigg EA. Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS One. 2011;6:e18793. doi: 10.1371/journal.pone.0018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, deYoung D, Henikoff S, Malik HS. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife. 2014;3:e03676. doi: 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert A, Uluocak P, Bastos RN, Mangat D, Graab P, Gruneberg U. PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J Cell Biol. 2014;206:833–842. doi: 10.1083/jcb.201406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell. 2009;20:1252–1267. doi: 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenoiu C, Wheelock MS, Funabiki H. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol Cell. 2013;52:734–745. doi: 10.1016/j.molcel.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghongane P, Kapanidou M, Asghar A, Elowe S, Bolanos-Garcia VM. The dynamic protein Knl1 - a kinetochore rendezvous. J Cell Sci. 2014;127:3415–3423. doi: 10.1242/jcs.149922. [DOI] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Kern DM, Kim T, Rigney M, Hattersley N, Desai A, Cheeseman IM. The outer kinetochore protein KNL-1 contains a defined oligomerization domain in nematodes. Mol Biol Cell. 2014;26:229–37. doi: 10.1091/mbc.E14-06-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol. 1999;1:514–521. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- Krenn V, Overlack K, Primorac I, van Gerwen S, Musacchio A. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr Biol. 2014;24:29–39. doi: 10.1016/j.cub.2013.11.046. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Sillje HH, Korner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011;10:M110 004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif KD, Rogers A, Cassiano J, Roth BL, Cimbora DM, McKinnon R, Peterson A, Douce TB, Robinson R, Dorweiler I, et al. Characterization of the cellular and antitumor effects of MPI-0479605, a small-molecule inhibitor of the mitotic kinase Mps1. Mol Cancer Ther. 2011;10:2267–2275. doi: 10.1158/1535-7163.MCT-11-0453. [DOI] [PubMed] [Google Scholar]
- Vleugel M, Hoogendoorn E, Snel B, Kops GJ. Evolution and function of the mitotic checkpoint. Dev Cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Vleugel M, Omerzu M, Groenewold V, Hadders MA, Lens SM, Kops GJ. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol Cell. 2015;57:824–835. doi: 10.1016/j.molcel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- Vleugel M, Tromer E, Omerzu M, Groenewold V, Nijenhuis W, Snel B, Kops GJ. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol. 2013;203:943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lischetti T, Nilsson J. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J Cell Sci. 2014;127:871–884. doi: 10.1242/jcs.139725. [DOI] [PubMed] [Google Scholar]
- Zhou L, Tian X, Zhu C, Wang F, Higgins JM. Polo-like kinase-1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep. 2014;15:273–281. doi: 10.1002/embr.201338080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.