Abstract
Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPARs) are the primary mediators for inter-neuronal communication and play a crucial role in higher brain functions including learning and memory. Our previous work demonstrated that AMPARs are subject to ubiquitination by the E3 ligase Nedd4, resulting in EPS15-mediated receptor internalization and Ubiquitin (Ub)–proteasome pathway (UPP)-dependent degradation. Protein ubiquitination is a highly dynamic and reversible process, achieved via the balance between ubiquitination and deubiquitination. However, deubiquitination of mammalian AMPARs and the responsible deubiquitinating enzymes remain elusive. In this study, we identify USP46 as the deubiquitinating enzyme for AMPARs. We find that AMPARs are subject to K63 type ubiquitination, and USP46 is able to deubiquitinate AMPARs in vivo and in vitro. In heterologous cells and neurons, expression of USP46 results in a significant reduction in AMPAR ubiquitination, accompanied by a reduced rate in AMPAR degradation and an increase in surface AMPAR accumulation. By contrast, knockdown of USP46 by RNAi leads to elevated AMPAR ubiquitination and a reduction in surface AMPARs at synapses in neurons. Consistently, miniature excitatory postsynaptic currents recordings show reduced synaptic strength in neurons expressing USP46-selective RNAi. These results demonstrate USP46-mediated regulation of AMPAR ubiquitination and turnover, which may play an important role in synaptic plasticity and brain function.
Keywords: AMPA receptor, deubiquitination, receptor internalization, trafficking, ubiquitination, USP46
Regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor (AMPAR) accumulation at synapses is the major molecular mechanism underlying the expression of synaptic plasticity, a process contributing to the execution of higher brain functions including learning and memory, and leading to neurological disorders and neurodegenerative diseases as well (Huganir and Nicoll 2013). The amount of functional AMPARs can be regulated by rapid trafficking of receptors to and from the synaptic surface via vesicle-mediated membrane insertion, internalization, and recycling (Malinow and Malenka 2002; Song and Huganir 2002; Newpher and Ehlers 2008; Wang et al. 2012). In addition to receptor translocation, a change in synaptic receptor levels can result from a change in AMPAR degradation and rate of synthesis, or a rebalance between these two processes (Schwarz et al. 2010; Lin et al. 2011).
Ubiquitination on certain proteins has been shown to regulate neuronal development and synaptic plasticity (Speese et al. 2003; Zhao et al. 2003; Yi and Ehlers 2007). Among membrane proteins, including neurotransmitter receptors, ubiquitination serves as a major signal to trigger endocytosis and direct the internalized protein to the lysosome and/or proteasome for degradation (Schwarz et al. 2010; Lin et al. 2011; Lin and Man 2013). Our previous study has shown that AMPARs are subject to Nedd4-mediated ubiquitination, leading to a reduction in cell-surface receptor expression and suppressed synaptic transmission (Lin et al. 2011; Lin and Man 2014). Additionally, a recent study has demonstrated that all AMPAR subunits are subject to ubiquitination, which is regulated by neuronal activity (Widagdo et al. 2015).
Ubiquitination is a reversible process mediated via the addition of ubiquitin by E3 ligases and the removal of ubiquitin moieties via deubiquitinating enzymes (DUB) (Nijman et al. 2005; Komander et al. 2009; Reyes-Turcu et al. 2009). In the human genome, there are an estimated 500–600 E3 ligases and less than one hundred DUBs identified thus far, suggesting the specification of DUBs on substrate selection (Komander et al. 2009; Kowalski and Juo 2012). DUBs are categorized into five families based on catalytic domains, including ubiquitin-specific proteases (USPs), ubiquitin c-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Josephins and JAB1/MPN/MOV34 metalloenzymes (JAMMs) (Komander et al. 2009). Among them, the ubiquitin c-terminal hydrolases, ubiquitin-specific proteases, OTU and Josephin families are Cys proteases and JAMM family members are zinc metallo-proteases. However, in mammalian neurons, the cellular process and molecular components involved in AMPAR deubiquitination remain largely unknown.
In this study, we identify USP46 as the DUB specific for AMPARs. We find that USP46 is enriched at the synapse and co-localizes with synaptic marker proteins. Expression of USP46 results in a significant reduction in AMPAR ubiquitination, accompanied by a decreased rate in AMPAR degradation and an increase in AMPAR synaptic accumulation. By contrast, knockdown of USP46 by shRNA leads to elevated AMPAR ubiquitination and a reduction in AMPAR protein amount in neurons. In line with changes in AMPAR synaptic localization, electrophysiological recordings show reduced miniature excitatory postsynaptic currents (mEPSCs) amplitude in neurons transfected with USP46 shRNA, indicating an important role for dynamic regulation in AMPAR ubiquitination in neuronal communication and brain function.
Materials and methods
Neuronal culture preparation, HEK cell culture and transfection
Primary cultured cortical and hippocampal neurons were prepared from embryonic day 18 rat embryos as previously described (Hou et al. 2008). E18 pregnant Sprague–Dawley rats were purchased from Charles River Laboratories Inc., Wilmington, MA, USA. Briefly, embryonic brain regions were dissected and digested with papain at 37°C. Dissociated neurons were seeded onto poly-l-lysine-coated coverslips at approximately 3 × 106 cells per 60 mm dish, each containing five coverslips. Neurons were maintained in Neurobasal medium (Gibco, Rockville, MD, USA) supplemented with 2% B27, 1% horse serum, 1% penicillin/streptomycin, and 0.4% l-glutamine for 2–3 weeks until use. One week after plating, 5-fluorodeoxyuridine (5 μM) was added to the media to inhibit glial growth. All cells were maintained in a humidified incubator containing 5% CO2. Transfections were performed at about 10– 11 days in vitro (DIV) and incubated for 4 h prior to media change. All transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Human embryonic kidney (HEK) 293A cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin and passaged at 100% confluency twice a week. Transfections were performed at approximately 50–70% confluency using Lipofectamine 2000. Three days following the transfection of neurons and 1 day for HEK cells, cells were fixed and immunostained or lysed for biochemical analysis. All procedures for animal study were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Boston University.
Biochemical analysis of protein ubiquitination
Cultured cortical neurons or HEK cells were rinsed with cold phosphate-buffered saline (PBS) and collected in 100–200 μL modified Radioimmunoprecipitation assay buffer (RIPA) lysis buffer [50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP40, 1% Sodium deoxycholate (SDOC) and 1% sodium dodecyl sulfate (SDS)] containing mini cOmplete protease inhibitors (Hoffmann-La Roche Grenzacherstrasse, Basel, Switzerland). The high SDS concentration was applied to avoid conventional protein–protein interaction. Lysates were further solubilized by sonication and 10 min incubation on ice followed by centrifugation for 10 min at 13 000 g. Supernatant was adjusted to 500 μL with RIPA lysis buffer and incubated overnight on rotation at 4°C with antibodies against glutamate receptor subunit 1 (GluA1) and protein A-Sepharose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Immunocomplexes were washed three times with ice-cold RIPA buffer without SDS (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP40, 1% SDOC), resuspended in 2X Laemmli buffer and denatured at 95°C for 10 min. Immunoprecipitates were separated by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with the appropriate antibodies. Antibodies used for immunoblotting were: Ubiquitin (1 : 1000; Abcam, Cambridge, MA, USA), beta-tubulin (1 : 5000; Sigma, St Louis, MO, USA), USP46 (1 : 1000; Abcam), PSD-95 (1 : 50; NeuroMab, Antibodies Incorporated, Davis, CA USA), Myc (1 : 1000; Santa Cruz Biotechnology), GluA1Ct (1 : 2000; homemade), Akt (1 : 1000; Abcam) and Nedd4 (1 : 1000; Abcam). Immunointensity of western blots was measured ImageJ: Wayne Rasband National Institute of Mental Health, MD, USA. For ubiquitination blots, smear signals above 100 kD were measured and quantified. GluA1 protein or GluA1 ubiquitination values were normalized to corresponding tubulin bands from the input, then normalized to controls prior to statistical analysis.
Immunocytochemistry on AMPAR surface and total expression
For immunostaining of surface AMPARs, transfected neurons were incubated with GluA1N antibodies (Millipore Corporation, Bedford, MA, USA) for 10 min at 37°C, washed once briefly with PBS, fixed for 10 min with fixation solution (4% sucrose, 4% Paraformaldehyde in PBS), washed twice briefly with PBS, and blocked for 1 h in 10% goat serum in PBS. Coverslips were then incubated in the dark for 1 h with Alexa Fluor-conjugated secondary antibodies (1 : 400; Life Technologies, Grand Island, NY, USA). Coverslips were washed three times for 5 min each in the dark to remove unbound secondary antibodies before being mounted with Prolong-Gold Antifade (Invitrogen).
For total AMPAR detection, cells were fixed and permeabilized with 0.3% Triton X-100 in Artificial cerebrospinal fluid (ACSF) for 10 min. Coverslips were then incubated for 1 h with primary antibodies GluA1Ct (1 : 400) or PSD-95 (1 : 5; NeuroMab) and USP46 (1 : 100; Abcam). Cells were then incubated with fluorescence-conjugated secondary antibodies.
AMPAR internalization assays
AMPAR internalization assays were performed as described previously (Lin et al. 2011). Briefly, hippocampal neurons were transfected at DIV 12 with dsRed and pcDNA3.1, as control, or dsRed and USP46. Two days after transfection, neurons were incubated with anti-GluA1 NT antibody (1 : 100) for 15 min on ice to label surface GluA1. Neurons were then washed and transferred to the incubator at 37°C with 50 μM glutamate for 15 min to trigger receptor endocytosis. Following fixation, the remaining surface-associated antibodies were blocked with horseradish peroxidase-conjugated secondary antibodies before permeabilization. Internalized antibody-bound AMPARs were then visualized following incubation with fluorescent secondary antibodies (1 : 400).
Synaptosomal membrane preparation
Cortical tissue dissected from adult rat brains was minced and homogenized in ice-cold solution (0.32 M sucrose, 1 mM NaHCO3, 1 mM MgCl2, 0.5 mM CaCl2). Lysates were then transferred to 15-mL conical tubes and further solubilized with the same buffer for 30 min at 4°C. Samples were centrifuged at 1400 g for 10 min and the supernatant (S1) was transferred to a new tube, and centrifuged at 13 800 g for 10 min. The pellet (P2) containing the synaptosome was resuspended in RIPA lysis buffer. Protein concentration was measured using the bicinchoninic acid protein determination kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) and samples of both lysates and P2 fraction were diluted to the same protein concentration with RIPA lysis buffer. 2× Laemmli buffer was then added and samples were denatured at 95°C for 10 min.
In vitro deubiquitination assay
Myc-USP46 and GFP-GluA1, with or without HA-ubiquitin (HA-Ubi), were transfected into HEK cells, respectively. Two days after transfection, Myc-USP46 and GFP-GluA1 were purified by immunoprecipitation with either Myc antibody or GluA1CT antibody in modified RIPA buffer (with 0.1% SDS). The in vitro deubiquitination assay was performed in deubiquitination buffer (100 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) containing mini cOmplete protease inhibitors (Roche). GFP-GluA1 (from cells co-transfected with HA-Ubi) was incubated with either boiled Myc-USP46 or native form of Myc-USP46 for 2 h at 37°C. 2 × Laemmli buffer was then added and samples were denatured at 95°C for 10 min prior to western analysis.
Virus constructs preparation and virus production
Full length rat USP46 and USP12 were PCR amplified to include the restriction sites BstBI and HpaI using the following oligonucleotides:
USP46 (5′-ATAAGAATTTCGAAATGGCATCAATGCAGAA G-3′; 5′-TATCAATCAAGAGAATAAGTTAACCG-3′)
USP12 (5′-ATAAGAATTTCGAAATGGCATCAATGCAGAA G-3′; 5′-TATCAGTCTCGGGACTGAGTTAACCG-3′).
USP46 and USP12 fragments from PCR were inserted into the pFUW vector with BstBI and HpaI sites within the multiple cloning site region. To package lentiviral particles, HEK293T cells at 60–70% confluency in 10-cm culture plates, were transfected for 4 h with 1.8 μg pCMV-VSVG (envelope vector), 1.8 μg pMDLg/pRRE (packaging vector), 1.8 μg pRSV-Rev (packaging vector), 4.6 μg of the target vector (either pFUW control or pFUW-Myc-USP46 or pFUW-Myc-USP12) using Lipofectamine 2000 (Invitrogen) in OPTI-MEM (Gibco). After 4 h, the transfection solution was replaced with fresh complete Dulbecco’s modified Eagle’s medium (10% Fetal bovine serum, 1% Pen/Strep). Approximately 24 h post-transfection, the cell culture medium was replaced with neuronal basal medium. About 48 h after medium change, culture medium was collected and cell debris was removed by centrifugation. Aliquots of the supernatant were restored in the freezer for future use. Virus-containing medium was applied at a final concentration of 20%.
shRNA sequences against USP46 and USP12 were designed based on the USP46 and USP12 coding sequences. The sense and antisense hairpin sequences were generated and cloned into pLKO.1-TRC vector (Addgene, Cambridge, MA, USA).
#1 shUSP46: Target sequence: GGGAACACTCACTAACGAAAC;
#2 shUSP46: Target sequence: CCAGAGCAGTTTCCGATCAAT;
#1 shUSP12: Target sequence: CCAGAACAGTTTCCGGTCAAC;
#2 shUSP12: Target sequence: AGAAGCAAAATGGCCGATTAC.
PCR products were digested and the fragments were inserted into the PLOK.1 TRC vector. All lentivirus vectors were confirmed by sequencing before virus production. Virus-containing medium was applied at a final concentration of 5%.
Image collection
Immunostained coverslips were mounted onto slides using ProlongGold Antifade reagent (Invitrogen) and kept in the dark for 4 h before imaging. Images were collected with an inverted fluorescence microscope at a 63× with an oil immersion objective (Zeiss Axiovert 200M, Carl Zeiss, Carl-Zeiss-Straϐe, Oberkochen, Germany). The exposure time for fluorescence signal was first set automatically by the software then adjusted manually so that the signals were within the full dynamic range. Either the glow scale look-up table or the histogram was used to monitor the saturation level. When analyzed using ImageJ software, images were manually thresholded to select GluA1 puncta for quantitative measurements.
Electrophysiology
Hippocampal neurons were transfected around DIV 10 with either enhanced GFP (EGFP) and pcDNA3.1, or EGFP together with USP46 or USP12 or shUSP46 or USP12, respectively. Total cDNA amounts were balanced by adding pcDNA3.1 in control transfections. Two days following transfection, a coverslip of neurons was transferred to a recording chamber with the extracellular solution containing 140 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 2.5 mM CaCl2, 11 glucose and 10 HEPES, pH 7.4, which was supplemented with tetrodotoxin (1 μM) to block action potentials, 2-amino-5-phosphonopetanoate (50 μM) to block NMDA receptors, and bicuculline (20 μM) to block GABAA receptor-mediated inhibitory synaptic currents. Whole-cell voltage-clamp recordings were made with patch pipettes filled with an intracellular solution containing 100 mM Cs-methanesulfonate, 10 mM CsCl, 10 mM HEPES, 0.2 mM EGTA, 4 mM Mg-ATP, 0.3 mM Na-GTP, 5 mM QX-314 and 10 mM Na-phosphocreatine, pH 7.4, with the membrane potential clamped at −70 mV. Recordings were performed on the same day with coverslips that were from the same batch of cells and transfected at the same time.
Data analysis
Ubi signals in western blots were analyzed, measured and quantified by ImageJ. The same area within each lane was selected and measured for intensity. Background was subtracted before comparison. A couple of independent experiments were performed as indicated in the text.
For immunostaining image analysis, 60–100 μm of at least three segments of secondary dendrites from different neurites were analyzed to represent one neuron. The intensity and density of GluA1 puncta (surface, total or internalized) was measured. The same threshold was used when measuring the same batch of images.
Results
UPS 46 is enriched at synapses and co-distributes with AMPARs
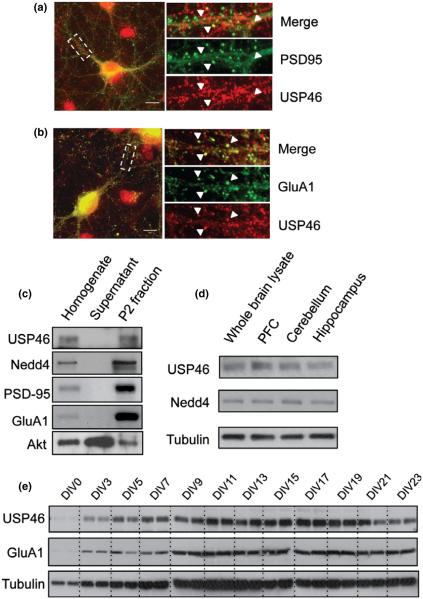
In the search for DUBs responsible for AMPAR deubiquitination, we found that an ortholog to the mammalian USP46 has been implicated in the regulation of glutamate receptor ubiquitination in C. elegans (Kowalski et al. 2011). We, therefore, wanted to explore whether USP46 is used for AMPAR deubiquitination in mammalian neurons. We first performed immunostaining to examine the subcellular distribution of USP46. In 2-week-old cultured rat hippocampal neurons, USP46 was labeled with specific antibodies together with AMPAR GluA1 subunits, or with a postsynaptic marker protein PSD-95. USP46 showed a punctate distribution pattern and was mostly co-localized with GluA1 or PSD95, indicating synaptic localization (Fig. 1a and b). To further investigate its synaptic distribution, we performed a synaptosomal P2 fraction purification assay from adult rat hippocampal tissue. Consistent with immunostaining results, western blots revealed that, similar to the known synaptic proteins including GluA1 and PSD95, USP46 was enriched in the synaptosomal P2 fractions compared with the lysates (Fig. 1c). As reported in our previous study (Lin et al. 2011), Nedd4 also showed preferential localization in the synaptosome. Next, we examined USP46 expression in different subbrain regions, including prefrontal cortex, cerebellum, and hippocampus. Using lysates from different brain regions from adult rats, equal amount of total proteins was loaded for western analysis. USP46 was expressed in all three regions with highest abundance in prefrontal cortex. By using cultured cortical neurons, we then analyzed the developmental profile in USP46 expression (Fig. 1e). We found that USP46 expression levels rapidly peaked at DIV 11 and decreased drastically after DIV 17 (Fig. 1e), a time window overlapping with synapse maturation.
Fig. 1.
UPS46 synaptic distribution and developmental expression. (a and b) Double staining of USP46 (Red) and post-synaptic protein PSD-95 (Green) or glutamate receptor subunit 1 (GluA1) (Green) in cultured DIV 14 hippocampal neurons. The selected region was enlarged for clarity (right). Arrowheads indicate co-localized puncta. Scale bar represents 20 lm. (c) USP46 and other proteins including Nedd4, PSD-95 and GluA1 are enriched in the synaptosomal P2 fraction purified from adult rat hippocampal tissue. In contrast, Akt is mainly distributed in the solubilized fraction. The same amount of protein (3 μg) was loaded for each lane. (d) USP46 expression in different brain regions. USP46 is relatively higher in prefrontal cortex (PFC). The AMPAR E3 ligase Nedd4 and tubulin was also probed. (e) USP46 expression time course was measured from cultured cortical neurons at varied days in vitro (DIV) after plating. USP46 amounts peaked in the 2nd and 3rd weeks. GluA1 and tubulin were also probed.
USP46 deubiquitinates AMPARs and regulates receptor protein stability
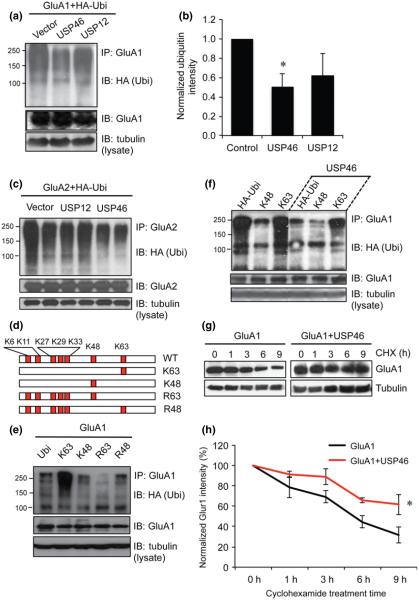
Our previous work and other studies have established that AMPARs are subject to ubiquitination modification (Schwarz et al. 2010; Lin et al. 2011). Because USP46 is involved in glutamate receptor deubiquitination in C. elegans (Kowalski et al. 2011) and it is enriched in synapses co-localizing with AMPARs in rat primary neurons as shown above, we questioned whether USP46 also serves as the DUB for mammalian AMPARs. We found that in the mammalian brain there exists another DUB, USP12, which shares 88% similarity with USP46 in amino acid sequence. It is intriguing to compare the role of USP46 together with USP12 to determine whether they share the same function. To determine the function in AMPAR deubiquitination, we co-transfected HA-Ubi, GluA1 and USP46 or USP12 in HEK293A cells. One day after transfection, cells were lysed within RIPA buffer. 1% SDS was included in the lysis buffer to eliminate regular protein–protein interactions, so that pure GluA1 could be immunoprecipitated. When ubiquitin signals on isolated GluA1 were examined by western blotting, we found a typical smear pattern in each sample, indicating the occurrence of GluA1 ubiquitination (Lin et al. 2011). Importantly, in cells co-transfected with USP46, GluA1 ubiquitination showed a significant reduction. Conversely, over-expression of USP12 showed no effect on the ubiquitination status of GluA1 (Fig. 2a and b, USP46: 0.50 ± 0.13, n = 3 independent experiments; p = 0.04; USP12: 0.62 ± 0.23, n = 3 independent experiments; p = 0.25). To examine the effect of USP46 on other AMPAR subunits, we did similar assays on GluA2. We found that USP46, but not USP12, led to similar deubiquitination on GluA2 (Fig. 2c). These findings suggest the specificity of USP46 on AMPAR deubiquitination.
Fig. 2.
USP46 specifically deubiquitinates AMPARs and stabilizes receptor protein stability. (a and b) Human embryonic kidney (HEK) cells were co-transfected with HA-ubiquitin (HA-Ubi), GFP-glutamate receptor subunit 1 (GluA1) together with either USP46 or USP12. GFP-GluA1 isolated by immunoprecipitation was probed for HA-ubiquitin. Ubiquitin signals (HA-Ubi) were reduced in cells expressing USP46, but not USP12. The same western blot was re-probed with GluA1 antibody to confirm the immunoprecipitation. Tubulin from cell lysates was probed to indicate equal amount of lysates (n = 3 independent experiments). Bar graphs represent Mean ± SEM, *p < 0.05, t-test. (c) Ubiquitination assays for GluA2. USP46, but not USP12, caused deubiquitination of GluA2. (d) An illustrative demonstration of wild-type ubiquitin, K48, K63, R48 and R63. Red boxes represent lysine residues within ubiquitin. (e) GluA1 is subject to K63-type ubiquitination. HEK cells were co-transfected with GFP-GluA1 and one among wild-type HA-Ubiquitin (HA-Ubi), ubiquitin with only one lysine (K63 or K48), or with single lysine mutation (R63 or K48). Lysine 63 in ubiquitin was sufficient (K63) and required (R63) for GluA1 ubiquitination. (f) USP46 recognizes K63-type ubiquitination on GluA1. USP46 was co-transfected with GFP-GluA1 and either wide-type HA-Ubiquitin, or HA-ubiquitin with only one lysine (K48 or K63). USP46 is absent from control group. Compared to control group, USP46 greatly reduces K63-type ubiquitination. (g) USP46 extends AMPAR half-life. HEK cells were transfected with GFP-GluA1 alone with together with USP46. Twenty four hours after transfection, cycloheximide (CHX; 1 μg/mL) was added to block protein synthesis. Cells were collected at later time points for protein amount analysis. Tubulin was probed as a loading control. (h) A plot of normalized protein amount shows reduced GluA1 turnover rate by USP46. n = 3 independent experiments. *p < 0.05, t-test.
K63-ubiquitin chain is the primary modification for GluA1 polyubiquitination
Given the high molecular weight of ubiquitinated AMPAR subunits, and that only four lysine residues at the subunit C-terminal can serve as ubiquitination targets, we concluded that AMPARs were conjugated with polyubiquitin chains (Lin et al. 2011; Lin and Man 2013). A polyubiquitin chain is formed by a sequential conjugation of one ubiquitin molecule to a lysine (K) residue on another ubiquitin unit. Because there are seven lysine residues in ubiquitin, including K6, K11, K27, K29, K33, K48, and K63, there can be at least seven different polyubiquitin chains. Two types of ubiquitin chains have been shown to be most commonly used in protein ubiquitination, that is., K48 and K63 chains. To examine the types of ubiquitin chains on GluA1, ubiquitination assays were performed using mutant ubiquitins (Fig. 2d). We transfected HEK cells with GluA1, together with an HA-tagged mutant ubiquitin that has all of the Ks mutated to R except only one K, either K48 or K63. Thus, compared to the control expressing wild-type HA-Ubi, a lack of ubiquitination in cells expressing a mutant ubiquitin will indicate that the remaining K is not used. To take an opposite strategy, we also used HA-Ubi that had only either K48 or K63 mutated to arginine (R48 or R63). Under this condition, a lack of ubiquitination indicates that the muted K is normally used for ubiquitination, and the mutated K is not important if typical ubiquitination remains. As shown in Fig. 2e, GluA1 showed a high level of ubiquitination when ubiquitin had only the K63 site (with all other 6 Ks mutated to R). Consistent with this, GluA1 ubiquitination was dramatically reduced in the ubiquitin K63R mutant (with all other Ks intact). However, the R48 mutant, with intact K63, showed normal ubiquitination intensity. To further confirm that the K63-ubiquitin chain is the major chain, we performed a ubiquitination assay with or without USP46 to examine if USP46 can specifically remove ubiquitin from the K63-ubiqutin chain. GluA1 was co-transfected with HA-tagged wild-type ubiquitin, K48, or K63. Or GluA1 and USP46 were co-transfected with the three components above, respectively. Our results show that K63 led to the highest ubiquitination level and the presence of USP46 greatly reduced the ubiquitination level both from wild-type ubiquitin and K63 but not from K48 (Fig. 2f). These results strongly indicate that the K63-ubiquitin chain is the primary form for GluA1 ubiquitination.
USP-mediated deubiquitination regulates AMPAR protein stability
Following ubiquitination, the substrate proteins are usually sorted to the degradation pathways. Thus, ubiquitination often leads to facilitated protein degradation. Considering deubiquitination as the opposing process counterbalancing ubiquitination, we expected that USP46 would stabilize AMPARs via inhibition of receptor ubiquitination. To this end, we performed a protein degradation assay to examine the protein degradation rates. HEK293A cells were transfected with GFP-GluA1 alone or together USP46. One day after transfection, cycloheximide (CHX, 0.5 mM) was added to the culture to inhibit protein synthesis and cell lysates were collected at different time points. GluA1 protein abundance was measured following western blotting (Fig. 2g and h). In the presence of USP46, the reduction rate of GluA1 was diminished compared with the control expressing only GluA1. About 9 h after CHX incubation, GluA1 levels were reduced by 70%, but only 40% in cells expressing USP46 (n = 3 independent experiments), indicating a suppression of GluA1 degradation by USP-mediated deubiquitination.
USP46 deubiquitinates AMPARs in vivo and in vitro
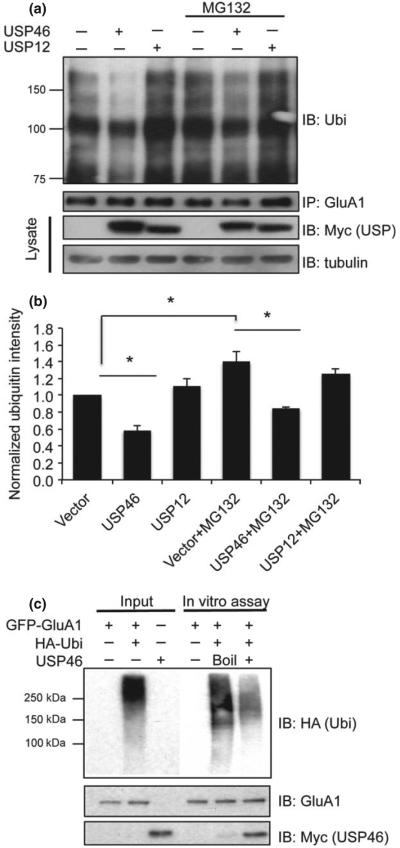
To confirm that USP46 can indeed deubiquitinate AMPARs in neurons, we cloned Myc-USP46 (and Myc-USP12) into a lentiviral vector so that a high infection rate could be achieved to allow biochemical analysis in neuronal cultures. Cultured cortical neurons at DIV 13 were incubated with virus-containing medium at a 20% final concentration, and ubiquitination assays were performed 4 days after infection. Because neurons have a low level of AMPAR ubiquitination during basal conditions (Lin et al. 2011), the proteasomal inhibitor MG132 (10 lM) was applied to some cultures for 8 h before cell collection. Endogenous GluA1 was isolated by immunoprecipitation as described previously, and ubiquitination was detected by western blotting. In infected cortical neurons, over-expression of USP46 significantly reduced the ubiquitination level of GluA1, whereas over-expression of USP12 showed no effect (Fig. 3a and b), again indicating the specificity of USP46 toward GluA1. As expected, application of the proteasome inhibitor MG132 significantly enhanced basal ubiquitination levels. In the presence of MG132, infection of USP46, but not USP12, significantly reduced the ubiquitination intensity on GluA1 (Fig. 3a and b) (Vector: 1.00; USP46: 0.58 ± 0.06, n = 3 independent experiments, p = 0.01; USP12: 1.11 ± 0.09, n = 3 independent experiments, p = 0.16; Vector+MG132: 1.34 ± 0.13, n = 3 independent experiments, p = 0.04; USP46 + MG132: 0.85 ± 0.01, n = 3 independent experiments, p = 0.02; USP12 + MG132: 1.26 ± 0.06, n = 3 independent experiments, p = 0.04). In addition, we found that over-expression of USP46 and USP12 did not alter the protein level of E3 ligases including Nedd4 and E6AP (Figure S1), suggesting that the reduced GluA1 ubiquitination was not likely due to a change in the ubiquitination process.
Fig. 3.
USP46 deubiquitinates glutamate receptor subunit 1 (GluA1) in vivo and in vitro. (a) Cultured DIV 13 cortical neurons were infected with viral Myc-USP46 or Myc-USP12 for 4 days. Cells were then incubated with proteasome inhibitor MG132 (10 lM) for 8 h and the endogenous GluA1 was isolated for ubiquitination assays. MG132 treatment increased the level of GluA1 ubiquitination. Expression of USP46, but not USP12, caused a significant reduction in GluA1 ubiquitination. (b) Quantification of Ubi signals. The same blot was reprobed for GluA1. Lysates were probed for USP46 and USP12 (Myc), as well as tubulin. n = 3 independent experiments. *p < 0.05, t-test. (c) USP46 deubiquitinates GFP-GluA1 in vitro. Myc-USP46 and GFP-GluA1 were purified, respectively, from transfected human embryonic kidney (HEK) cells. Co-incubation of active Myc-USP46 and GFP-GluA1 resulted in a reduction in GluA1 ubiquitination.
To further confirm a direct role for USP46 on GluA1 deubiquitination, we performed in vitro deubiquitination assays. Myc-USP46 and GFP-GluA1 (with or without HA-Ubi) were over-expressed in HEK cells and then isolated by immunoprecipitation. GFP-GluA1 (from cells co-expressing HA-Ubi) was then incubated with either boiled or native Myc-USP46 at 37°C for 2 h. Co-incubation of GFP-GluA1 and Myc-USP46 led to a remarkable decrease in GluA1 HA-Ubi intensity, confirming that USP46 was able to directly remove the ubiquitin chain from GluA1 (Fig. 3c).
Endogenous USP46 suppresses AMPAR ubiquitination in neurons
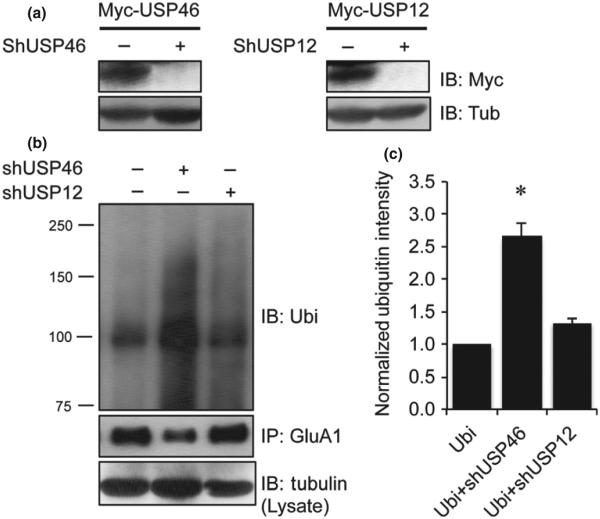
Following examination of the role of USP46 by exogenous over-expression, we wanted to determine whether the endogenous USP46 is effective under normal conditions. To this end, we generated shRNA viral constructs specifically against USP46 or USP12, based on their coding region sequences. To test the effectiveness, we infected HEK293A cells with the virus shRNA (using 10% medium) together with viruses of Myc-USP46 (5% medium) or Myc-USP12 (5% medium). The shRNA completely blocked the expression of USP46 and USP12, respectively (Fig. 4a). In cortical neurons infected with similar amounts of virus, we performed ubiquitination assays by immunoprecipitation of GluA1. Our results showed that USP46 knock down significantly enhanced ubiquitination intensity compared to both vector control and USP12 knock down (Fig. 4b and c) (Ubi: 1.00 ± 0.10, n = 3 independent experiments; Ubi+shUSP46: 2.66 ± 0.21, n = 3 independent experiments, p = 0.04 (vs. Ubi); Ubi+shUSP12: 1.33 ± 0.06, n = 3 independent experiments, p = 0.06). These findings strongly indicate that endogenous USP46 indeed dynamically regulates AMPAR ubiquitination.
Fig. 4.
Knockdown of USP46 enhances AMPAR ubiquitination. (a) Confirmation of the effectiveness of viral shRNAs against USP46 and USP12. shUSP46 or shUSP12 is a mixture of two viruses containing two distinct shRNA sequences. HEK293A cells were infected with viral Myc-USP46 (5%) or Myc-USP12 (5%) alone, or together with specific viral shRNAs (shUSP46 or shUSP12, 5% each). Cell lysates prepared 2 days after infection were subject to western analysis. The viral shRNAs completely blocked the expression of USP46 and USP12. (b and c) Cultured DIV 13 cortical neurons were infected with viral shUSP46 or shUSP12, and endogenous glutamate receptor subunit 1 (GluA1) ubiquitination was examined 4 days after infection by probing ubiquitin signals on immunoprecipitated GluA1. GluA1 ubiquitination was significantly enhanced in neurons infected with shUSP46. n = 3 independent experiments. *p < 0.05, t-test.
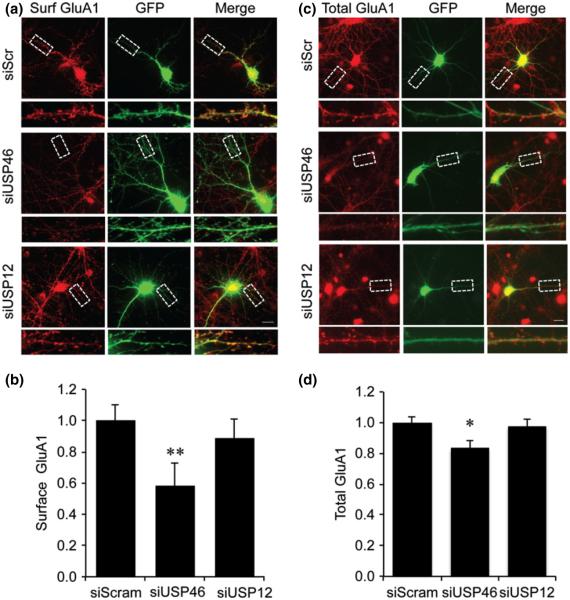
Knockdown of USP46 reduces surface AMPARs in cultured hippocampal neurons
In order to examine the effect of USP46 on AMPA receptor expression and subcellular distribution in vivo, we knocked down USP46 or USP12 by siRNA in DIV 14 cultured hippocampal neurons. A scrambled siRNA was used as a control. Two days after transfection, surface GluA1 was detected by live labeling with antibodies against the extracellular N-terminal of GluA1. A dramatic reduction in surface GluA1 was observed in neurons expressing siUSP46. Consistent with the modest effect of USP12 on AMPAR ubiquitination, no significant changes in surface GluA1 were detected in neurons expressing siUSP12 (Fig. 5a and b). (siScram: 1 ± 0.1, n = 10 cells; siUSP46: 0.58 ± 0.15, n = 10 cells, p = 0.005; siUSP12: 0.89 ± 0.12, n = 6 cells, p = 0.46). In parallel, we also performed immunostainings under permeant conditions so that both surface and intracellular GluA1 was labeled. Compared with the control, total GluA1 was markedly reduced in neurons transfected with siUSP46, but not siUSP12 (Fig. 5c and d) (siScram: 1 ± 0.04, n = 15 cells; siUSP46: 0.83 ± 0.05, n = 16 cells, p = 0.003; siUSP12: 0.98 ± 0.05, n = 11 cells, p = 0.70). These results indicate that USP46-mediated changes in ubiquitination affect AMPAR accumulation and surface expression at the surface sites.
Fig. 5.
Knockdown of USP46 reduces both surface and total AMPARs in hippocampal neurons. Cultured DIV 14 hippocampal neurons were transfected with a scrambled control siRNA (siScramble), or siRNAs specific for USP46 (siUSP46) or USP12 (siUSP12), and immunostained 2 days later. (a and b) To detect surface AMPARs, live cells were incubated with glutamate receptor subunit 1 (GluA1) NT antibodies. Following wash and fixation, surface AMPARs were visualized with fluorescence secondary antibodies. A selected region in each panel was enlarged for clarity. Measurement of GluA1 puncta intensity showed significantly reduction in siUSP46 neurons (siScram, n = 10 cells; siUSP46, n = 10 cells; siUSP12, n = 6 cells) *p < 0.05, **p < 0.01, t-test. (c and d) Transfected cells were immunostained following fixation and permeabilization to label total AMPARs. The intensity of total GluA1 was significantly reduced in USP46 knockdown neurons (siScram, n = 15 cells; siUSP46, n = 16 cells; siUSP12, n = 11 cells) *p < 0.05, t-test. Scale bar represents 20 μm.
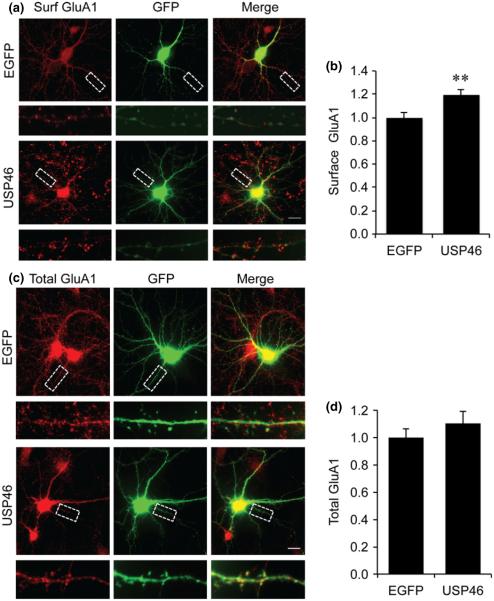
USP46 over-expression affects AMPAR surface expression and surface trafficking
We showed that in HEK cells USP46 over-expression negatively regulates the turnover rate of GluA1 and thus stabilizes GluA1 from degradation. To further confirm this in neurons, we co-transfected DIV 14 cultured hippocampal neurons with Myc-USP46 or Myc-USP12, together with EGFP to visualize transfected neurons. Two days after transfection, neurons were either incubated live with antibodies against GluA1 extracellular N-terminal to label surface AMPARs, or fixed and permeabilized to probe total GluA1 protein levels. We found that the surface GluA1 was significantly enhanced by USP46 over-expression. Quantification indicated a significant difference between EGFP control and cells with USP46 over-expression (Fig. 6a and b) (EGFP: 1 ± 0.05, n = 9 cells; USP46: 1.20 ± 0.05, n = 10 cells, p = 0.002). In contrast, no significant changes were detected in total GluA1 in neurons expressing either USP46 or USP12 (Fig. 6c and d) (EGFP: 1.0 ± 0.07, n = 9 cells; USP46: 1.08 ± 0.09, n = 9 cells, p = 0.20).
Fig. 6.
Over-expression of USP46 increases surface AMPAR expression. (a) Cultured DIV 14 hippocampal neurons were transfected with GFP as control or co-transfected with USP46. Surface glutamate receptor subunit 1 (GluA1) was visualized by live application of antibodies against GluA1 N-terminal. The selected region was enlarged for clarity (bottom). (b) Quantification of the surface synaptic GluA1 puncta intensity showed USP46 over-expression significantly enhanced surface GluA1 level (enhanced GFP, EGFP, n = 9 cells; USP46, n = 10 cells) **p < 0.01, t-test. (c) Effect of USP46 on total GluA1. Following the same transfection, hippocampal neurons were stained under permeant conditions for GluA1. The selected region was enlarged for clarity (bottom). (d) Quantification of the total GluA1 puncta intensity. No significant changes were detected in total GluA1 (EGFP, n = 9 cells; USP46, n = 9 cells). Scale bar represents 20 μm
USP46 over-expression suppresses AMPAR internalization
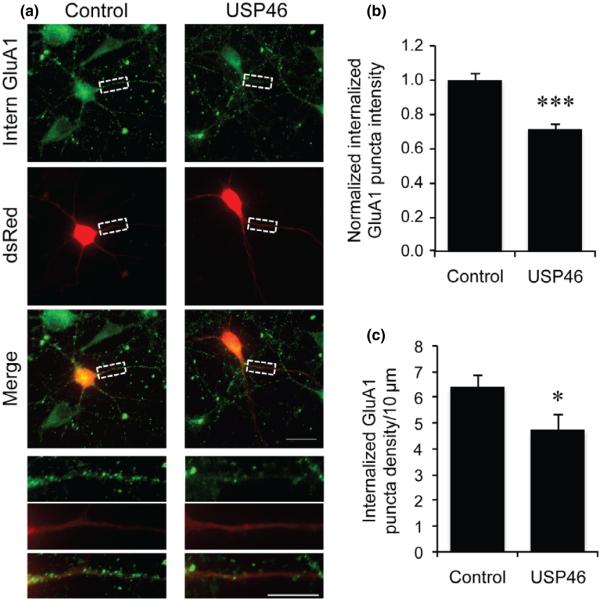
Ubiquitination plays an important role in the control of receptor trafficking. Our previous studies show an effect in AMPAR internalization (Lin et al. 2011; Lin and Man 2014). To determine the role of USP46 in AMPAR trafficking, we performed receptor internalization assays in cultured hippocampal neurons. DIV 12 neurons were transfected with dsRed alone or dsRed and USP46 for 2 days, and the surface AMPARs were labeled with antibodies against the GluA1 N-terminus. Receptor endocytosis was then triggered by a 15-min incubation with glutamate (50 μM). After fixation, the remaining surface GluA1 antibodies were blocked by a non-fluorescent secondary antibody, and the internalized AMPARs were then visualized by application of fluorescent secondary antibodies following cell permeabilization. Consistent with the role of AMPAR deubiquitination, over-expression of USP46 markedly reduced the intensity of internalized GluA1 (Fig. 7a and b) (Control: 1.00 ± 0.04, n = 12 neurons; USP46: 0.71 ± 0.03, n = 12 neurons, p < 0.001). Internalized GluA1 puncta density was also significantly reduced by USP46 over-expression (Fig. 7c) (Control: 6.42 ± 0.40 puncta per 10 μm, n = 12 neurons; USP46: 4.73 ± 0.61 puncta per 10 μm, n = 12 neurons, p = 0.03).
Fig. 7.
Effect of USP46 on AMPAR internalization. (a) Cultured DIV 12 hippocampal neurons were co-transfected with dsRed and pcDNA3.1 as control or dsRed and USP46. GluA1Nt antibody was applied to label surface glutamate receptor subunit 1 (GluA1), followed by application of glutamate (50 μM) to trigger internalization. After blocking of remaining surface GluA1, the internalized GluA1 was revealed by fluorescence secondary antibody. A selected region in each panel was enlarged for clarity. (b) Measurement of internalized GluA1 puncta intensity showed significant reduction in neurons over-expressing USP46 (Control, n = 12 cells; USP46, n = 12 cells) *p < 0.05, **p < 0.01, t-test. (c) Measurement of internalized GluA1 puncta density showing significantly reduction in USP46 transfected neurons (Control, n = 12 cells; USP46, n = 12 cells) *p < 0.05, ***p < 0.001, t-test. Scale bar represents 20 μm; scale bar represents 10 μm in enlarged picture.
USP46 activity regulates AMPAR-mediated excitatory synaptic transmission
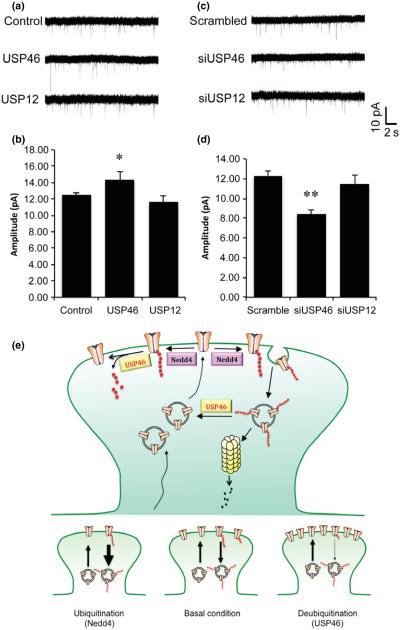
We found that USP46 suppressed AMPAR ubiquitination leading to an increase in receptor protein amount as well as in receptor synaptic localization. To examine the functional role of USP46 on synaptic transmission, we carried out whole-cell patch-clamp recordings of AMPAR-mediated miniature excitatory postsynaptic currents (mEPSCs). Hippocampal neurons were transfected at DIV 12 and recordings were performed 2 days later. Compared to control cells, USP46 over-expression induced a moderate increase in mEPSC amplitude, whereas no change was detected in neurons transfected with USP12 (Fig. 8a and b) (Control: 1.00 ± 0.03, n = 6 cells; USP46: 1.18 ± 0.07, n = 5 cells, p = 0.03; USP12: 0.93 ± 0.07, n = 5 cells, p = 0.30). On the contrary, knockdown of USP46, but not USP12, by siRNA transfection caused a significant reduction in mEPSC amplitude (Fig. 8c and d) (siScram: 1 ± 0.04, n = 6; siUSP46: 0.68 ± 0.05, n = 5, p = 0.001; siUSP12: 0.93 ± 0.07, n = 5, p = 0.42). Thus, synaptic activity is regulated by USP46, presumably via ubiquitination-mediated alterations in AMPAR synaptic expression.
Fig. 8.
USP46 regulates AMPAR mediated synaptic currents. DIV 12 hippocampal neurons were transfected with USP46 or UPS12 for overexpression; or with respective siRNA to knock down USP46 or USP12. Neurons were co-transfected with GFP as an indicator for cell selection during recordings. Two days after transfection, whole-cell recordings were performed to measure the mEPSCs. (a and c) Representative mEPSCs were shown. (b) Over-expression of USP46 caused a significant increase in mEPSC amplitude (Control, n = 6 cells; USP46, n = 5 cells; USP12, n = 5 cells). *p < 0.05, t-test. (d) Knockdown of USP46, but not USP12, resulted in a significant reduction in mEPSC amplitude (siScramble, n = 6 cells; siUSP46, n = 5 cells; siUSP12, n = 5 cells). **p < 0.01, t-test. (e) A schematic illustration of the role of USP46 in AMPAR ubiquitination. Following vesicle-mediated insertion of AMPARs onto the plasma membrane, AMPARs are subject to ubiquitination modification. The E3 ligase Nedd4 conjugates polyubiquitins to the intracellular C-terminals of AMPAR subunits, resulting in receptor internalization and degradation in the proteasome. USP46 specifically removes the ubiquitin chains from AMPARs and counteracts with Nedd4 to prevent receptors from ubiquitination-mediated internalization and degradation.
Discussion
AMPARs are the primary mediator for excitatory synaptic transmission in the brain. Given that the abundance of synaptic AMPARs determines the strength of synaptic transmission, regulation of AMPAR expression at the synaptic domain serves as one of the most important mechanisms underlying synaptic plasticity and brain functions (Huganir and Nicoll 2013). Post-translationally, AMPAR homeostasis is controlled by receptor protein stability and turnover. Recently, it has been shown that AMPARs, including both GluA1 (Schwarz et al. 2010; Lin et al. 2011) and GluA2 subunits (Lussier et al. 2011), can be modified by ubiquitination. Specifically, GluA1 is subjected to polyubiquitination to facilitate internalization (Lin et al. 2011). However, whether and how AMPARs can be deubiquitinated remain poorly understood. In this study, we show that USP46 is the DUB enzyme that specifically deubiquitinates AMPARs both in vitro and in vivo. USP46 over-expression leads to a reduction in AMPAR ubiquitination, whereas knockdown of USP46 by either siRNAs or shRNAs causes a significant increase in AMPAR ubiquitination. Consistent with our previous work indicating that ubiquitination facilitates AMPAR degradation, suppressing ubiquitination by USP46 slows down the degradation and prolongs the half-life of AMPARs. More relevant to neuronal function, USP46 activity results in changes in AMPAR accumulation and surface expression at synapses. In line with this finding, synaptic mEPSC amplitude is also regulated by USP46, indicating a role of the deubiquitination process in synaptic plasticity. We find an increase in synaptic GluA1 by USP46 over-expression, and vice versa. Thus, in counterbalancing the effect of the E3 ligase Nedd4, USP46 should play a crucial role in the maintenance of AMPAR proteostasis.
USP46 is the mammalian ortholog of a DUB identified in C. elegans shown to be able to suppress ubiquitination in glutamate receptors (Kowalski et al. 2011). USP46 and its C. elegans ortholog share 60% identity (and 71% similarity) in amino acid sequence. USP46 is a 366 amino acid protein containing a catalytic core, but lacking subdomains or motifs (Nijman et al. 2005). Within the same family, USP12 shares 88% sequence identity with USP46. They both possess the same USP catalytic triad (cysteine, histidine, and aspartic acid) within the catalytic core domain that occupies almost the full length of the proteins (Cohn et al. 2009). The DUB activity of USP46 and USP12 has been proven in vitro (Quesada et al. 2004; Cohn et al. 2009; Kee et al. 2010). In C. elegans, USP46 and USP12 both bind to USP1-associated factor 1 (UAF1/WDR48) and the binding dramatically enhances the activity of USP12 and USP46 (Cohn et al. 2007, 2009; Dahlberg and Juo 2014). Our findings indicate that USP46 function is well conserved among species. Interestingly, although USP12 shares high similarity with USP46, USP12 does not show significant effect in AMPAR deubiquitination, suggesting enzyme specificity to a target. It remains elusive as to which domain or sequence in USP46 and USP12 determines the substrate specificity. We observed that USP46 is able to deubiquitinate not only GluA1 but also GluA2, indicating a relative stringency in contrast to the distinct E3 ligases that are employed for GluA1 and GluA2 (Lin et al. 2011; Lussier et al. 2011). Indeed, it has been estimated that in the human genome there are 600 E3 ubiquitin ligases but only about 80 DUBs (Komander et al. 2009; Kowalski and Juo 2012). A recent study revealed that USP8 and Nedd4 inversely regulate surface AMPAR levels, suggesting the role of DUBs on regulating synaptic protein levels and synaptic strength (Scudder et al. 2014).
It is somewhat surprising to observe a high enrichment of USP46 in the synaptosomal P2 fraction, which is supported by immunostaining, showing its co-localization with synaptic proteins including GluA1 and PSD-95. Given the fact that no interacting domains or motifs have been identified on USP46 (Komander et al. 2009), it is intriguing about the molecular basis for its extensive residency in the synaptic compartment. Expression profiling during development reveals that USP46 levels reached a peak during DIV 11–19, a time window well overlapping with synaptogenesis and glutamate receptor surface expression, underscoring the potential importance of USP46 not only in AMPAR turnover, but probably also in synaptic formation or neuronal morphogenesis (Drinjakovic et al. 2010). Our results show that USP46 activity regulates the efficacy of synaptic transmission, presumably resulting from ubiquitination-mediated alteration in synaptic AMPAR accumulation. Dynamic trafficking regulates the amount of synaptic AMPARs; rapid changes in membrane insertion and internalization of AMPARs serve as the fundamental mechanism for neuronal plasticity (Zhao et al. 2003; Shepherd and Huganir 2007; Widagdo et al. 2015). Ubiquitinated surface AMPARs are marked for efficient endocytosis, leading to inhibited synaptic transmission (Lin et al. 2011). A recent study showed that GluA1 and GluA2 intracellular sorting and degradation were ubiquitination-dependent, suggesting a role for AMPAR ubiquitination on internalization in physiological conditions (Widagdo et al. 2015). Nedd4 expression is markedly increased in human brains of degenerative diseases including Alzheimer’s, Parkinson’s, and Huntington’s diseases, which is implicated in the ubiquitination and degradation of IGF receptors and neuronal cell death (Kwak et al. 2012). The effect of Nedd4-mediated ubiquitination and USP46-dependent deubiquitination on AMPARs in these diseases remains unknown. Our results show that reversing the ubiquitination modification by USP46 leads to AMPAR accumulation and enhanced synaptic strength, which may have functional consequences on brain function. Consistent with this, animal behavior studies have indicated that USP46 can affect brain function. A USP46 mutant with one lysine codon deletion has been identified in mice that showed extremely low immobility time in the tail suspension test and forced swimming test (Tomida et al. 2009). In sequence-based large scale screening studies, researchers showed that one Single nucleotide polymorphism within the USP46 coding region is highly associated with schizophrenia in a Caucasian population (Need et al. 2009). More recently, USP46 binding partners, WDR-20 and WDR-48, have been identified in C. elegans, and loss of function of these molecules affects normal locomotion behavior (Dahlberg and Juo 2014). The USP46-linked pathology may, at least in part, result from dysregulation in AMPAR stability and synaptic localization.
Supplementary Material
Acknowledgments
We thank Dr Peter Juo at Tufts University for providing cDNA constructs, and Man Lab members for helpful discussion. This work was supported by NIH Grant R01 MH079407 (HYM).
All experiments were conducted in compliance with the ARRIVE guidelines.
Abbreviations used
- AMPAR
alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- DUB
deubiquitinating enzyme
- EGFP
enhanced GFP
- GFP
green fluorescent protein
- GluA1
glutamate receptor subunit 1
- HA
hemagglutinin
- HEK
human embryonic kidney
- Nedd4
neural precursor cell expressed, developmentally down-regulated 4
- PSD95
postsynaptic density protein 95
- UPP
ubiquitin–proteasome pathway
- USP12
ubiquitin-specific peptidase 12
- USP46
ubiquitin-specific peptidase 46
Footnotes
Conflict of interest disclosure
The authors have no conflicts of interest to declare.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site:
Figure S1. Over-expression of USP46 or USP12 does not affect the expression of E3 ligases.
References
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg CL, Juo P. The WD40-repeat proteins WDR-20 and WDR-48 bind and activate the deubiquitinating enzyme USP-46 to promote the abundance of the glutamate receptor GLR-1 in the ventral nerve cord of Caenorhabditis elegans. J. Biol. Chem. 2014;289:3444–3456. doi: 10.1074/jbc.M113.507541. doi: 10.1074/jbc.M113.507541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinjakovic J, Jung H, Campbell DS, Strochlic L, Dwivedy A, Holt CE. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Zhang D, Jarzylo L, Huganir RL, Man HY. Homeostatic regulation of AMPA receptor expression at single hippocampal synapses. Proc. Natl Acad. Sci. USA. 2008;105:775–780. doi: 10.1073/pnas.0706447105. doi: 10.1073/pnas.0706447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D’Andrea AD. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J. Biol. Chem. 2010;285:11252–11257. doi: 10.1074/jbc.M109.095141. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kowalski JR, Juo P. The role of deubiquitinating enzymes in synaptic function and nervous system diseases. Neural. Plast. 2012;2012:892749. doi: 10.1155/2012/892749. doi: 10.1155/2012/892749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J. Neurosci. 2011;31:1341–1354. doi: 10.1523/JNEUROSCI.4765-10.2011. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YD, Wang B, Li JJ, Wang R, Deng Q, Diao S, Liao FF. Upregulation of the E3 ligase NEDD4-1 by oxidative stress degrades IGF-1 receptor protein in neurodegeneration. J. Neurosci. 2012;32:10971–10981. doi: 10.1523/JNEUROSCI.1836-12.2012. doi: 10.1523/JNEUROSCI.1836-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Man HY. Ubiquitination of neurotransmitter receptors and postsynaptic scaffolding proteins. Neural. Plast. 2013;2013:432057. doi: 10.1155/2013/432057. doi: 10.1155/2013/432057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Man HY. Endocytic adaptor epidermal growth factor receptor substrate 15 (Eps15) is involved in the trafficking of ubiquitinated alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptors. J. Biol. Chem. 2014;289:24652–24664. doi: 10.1074/jbc.M114.582114. doi: 10.1074/jbc.M114.582114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, Man HY. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J. Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier MP, Nasu-Nishimura Y, Roche KW. Activity-dependent ubiquitination of the AMPA receptor subunit GluA2. J. Neurosci. 2011;31:3077–3081. doi: 10.1523/JNEUROSCI.5944-10.2011. doi: 10.1523/JNEUROSCI.594410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58:472–497. doi: 10.1016/j.neuron.2008.04.030. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, Lopez-Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. doi: 10.1146/annurev. biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Hall BJ, Patrick GN. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 2010;30:16718–16729. doi: 10.1523/JNEUROSCI.3686-10.2010. doi: 10.1523/JNEUROSCI.3686-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudder SL, Goo MS, Cartier AE, Molteni A, Schwarz LA, Wright R, Patrick GN. Synaptic strength is bidirectionally controlled by opposing activity-dependent regulation of Nedd4-1 and USP8. J. Neurosci. 2014;34:16637–16649. doi: 10.1523/JNEUROSCI.2452-14.2014. doi: 10.1523/JNEUROSCI.2452-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 2003;13:899–910. doi: 10.1016/s0960-9822(03)00338-5. [DOI] [PubMed] [Google Scholar]
- Tomida S, Mamiya T, Sakamaki H, Miura M, Aosaki T, Masuda M, Ebihara S. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 2009;41:688–695. doi: 10.1038/ng.344. doi: 10.1038/ng.344. [DOI] [PubMed] [Google Scholar]
- Wang G, Gilbert J, Man HY. AMPA receptor trafficking in homeostatic synaptic plasticity: functional molecules and signaling cascades. Neural. Plast. 20122012:825364. doi: 10.1155/2012/825364. doi: 10.1155/2012/825364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Chai YJ, Ridder MC, Chau YQ, Johnson RC, Sah P, Anggono V. Activity-dependent ubiquitination of GluA1 and GluA2 regulates AMPA receptor intracellular sorting and degradation. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.015. doi: 10.1016/j.celrep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol. Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hegde AN, Martin KC. The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Curr. Biol. 2003;13:887–898. doi: 10.1016/s0960-9822(03)00332-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.