Abstract
The evolutionary relationships between Peromyscus, Habromys, Isthmomys, Megadontomys, Neotomodon, Osgoodomys, and Podomys are poorly understood. In order to further explore the evolutionary boundaries of Peromyscus and compare potential taxonomic solutions for this diverse group and its relatives, we conducted phylogenetic analyses of DNA sequence data from alcohol dehydrogenase (Adh1-I2), beta fibrinogen (Fgb-I7), interphotoreceptor retinoid-binding protein (Rbp3), and cytochrome-b (Cytb). Phylogenetic analyses of mitochondrial and nuclear genes produced similar topologies although levels of nodal support varied. The best-supported topology was obtained by combining nuclear and mitochondrial sequences. No monophyletic Peromyscus clade was supported. Instead, support was found for a clade containing Habromys, Megadontomys, Neotomodon, Osgoodomys, Podomys, and Peromyscus suggesting paraphyly of Peromyscus and confirming previous observations. Our analyses indicated an early divergence of Isthmomys from Peromyscus (approximately 8 million years ago), whereas most other peromyscine taxa emerged within the last 6 million years. To recover a monophyletic taxonomy from Peromyscus and affiliated lineages, we detail 3 taxonomic options in which Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys are retained as genera, subsumed as subgenera, or subsumed as species groups within Peromyscus. Each option presents distinct taxonomic challenges, and the appropriate taxonomy must reflect the substantial levels of morphological divergence that characterize this group while maintaining the monophyletic relationships obtained from genetic data.
Key Words: Habromys, Megadontomys, Neotomodon, Osgoodomys, Peromyscus, phylogeny, Podomys, species group, systematics, taxonomy
What is Peromyscus? More than 100 years since Osgood’s (1909) monograph the question remains unsolved. A historical perspective and overview of the taxonomic challenges affiliated with Peromyscus are provided in Bradley et al. (2007), Carleton (1980, 1989), and Miller and Engstrom (2008). At conflict is the taxonomic status of Habromys, Isthmomys, Megadontomys, Neotomodon, Osgoodomys, and Podomys. At various times, these taxa have been recognized at the generic (sensu stricto) or subgeneric (sensu lato) level, though most major classifications generally fall into 1 of 2 categories. No single historical classification fits perfectly into the sensu stricto or sensu lato categories, though most major classifications tend to reflect one interpretation over the other. Carleton (1980, 1989) and Musser and Carleton (2005) are most closely aligned with a Peromyscus (sensu lato) taxonomy, whereas Hooper (1968) is a variation of a Peromyscus (sensu stricto) classification. Most current classifications recognize Peromyscus (sensu stricto).
Bradley et al. (2007) completed the most comprehensive molecular study of Peromyscus and its allies in which DNA sequences from the entire mitochondrial cytochrome-b gene (Cytb) were examined. They found 5 genera (Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys) to be embedded within a monophyletic clade containing Peromyscus (sensu stricto); Isthmomys was sister to Reithrodontomys and basal to this group. In order to recognize Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys as genera, Bradley et al. (2007) stated that at least 5 additional genera would have to be recognized to accommodate strongly supported clades under the rules of monophyly and phylogenetic principles.
Miller and Engstrom (2008) added to the molecular data set by obtaining DNA sequences from Cytb as well as 2 nuclear genes: interphotoreceptor retinoid-binding protein (Rbp3) and growth hormone receptor (Ghr). Their results were similar to Bradley et al. (2007) in that Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys were placed inside of Peromyscus (sensu stricto) and Isthmomys was sister to Reithrodontomys. Further, Miller and Engstrom (2008) agreed with Bradley et al. (2007) that additional groups would have to be elevated to avoid paraphyly in a sensu stricto interpretation of Peromyscus.
The primary objective in this study was to examine the phylogenetic relationships within the genus Peromyscus using a combination of mitochondrial and nuclear markers. Although the studies of Bradley et al. (2007) and Miller and Engstrom (2008) recovered paraphyly within Peromyscus, each study had limitations that impact phylogenetic interpretation. Bradley et al. (2007) had a more broad taxonomic sampling scheme but was based on a single genetic marker (Cytb). Miller and Engstrom (2008) lacked representation of some species groups but included multiple genetic markers. Herein, we seek to expand upon these molecular data sets by including representatives from all species groups and to examine DNA sequences from 4 markers, 2 of which were not used in the previous studies: intron 2 of the alcohol dehydrogenase (Adh1-I2) and intron 7 of the beta fibrinogen gene (Fgb-I7). We selected these markers based on their previous use in rodent phylogenetics (Amman et al. 2006; Longhofer and Bradley 2006; Reeder and Bradley 2007). We combined nuclear and mitochondrial DNA sequence data from Adh1-I2, Fgb-I7, Rbp3, and Cytb to test the monophyly of Peromyscus (sensu stricto versus sensu lato) and to ascertain whether Habromys, Isthmomys, Megadontomys, Neotomodon, Osgoodomys, and Podomys are paraphyletic within Peromyscus. The genetic evidence presented herein support the need for a formal taxonomic revision of Peromyscus. Below we identify 3 potential taxonomic solutions that are consistent with the evidence in hand.
Materials and Methods
Samples.
Tissue samples obtained from individuals collected in naturally occurring populations or through museum loans were used to generate DNA sequences for the 4 genetic markers described below. In some cases, DNA sequences were obtained directly from GenBank. A single representative was examined from the following taxa: Baiomys, Habromys, Isthmomys, Megadontomys, Neotoma, Neotomodon, Ochrotomys, Onychomys, Osgoodomys, and Podomys; 4 representatives were included for Reithrodontomys. For the subgenus Peromyscus, 2 representatives of each of the species groups were examined (except the crinitus, furvus, hooperi, megalops, and melanophrys species groups—1 sample each). Likewise, for the subgenus Haplomylomys, 1 sample each from the californicus and eremicus species groups were examined. An attempt was made to obtain mitochondrial and nuclear sequences from a single individual, but in a few instances, this was not possible. In these cases, sequences from conspecific individuals within close geographic proximity were used to complete the data set. Concatenation of sequence data from conspecifics to represent a composite species rather than a single individual has been successfully used in various taxa (Campbell and Lapointe 2009; Townsend et al. 2011; Haddrath and Baker 2012). This strategy guaranteed at least 1 individual per species group was sampled. Specimens used in this study are listed in Table 1.
Table 1.
Specimens examined in this study are listed by taxon and genetic marker (Adh1-I2—intron 2 of alcohol dehydrogenase, Cytb—cytochrome-b, Fgb-I7—intron of the beta-fibrinogen, and Rbp3—interphotoreceptor retinoid-binding protein) and grouped by tribe, genus, and species group. GenBank accession (left of slash) and museum catalog (right of slash) numbers are given for each specimen. Museum acronyms are as follows: ASNHC (Angelo State Natural History Collection), BYU (Brigham Young University), CNMA (Colección Nacional de Mamíferos, Universidad Nacional Autónoma de México), PGSC (Peromyscus Genetic Stock Center), ROM (Royal Ontario Museum), TCWC (Texas Cooperative Wildlife Collection), and TTU (Museum of Texas Tech University). If museum catalog numbers were unavailable, specimens were referenced with the corresponding collector’s numbers or TK (special number of the Museum of Texas Tech University).
| Taxon | Adh1-I2 | Cytb | Fgb-I7 | Rbp3 |
|---|---|---|---|---|
| Tribe Baiomyini | ||||
| Baiomys | ||||
| B. taylori | AY994205/TTU82642 | AF548469/TTU54633 | AY274213/TTU54633 | EF989838/ROM114886 |
| Tribe Ochrotomyini | ||||
| Ochrotomys | ||||
| O. nutalli | JX910114/TCWC31929 | AY195798/TCWC31929 | AY274203/TCWC31929 | EF989862/ROM113008 |
| Tribe Neotomini | ||||
| Neotoma | ||||
| N. mexicana | AY817646/TTU79129 | AF294345/TTU79129 | AY274200/TTU79129 | JX910120/TTU79129 |
| Tribe Reithrodontomyini | ||||
| Habromys | ||||
| H. ixtlani | AY994239/TK93160 | DQ000482/TK93160 | FJ214701/TTU82703 | EF989842/CNMA29849 |
| Isthmomys | ||||
| I. pirrensis | FJ214668/TTU39162 | FJ214681/TTU39162 | FJ214692/TTU39162 | EF989846/ROM116308 |
| Neotomodon | ||||
| N. alstoni | AY994210/TK45309 | AY195796/TK45302 | AY274202/TK45309 | EF989851/ASNHC1595 |
| Megadontomys | ||||
| M. thomasi | AY994208/TK93388 | AY195795/TK93388 | FJ214693/TK93388 | EF989849/CNMA29186 |
| Onychomys | ||||
| O. arenicola | JX910115/TTU67559 | AY195793/TTU67559 | AY274204/TTU67559 | EF989855/ROM114904 |
| Osgoodomys | ||||
| O. banderanus | AY994209/TK45952 | DQ000473/TK45952 | FJ214694/TK45952 | EF989857/ASNHC2664 |
| Peromyscus | ||||
| aztecus group | ||||
| P. evides | FJ214670/TTU82696 | FJ214685/TTU82696 | FJ214700/TTU82696 | JX910121/TTU82696 |
| P. spicilegus | AY994234/TK45255 | FJ214669/TK45255 | FJ214695/TK45255 | JX910122/TK47888 |
| boylii group | ||||
| P. boylii | AY994227/TTU82688 | AF155388/TTU81702 | AY274208/TTU81702 | EF989871/ASNHC3449 |
| P. levipes | AY994224/TK47819 | DQ000477/TK47819 | FJ214707/TTU105150 | JX910123/TK47819 |
| californicus group | ||||
| P. californicus | AY994211/TTU83292 | AF155393/TTU81275 | FJ214697/TTU83291 | EF989873/PGSCIS1590 |
| crinitus group | ||||
| P. crinitus | AY994213/DSR6171 | FJ214684/TK119629 | FJ214698/TK119629 | EF989874/BYU16629 |
| eremicus group | ||||
| P. eremicus | AY994212/TTU81850 | AY322503/TTU83249 | FJ214699/TTU83249 | EF989876/BYU17952 |
| furvus group | ||||
| P. furvus | JX910116/FXG1168 | AF271032/CNMA32298 | JX910113/FXG1168 | JX910124/FXG1168 |
| hooperi group | ||||
| P. hooperi | FJ214672/TTU104425 | DQ973103/TTU104425 | FJ214704/TTU104425 | JX910125/TTU104425 |
| leucopus group | ||||
| P. gossypinus | FJ214671/TTU80682 | DQ973102/TTU80682 | FJ214702/TTU80682 | JX910126/TTU80682 |
| P. leucopus | AY994240/TTU75694 | DQ000483/TTU101645 | FJ214706/TTU101645 | EF989880/ROM101861 |
| maniculatus group | ||||
| P. maniculatus | AY994242/TTU97830 | DQ000484/TTU38739 | FJ214708/TTU97830 | EF989884/ROM98941 |
| P. melanotis | FJ214673/TK70997 | AF155398/TK70997 | FJ214711/TK70997 | EF989891/PGSC25 |
| megalops group | ||||
| P. megalops | AY994217/TTU82712 | DQ000475/TTU82712 | FJ214709/TTU82712 | JX910127/TTU82712 |
| melanophrys group | ||||
| P. melanophrys | AY994216/TTU75509 | AY322510/TTU75509 | FJ214710/TTU75509 | EF989890/PGSCXZ1073 |
| mexicanus group | ||||
| P. mexicanus | AY994236/TTU97013 | JX910118/TTU105005 | AY274210/TTU82759 | EF989895/ROM113250 |
| P. nudipes | AY994238/TTU96972 | FJ214687/TTU96972 | FJ214713/TTU96972 | EF989893/ROM113216 |
| truei group | ||||
| P. attwateri | AY994220/TTU55688 | AF155384/TTU55688 | AY274207/TTU55688 | JX910128/TTU55688 |
| P. gratus | AY994218/TK46354 | AY376421/TK46354 | FJ214703/TK46354 | JX910129/TK46354 |
| Unknown group | ||||
| P. ochraventer | FJ214676/TTU104930 | JX910119/TTU104930 | FJ214715/TTU104930 | JX910130/TTU104930 |
| P. pectoralis | AY994221/TK48645 | AY376427/TK48642 | FJ214716/TK48645 | JX910131/TK48645 |
| Podomys | ||||
| P. floridanus | AY994214/TTU97867 | DQ973109/TTU97867 | FJ214723/TTU97867 | EF989878/TTU97866 |
| Reithrodontomys | ||||
| R. fulvescens | AY994207/TTU54898 | AF176257/TTU54898 | AY274211/TTU54898 | EF989901/ASNHC3465 |
| R. sumichrasti | JX910117/TTU54952 | AF176256/TTU54952 | AY274212/TTU54952 | EF989924/ROM98383 |
| R. megalotis | AF176248/TTU40942 | AF176248/ TTU40942 | KJ697789/TTU40942 | EF989909/ASNHC2133 |
| R. mexicanus | KJ697791/TTU85234 | AY859453/ROM101508 | — | EF989911/ROM98468 |
Dna Isolation and Pcr.
DNA was isolated from liver samples (0.1g) using 2 methods. Mitochondrial DNA was extracted and purified using a Wizard Miniprep kit (Promega, Madison, Wisconsin), whereas total genomic DNA was extracted from liver using DNeasy Blood and Tissue kits (Qiagen, Valencia, California) following the method of Smith and Patton (1999). The complete mitochondrial Cytb gene (1,143bp) was amplified following methods outlined in Bradley et al. (2007) and Tiemann-Boege et al. (2000) using primers MVZ05 (Smith and Patton 1993), H15915 (Irwin et al. 1991), and CB40 (Hanson and Bradley 2008). Intron 2 of the alcohol dehydrogenase gene (Adh1-I2, 598bp) was amplified following the methods of Amman et al. (2006) using primers 2340-I, 2340-II, Exon II-F, and Exon III-R. The complete intron of the beta-fibrinogen gene (Fgb-I7, 674bp) was amplified following the methods of Carroll et al. (2005) and Wickliffe et al. (2003) using primers Fgb-17U-Rattus, Fgb-17L-Rattus (Wickliffe et al. 2003), B17-mammU, and B17-mammL (Matocq et al. 2007). Exon I of interphotoreceptor retinoid-binding protein gene (Rbp3, 924bp) was amplified following the methods of Chambers et al. (2009) and Jansa and Voss (2000) using primers A and B (Stanhope et al. 1992).
Sequencing.
PCR products were purified using the QIAquick PCR Purification kit (Qiagen) or ExoSAP-IT (USB Products, Cleveland, Ohio) and PCR amplicons were sequenced using ABI Prism Big Dye Terminator v3.1 ready reaction mix (Applied Biosystems, Foster City, California). Nucleotide sequences were resolved on an ABI 3100 Avant automated sequencer (Applied Biosystems) with the following primers: Cytb—PERO3′ and 752R (Tiemann-Boege et al. 2000), CWE-1 and 400F (Edwards et al. 2001), and 700L and WDRAT400R (Peppers and Bradley 2000); Adh1-I2—Exon II-F, Exon III-R, Adh350F, and Adh350R (Amman et al. 2006); Fgb-I7—Fgb-17U-Rattus and Fgb-17L-Rattus (Wickliffe et al. 2003) and bFIB-I7U and bFIB-I7L (Carroll et al. 2005); and Rbp3—A, B, and D (Stanhope et al. 1992), E2 (Weksler 2003), and 125F (DeBry and Sagel 2001). Sequencher 5.0 software (Gene Codes Corporation 2013) was used to align and proof individual sequencing reads into contigs representing each gene. Conflicting base calls were verified against the associated chromatograms. For nuclear intron sequences, all heterozygous sites were designated following the International Union of Biochemistry polymorphic code. All DNA sequences were deposited in GenBank and accession numbers are provided in Table 1.
Phylogenetic Analysis.
Nucleotide positions were treated as unordered, discrete characters with 6 possible states: A, C, G, T, gaps (−), or missing (?) for each marker. For nuclear intron sequences, polymorphic sites were designated following the International Union of Biochemistry polymorphic code. However, because these polymorphisms could be the result of heterozygosity or sequencing error, to be conservative, these nucleotides were excluded from downstream analyses. Alignment of Adh1-I2 and Fgb-I7 sequences required hypothesized gaps (inserted based on homology) to represent insertion or deletion events, but gaps were not included in the phylogenetic analysis. Analyses were conducted using 3 data sets: nuclear (Adh1-I2, Fgb-I7, and Rbp3), mitochondrial (Cytb), and combined (Adh1-I2, Cytb, Fgb-I7, and Rbp3). Neotoma mexicana was used as the outgroup taxon for all analyses (Bradley et al. 2004b).
MrModeltest and the Akaike information criterion (AIC—Nylander 2004) were used to estimate the most appropriate model of evolution for each gene region. Bayesian inference (BI) was conducted to estimate a phylogeny and generate posterior probability values for the mitochondrial, nuclear, and combined data sets using MRBAYES v3.2.1 (Ronquist et al. 2012). Each analysis included the appropriate model identified by MrModeltest (Nylander 2004), 2 simultaneous runs of 4 Markov-chains, 10×109 generations, and a sample frequency of every 1,000th generation. The number of invariable sites and gamma distribution were estimated from the data. After a visual inspection of likelihood score distributions in Tracer v1.5 (Drummond and Rambaut 2008), the first 10,000 trees were discarded and a consensus tree (50% majority rule) was constructed from the remaining trees. Values ≥ 95% were viewed as supportive following Alfaro et al. (2003), Douady et al. (2003), and Huelsenbeck et al. (2002). For ML analyses, RaxML (Stamatakis et al. 2005) was used to generate trees from each data set. In these analyses, the GTR+G substitution model was used since the less parameter rich HKY+G model, identified by MrModeltest (Nylander 2004) as the most appropriate model, is unavailable. Nodal support was estimated with 10,000 bootstrap replicates using the “fast bootstrapping” option (Felsenstein 1985).
Topological Tests.
Maximum likelihood (ML) trees from RAxML (Stamatakis et al. 2005) were used to test the difference between competing taxonomic hypotheses. Site likelihood scores generated in RAxML (Stamatakis et al. 2005) were used to score several constrained topologies. P-values were generated in Consel (Shimodaira and Hasegawa 2001) for each topology using the approximately unbiased test (Shimodaira 2002). In particular, Peromyscus (sensu stricto) versus (sensu lato) were tested against the ML topology from the combined data analysis as well as other alternative hypothetical taxonomic groupings.
Molecular Dating.
BEAST v1.7 (Drummond et al. 2012) was used to estimate divergence dates for the sampled taxa. Sequence data from each gene were used in the analysis but were partitioned to allow modeling of each data set. Models of substitution were the same as those used in previous Bayesian analyses (see above). The program MEGA 5.05 (Tamura et al. 2011) was used to determine whether to accept or reject a strict molecular clock for each data set. Given all data sets contained a single individual from each species sampled, a Yule tree prior was chosen for the BEAST analysis. Fossil limits were used to calibrate the leucopus/maniculatus group (~0.3 million years ago [mya]—Dalquest 1962; Karow et al. 1996) and Reithrodontomys (~1.8 mya—Cassiliano 1999). To account for the uncertainty in the fossil record, a prior lognormal distribution was used for both calibrations with means and standard deviations adjusted to create an upper bound of 14.8 mya to reflect the closest dated fossil outside of the taxa sampled (Behrensmeyer and Turner 2013). Test runs of 2.5×107 generations with a 10% burn-in were used to optimize for the final analysis. Bayes factors (Kass and Raftery 1995; Suchard et al. 2001) were calculated to compare the results of test runs to determine final parameters. Two final runs of 1.0×108 generations were analyzed with log and tree files combined for final divergence date estimates. Results were examined for sufficient mixing, convergence stability, and effective sample size > 200 for all parameters using the program Tracer.
Genetic Divergence.
To compare rates of genetic divergence between taxa recognized at various taxonomic ranks, Kimura 2-parameter (K2P—Kimura 1980) genetic distance values were compared among currently recognized genera (Habromys, Isthmomys, Megadontomys, Neotomodon, Osgoodomys, and Podomys). The K2P model was selected based on its utility as a distance metric in rodent phylogenetics (Bradley and Baker 2001).
Results
Phylogenetic Analyses.
Twenty-seven species of Peromyscus (sensu lato) and 8 additional taxa (outgroup and reference samples), representing taxonomic diversity within the Neotominae, were sampled for the nuclear introns Adh1-I2 and Fgb-I7, nuclear exon Rbp3, and the mitochondrial gene Cytb. The entire 560bp of Adh1-I2, 590bp of Fgb-I7, and 921bp of Rbp3 were analyzed for 23 of the 27 Peromyscus (sensu lato) species. Full gene sequences were not available for Peromyscus californicus (Rbp3: 833 of 921bp), P. eremicus (Rbp3: 907 of 921bp), P. furvus (Cytb: 540 of 1,143bp), or P. ochraventer (Rbp3: 914 of 921bp). To analyze the most complete data set possible, other gene sequences available through GenBank, including Ghr, were not used due to lack of sequence data for many taxa. MrModeltest (Nylander 2004) identified the substitution models HKY+G for Adh1-I2 (AIC = 6577.2827, −lnL = 3283.6414, G = 1.1092), GTR+G for Fgb-I7 (AIC = 6931.1782, −lnL = 3456.5891, G = 1.3322), and GTR+I+G for Rbp3 (AIC = 6213.5400, −lnL = 3096.7700, I = 0.4097, G = 0.8473) as the best-fit models.
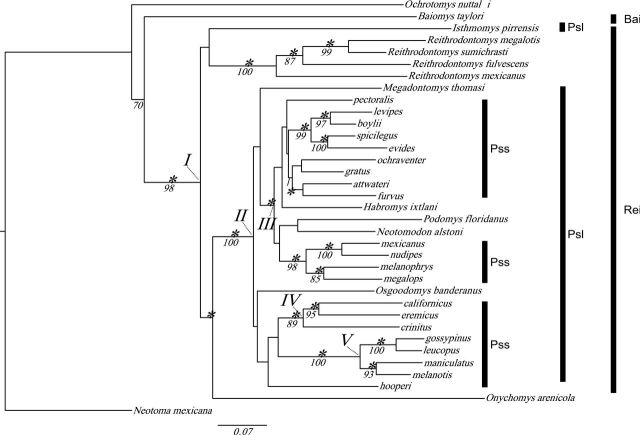
Individual Adh1-I2, Cytb, Fgb-I7, and Rbp3 sequences were combined to generate a single concatenated sequence (3,283bp). The ML phylogeny (−lnL = 22631.507197) with bootstrap values and Bayesian clade probability values are shown in Fig. 1. Specific placement of some taxa varied between the ML and BI analyses, specifically in the placement of Megadontomys thomasi, Osgoodomys banderanus, and P. hooperi. Despite uncertain placement of these taxa, placement of terminal taxa within well-supported nodes did not conflict between markers. Significant nodal support was obtained throughout the phylogeny with most basal and terminal nodes garnering support. Middle regions of the phylogeny were less likely to receive nodal support. Both BI and ML analyses were unable to recover a monophyletic Peromyscus (sensu stricto) or Peromyscus (sensu lato) clade.
Fig. 1.
Phylogenetic tree obtained from maximum likelihood analysis of the combined mitochondrial cytochrome-b gene (Cytb) and 3 nuclear genes (alcohol dehydrogenase—Adh1-I2, beta fibrogen—Fgb-I7, and interphotoreceptor retinoid-binding protein—Rbp3). Taxonomic groups of interest are designated as follows: Pss (Peromyscus [sensu stricto]), Psl (Peromyscus [sensu lato]), Rei (Reithrodontomyini), and Bai (Baiomyini). Nodal support values are superimposed on the maximum likelihood tree topology. Support values are as follows: 10,000 bootstrap replicates of the maximum likelihood analysis (below branch) and clade probability values for the Bayesian inference analysis (above branch). Statistically significant clade probability values (≥ 0.95) are designated with an asterisk. All bootstrap support values ≥ 50 are shown. For members of Peromyscus (sensu stricto) only, species epithets are given. Peromyscus (sensu lato) affiliated genera are indicated in bold. Major nodes are indicated with roman numerals.
Constrained topologies reflecting various taxonomic groupings were tested using the approximately unbiased test in CONSEL (Shimodaira and Hasegawa 2001) with 10,000 replicates per test. A generalized Peromyscus (sensu lato) was unable to be rejected (P = 0.406), but Peromyscus (sensu stricto) was strongly rejected (P = 0.014) by the approximately unbiased test. An additional taxonomic scheme uniting Peromyscus (sensu lato), but excluding Isthmomys pirrensis, was unable to be rejected (P = 0.556).
Molecular Dating.
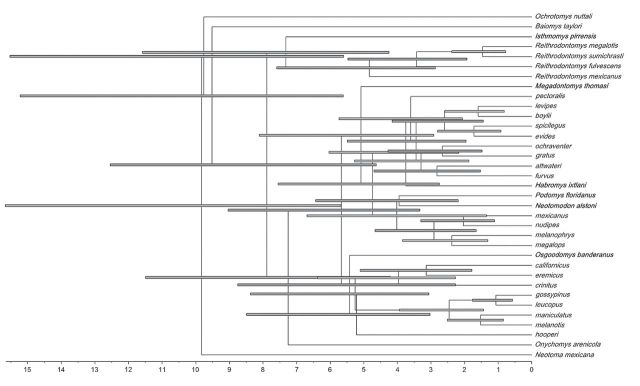
Molecular clock tests (Tamura et al. 2011) indicated a strict molecular clock for the Cytb and Fgb-I7 data sets but a relaxed molecular clock for the Adh1-I2 and Rbp3 data sets. BEAST analyses estimated a Yule birth rate of 0.23 (95% highest posterior density [HPD]: 0.11–0.37). Mean rates of evolution (as substitutions per site per million years) were 0.007 for Adh1-I2 (95% HPD: 0.004–0.01), 0.06 for Cytb (95% HPD: 0.03–0.09), 0.006 for Fgb-I7 (95% HPD: 0.003–0.009), and 0.003 for Rbp3 (95% HPD: 0.001–0.004). Divergence date estimates (Fig. 2) suggested that the split of Reithrodontomyini and Baiomyini began approximately 9.56 mya (95% HPD: 5.65–15.27), during the Late Miocene. The split between the Isthmomys/Reithrodontomys clade and Onychomys/Peromyscus (senus lato) clade was estimated to occur approximately 7.93 mya (95% HPD: 4.67–12.59), also in the Late Miocene. In addition, the divergence of Peromyscus (sensu lato) was estimated to occur during the Late Miocene but near the Miocene/Pliocene boundary (approximately 5.71 mya, 95% HPD: 3.37–9.08). However, most species-level divergence within Peromyscus (sensu lato) occurred during the Blancan North American land mammal age (1.8–4.9 mya).
Fig. 2.
Maximum clade credibility tree showing divergence date estimates based on a combined analysis of the mitochondrial cytochrome-b gene (Cytb) and 3 nuclear genes (alcohol dehydrogenase—Adh1-I2, beta fibrogen—Fgb-I7, and interphotoreceptor retinoid-binding protein—Rbp3). Divergence date estimates are indicated along the x-axis in millions of years. Error bars represent the 95% highest posterior density for node height. Peromyscus (sensu lato) affiliated genera are indicated in bold.
Genetic Distances.
K2P (Table 2) genetic distances were used to compare taxa and provide additional information on the phylogenetic utility of each marker. Data obtained from comparisons of Isthmomys, Onychomys, and Reithrodontomys to other genera and species groups indicated the highest levels of genetic divergence among taxa. Comparison of the 5 genera (Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys) to all other genera resulted in genetic distances ranging from 2.58% (Rbp3) to 15.4% (Cytb). Values obtained from comparisons of these 5 genera to currently recognized species groups within Peromyscus (sensu stricto) ranged from 1.9% (Rbp3) to 14.62% (Cytb) and were similar in magnitude to comparisons of species groups to each other. By comparing the genetic distance between all taxa examined for each respective marker, their relative rates of evolution could be compared. Overall, Adh1-I2 and Fgb-I7 exhibited rates of evolution slower than Cytb; however, Rbp3 was substantially slower than all of the other markers.
Table 2.
Estimated genetic distances (K2P—Kimura 1980) for selected taxonomic groups based on sequences from the 4 genetic markers (Adh1-I2—intron 2 of alcohol dehydrogenase, Cytb—cytochrome-b, Fgb-I7—intron of the beta-fibrinogen, and Rbp3—interphotoreceptor retinoid-binding protein) examined in this study.
| Taxon | Adh1-I2 | Cytb | Fgb-I7 | Rbp3 |
|---|---|---|---|---|
| Reithrodontomys and Onychomys versus all other groups | 8.6/13.07 | 18.0/19.7 | 7.5/7.5 | 4.3/2.7 |
| Isthmomys versus all other groups | 14.0 | 16.5 | 8.9 | 3.6 |
| Habromys versus other "genera" | 5.2 | 15.0 | 6.0 | 2.0 |
| Megadontomys versus other "genera" | 5.2 | 15.0 | 6.7 | 2.9 |
| Neotomodon versus other "genera" | 5.3 | 14.7 | 6.1 | 2.6 |
| Osgoodomys versus other "genera" | 4.6 | 15.8 | 6.0 | 2.6 |
| Podomys versus other "genera" | 6.2 | 16.3 | 6.6 | 2.8 |
| Habromys versus Peromyscus (sensu stricto) | 4.0 | 14.0 | 5.2 | 1.3 |
| Megadontomys versus Peromyscus (sensu stricto) | 4.0 | 14.4 | 5.7 | 2.2 |
| Neotomodon versus Peromyscus (sensu stricto) | 3.19 | 13.9 | 5.2 | 2.0 |
| Osgoodomys versus Peromyscus (sensu stricto) | 3.5 | 15.1 | 5.0 | 2.0 |
| Podomys versus Peromyscus (sensu stricto) | 5.0 | 15.7 | 5.7 | 2.2 |
| All species groups versus each other | 4.4 | 14.2 | 5.1 | 1.6 |
| aztecus species group versus other species groups | 4.7 | 14.3 | 5.0 | 1.2 |
| boylii species group versus other species groups | 3.5 | 13.5 | 3.7 | 1.2 |
| californicus species group versus other species groups | 4.3 | 15.0 | 6.8 | 2.7 |
| crinitus species group versus other species groups | 4.4 | 14.2 | 5.8 | 1.8 |
| eremicus species group versus other species groups | 5.4 | 14.7 | 6.8 | 1.7 |
| furvus species group versus other species groups | 3.3 | 12.7 | 4.4 | 1.4 |
| hooperi species group versus other species groups | 5.9 | 12.7 | 4.4 | 1.4 |
| leucopus species group versus other species groups | 4.4 | 14.3 | 5.9 | 2.1 |
| maniculatus species group versus other species groups | 5.9 | 14.0 | 6.3 | 2.0 |
| megalops species group versus other species groups | 3.6 | 13.9 | 4.7 | 1.3 |
| melanophrys species group versus other species groups | 3.8 | 13.7 | 3.8 | 1.8 |
| mexicanus species group versus other species groups | 4.2 | 15.3 | 4.0 | 1.2 |
| truei species group versus other species groups | 3.9 | 14.2 | 4.5 | 1.3 |
Discussion
Use of a combined data set often increases resolution at different hierarchical levels with one data set providing resolution at deep nodes and others resolving shallow nodes. More quickly evolving mitochondrial markers tend to depict more resolution at terminal nodes, whereas nuclear markers generally resolve relationships at the base of a phylogeny. Therefore, combined data sets are often advantageous in studies whose phylogenetic relationships have been debated due to inconsistencies among studies or data sets. In addition, increasing the number of characters allows phylogenetic signal to assert itself over noise (homoplasy), resulting in a more accurate estimate of relationships.
Of the data analyzed, the combined data set provided the greatest resolution and nodal support (Fig. 1). Additionally, the combined data set provided resolution at several levels throughout the topology. Therefore, we use the topology from the BI analysis, as well as statistical support from ML analyses (Fig. 1) of the combined data set, to discuss the phylogenetic relationships of Peromyscus. We begin by discussing Peromyscus using the current taxonomy based on the sensu stricto framework unless indicated otherwise (Carleton 1980; Musser and Carleton 2005).
Clade I contains members of Peromyscus (sensu lato), Onychomys, and Reithrodontomys (Fig. 1). This relationship agrees with a Reithrodontomyini tribal definition as proposed by Miller and Engstrom (2008) and Musser and Carleton (2005) as well as relationships recovered in Bradley et al. (2004b, 2007), Carleton (1980), McKenna and Bell (1997), and Reeder and Bradley (2004, 2007). The pairing of Isthmomys and Reithrodontomys in the combined analyses garnered no statistical support but generally agrees with analyses using allozymic (Rogers et al. 2005) and multiple combinations of DNA sequence data (Bradley et al. 2007; Miller and Engstrom 2008).
Clade II most closely represents Peromyscus (sensu lato) as interpreted by Hooper (1968), with the exclusion of Isthmomys. Analyses of the nuclear and combined data sets each formed a well-supported clade similar to clade II. Although some basal branching patterns within this clade receive little to no support, it is apparent that the taxa recognized as separate genera (Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys) by Carleton (1980, 1989) and Musser and Carleton (2005) are embedded within an assemblage containing Peromyscus (sensu stricto). Subclades within clade II support Peromyscus (sensu stricto) paraphyly.
Clade III contains the majority of peromyscine species examined and generally agrees with the findings of Bradley et al. (2007). However, the current study differs from Bradley et al. (2007) in the placement of certain taxonomic groups although the terminal branching patterns are similar. Two strongly supported subclades recovered composed of the aztecus (Peromyscus evides and P. spicilegus) and boylii (Peromyscus boylii and P. levipes) species groups (Rennert and Kilpatrick 1986, 1987; Sullivan et al. 1991, 1997; Sullivan and Kilpatrick 1991; Tiemann-Boege et al. 2000; Bradley et al. 2004b) as well as a clade containing the megalops (Peromyscus megalops), melanophrys (Peromyscus melanophrys), and mexicanus (Peromyscus mexicanus and P. nudipes) species groups, which agrees with the study by Bradley et al. (2007). Clade IV depicts the relationship among P. californicus, P. crinitus, and P. eremicus, and P. californicus and P. eremicus are members of the subgenus Haplomylomys, whereas P. crinitus is the sole member of the crinitus species group. The relationship between these 2 species groups was supported in the combined Bayesian analysis. Clade V included members of the leucopus (Peromyscus gossypinus and P. leucopus) and maniculatus (Peromyscus maniculatus and P. melanotis) species groups. Strong support existed for the sister relationship between these species groups.
Several clades did not receive support in any of the analyses. This includes the unresolved placement of M. thomasi, Neotomodon alstoni, O. banderanus, and Podomys floridanus although their inclusion within Clade II is supported. In addition, no support was recovered for a monophyletic group containing all members of the P. attwateri (truei species group) or for the relationships of several species groups (e.g., hooperi). A sister relationship between Reithrodontomys and Peromyscus (sensu lato, excluding Isthmomys) received strong support.
The origin of Peromyscus (sensu lato) began approximately 8 mya (Fig. 2); however, the radiation of Peromyscus (sensu lato) excluding Isthmomys appears to have been focused around 5.71 mya (95% HPD: 3.37–9.08). During this time, Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys originated as well as several major Peromyscus lineages including Haplomylomys (P. californicus, P. eremicus, and P. crinitus), the mexicanus (P. mexicanus and P. nudipes), and boylii (P. boylii and P. levipes) species groups, and P. pectoralis. These lineages emerged between the minimum and maximum dates from León-Paniagua et al. (2007) and are further evidence for an origination date of Peromyscus (sensu lato), excluding Isthmomys, around 6 mya followed by a rapid diversification.
Estimation of the genetic distance values among selected taxa (genera and species groups) allowed for a gross-level comparison of genetic divergence among said groups (Table 2). For example, Isthmomys, Onychomys, and Reithrodontomys depicted substantially higher levels of genetic divergence than any other comparison. Comparisons of genetic divergence among the 5 genera (Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys) to other currently recognized species groups within Peromyscus (sensu stricto) produced values similar in magnitude to comparisons of the species groups to each other.
Although several recent studies have focused on developing phylogenies for Peromyscus (sensu lato and sensu stricto) and its affiliated genera, for a variety of reasons, none have been able to offer unambiguous taxonomic recommendations. First, Peromyscus is a large genus with new species still being described (Bradley et al. 2004a, 2014); complete taxonomic sampling is often difficult. Second, because several character systems have been studied and analyzed, it presents a challenge to resolve discrepancies when there are conflicting data. Third, Peromyscus may have undergone a rapid radiation that makes it difficult to reconstruct phylogenetic relationships (Fig. 1) with the available data. Fourth, and perhaps most important, is the occurrence of genetic conservation between taxa that exhibit substantial levels of morphological differences. For example, Carleton’s (1980) decision to elevate Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys to generic status was based on the occurrence of substantial morphological differentiation among taxa; yet these same taxa do not exhibit comparable levels of genetic divergence. Other morphological studies involving the glans penes and bacula (Hooper 1958; Hooper and Musser 1964) also have indicated high levels of morphological divergence among these same genera. Although recent genetic studies (i.e., herein; Rogers et al. 2005; Miller and Engstrom 2008) did not examine genes coding for the external morphological characters examined in Carleton’s (1980) study, one would assume a concomitant rate of evolution. Resolving the incongruence between morphological and genetic data may be an exciting development of its own. A cursory analysis of the molecular topology produced herein and the morphological characters analyzed in Carleton (1980) failed to recover a fixed, derived character that unites Peromyscus.
No phylogenetic analysis, herein, recovered a clade that corresponded to Peromyscus (sensu stricto). In our analyses, Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys continually were placed inside of Peromyscus (sensu stricto), producing a paraphyletic assemblage. When the molecular topology was constrained to reflect a Peromyscus (sensu stricto) framework, a significantly worse (P < 0.05) topology was recovered. Further, we were unable to reject a Peromyscus (sensu lato) relationship. When these results are combined with available genetic data (Reeder and Bradley 2004, 2007; Rogers et al. 2005; Reeder et al. 2006; Bradley et al. 2007; Miller and Engstrom 2008), it is clear that Peromyscus (sensu stricto) as currently recognized should be abandoned based on its paraphyletic nature. Providing a new, more accurate Peromyscus taxonomy is difficult because its placement can be interpreted in multiple ways due to the paraphyletic inclusion of Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys in most analyses.
Similarly, Peromyscus (sensu lato) requires revision, as one of its members (Isthmomys) forms a paraphyletic assemblage with Reithrodontomys or groups outside of a monophyletic Peromyscus. However, the Peromyscus (sensu lato) moniker can be recovered by simply removing Isthmomys and recognizing it as a separate genus (regardless of its affinities outside of Peromyscus), following Bradley et al. (2007), Miller and Engstrom (2008), and Rogers et al. (2005). BI and ML analyses both recovered topologies that supported exclusion of Isthmomys from a Peromyscus/Onychomys clade, yet an approximately unbiased topology test did not produce a significantly worse topology when monophyly was enforced on a Peromyscus (sensu lato) and Isthmomys clade. The inability of the approximately unbiased topology test to support exclusion of Isthmomys from Peromyscus (sensu lato) is likely due to the inclusion of only 4 of more than 20 species of Reithrodontomys and a single representative for Isthmomys. It is expected that increased sampling will further support the exclusion of Isthmomys from Peromyscus (sensu lato). Even with the removal of Isthmomys, paraphyly in the subgenera Peromyscus and Haplomylomys produced by the inclusion of Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys would remain problematic as discussed above.
It is possible to resolve monophyly of Peromyscus with taxonomies that broadly recognize groups at the generic, subgeneric, or species group level. Monophyletic clades from within Peromyscus (sensu stricto), as well as Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys, could each be recognized as genera. Similarly, Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys could be subsumed to subgenera within Peromyscus. Finally, Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys could be subsumed to species groups within Peromyscus. Each option presents specific taxonomic challenges that are discussed below and summarized in Table 3.
Table 3.
Three potential taxonomic solutions for Habromys, Isthmomys, Megadontomys, Neotomodon, Osgoodomys, Peromyscus, and Podomys. Generic designations were identified by supported monophyletic clades within Fig. 1. Only species included in phylogenetic analyses are presented.
| Generic taxonomy | Subgeneric taxonomy | Species group taxonomy |
|---|---|---|
| Genus Isthmomys | Genus Isthmomys | Genus Isthmomys |
| I. pirrensis | I. pirrensis | I. pirrensis |
| Genus Habromys | Genus Peromyscus | Genus Peromyscus |
| H. ixtlani | Subgenus Habromys | Species group lepturus |
| Genus Megadontomys | H. ixtlani | H. ixtlani |
| M. thomasi | Subgenus Megadontomys | Species group thomasi |
| Genus Neotomodon | M. thomasi | M. thomasi |
| N. alstoni | Subgenus Neotomodon | Species group alstoni |
| Genus Osgoodomys | N. alstoni | N. alstoni |
| O. banderanus | Subgenus Osgoodomys | Species group banderanus |
| Genus Podomys | O. banderanus | O. banderanus |
| P. floridanus | Subgenus Podomys | Species group floridanus |
| Genus Haplomylomys | P. floridanus | P. floridanus |
| P. californicus | Subgenus Haplomylomys | Species group californicus |
| P. crinitus | P. californicus | P. californicus |
| P. eremicus | P. crinitus | Species group crinitus |
| Genus Peromyscus | P. eremicus | P. crinitus |
| P. gossypinus | Subgenus Peromyscus | Species group eremicus |
| P. leucopus | P. gossypinus | P. eremicus |
| P. maniculatus | P. leucopus | Species group aztecus |
| P. melanotis | P. maniculatus | P. evides |
| New Genus A | P. melanotis | P. spicilegus |
| P. megalops | New Subgenus A | Species group boylii |
| P. melanophrys | P. megalops | P. boylii |
| P. mexicanus | P. melanophrys | P. levipes |
| P. nudipes | P. mexicanus | Species group furvus |
| New Genus B | P. nudipes | P. furvus |
| P. boylii | New Subgenus B | Species group hooperi |
| P. evides | P. boylii | P. hooperi |
| P. levipes | P. evides | Species group leucopus |
| P. spicilegus | P. levipes | P. gossypinus |
| New Genus C | P. spicilegus | P. leucopus |
| P. attwateri | New Subgenus C | Species group maniculatus |
| P. furvus | P. attwateri | P. maniculatus |
| P. gratus | P. furvus | P. melanotis |
| P. ochraventer | P. gratus | Species group megalops |
| New Genus D | P. ochraventer | P. megalops |
| P. pectoralis | New Subgenus D | Species group melanophrys |
| New Genus E | P. pectoralis | P. melanophrys |
| P. hooperi | New Subgenus E | Species group mexicanus |
| P. hooperi | P. mexicanus | |
| P. nudipes | ||
| Species group truei | ||
| P. attwateri | ||
| P. gratus | ||
| P. ochraventer | ||
| Species group pectoralis | ||
| P. pectoralis |
By retaining Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys at the generic level, the paraphyly within Peromyscus must be resolved by elevating monophyletic clades to the generic level (Table 3). Unfortunately, many of these clades originate at unsupported nodes within the phylogeny produced herein (Fig. 1). Further studies may be better able to resolve these relationships. Based on the current phylogeny, the elevation of a minimum of 2 new genera would be necessary to resolve paraphyly within Peromyscus (Genus A—P. pectoralis, P. levipes, P. boylii, P. spicilegus, P. evides, P. ochraventer, P. gratus, P. attwateri, and P. furvus; Genus B—P. mexicanus, P. nudipes, P. melanophrys, and P. megalops). This option is unfeasible due to lack of statistical support in the phylogeny. A 2-genus option will need to be continually evaluated as new data and data types become available. If genera are designated only at supported monophyletic nodes, then up to 4 new genera would require elevation from Peromyscus (Genus A—P. megalops, P. melanophyrs, P. mexicanus, and P. nudipes; Genus B—P. evides, P. boylii, P. levipes, and P. spicilegus; Genus C—P. attwateri, P. furvus, P. gratus, and P. ochraventer; Genus Peromyscus—P. gossypinus, P. leucopus, P. maniculatus, and P. melanotis) with uncertain placement of P. hooperi and P. pectoralis. Using the subgeneric option, a genus taxonomically similar to Peromyscus can be retained by subsuming Habromys, Megadontomys, Neotomodon, Osgoodomys, and Podomys (Table 3). The elevation of subgenera within Peromyscus would be necessary, and newly elevated subgenera would be similar in species content to the genera created using the generic option. Finally by removing higher taxonomic ranks (genus or subgenus), paraphyletic assemblages can be resolved while continuing to recognize morphological variation and account for clades identified with genetic data (Table 3). Species groups have proven to be valuable units to study evolution within Peromyscus (Riddle et al. 2000; Bradley et al. 2004b; Durish et al. 2004) and perhaps their usage would serve as a viable solution until the phylogenetic relationships of unresolved taxa are determined. Additionally, monophyly of most species groups has been somewhat resolved (except mexicanus and furvus—Bradley et al. 2007). The species group option, however, fails to recognize degrees of morphological variation that the generic and subgeneric options could offer if additional subrankings were established. Taxonomic changes would still be required in the recognition of the species groups; however, this option requires minimal changes relative to recognizing additional genera or subgenera.
In developing a revised classification for Peromyscus, standards must be agreed upon that designate distinction at a genetic level yet accommodate morphological variation. Some of these standards already are understood such as achieving monophyly and cohesion within the group. However, determining how much variation warrants generic recognition is difficult. For example, Helgen et al. (2009) and Weksler (2003) recently revised the genera formerly recognized as Spermophilus and Oryzomys, respectively. Their revisions produced monophyly and clarification of groups by the naming of additional genera to accommodate monophyletic clades produced in their analyses. Based on the data herein, it is clear that the current taxonomy of Peromyscus (sensu stricto) should be abandoned as well. However, to resolve the paraphyly within Peromyscus, at least 3 different taxonomic options are available and should be considered. More diverse data types, including morphology, karyology, and ecology, as well as additional genetic data, will be required to develop the taxonomy that properly recognizes the diversity and distinction within Peromyscus.
Acknowledgments
We thank D. S. Rogers, M. D. Engstrom, and J. R. Miller for the generation of many of the sequences available in GenBank. We thank J. Light (Texas Cooperative Wildlife Collection, Texas A&M University) and R. J. Baker (Natural Science Research Laboratory, Museum, Texas Tech University) for kindly providing tissue loans. H. R. Huynh, M. R. Mauldin, N. O. Ordóñez-Garza, and E. K. Roberts provided helpful comments on previous versions of this manuscript. This research was supported in part by grants from the National Institutes of Health (DHHS A141435-01 to RDB) and the National Science Foundation (MCB-0841821 and DEB-1020865 to RNP). Additional support was provided by the Mississippi Agricultural and Forestry Experiment Station.
Literature Cited
- Alfaro M. E., Zoller S., Lutzoni F. 2003. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molecular Biology and Evolution 20:255–266. [DOI] [PubMed] [Google Scholar]
- Amman B. R., Hanson J. D., Longhofer L. K., Hoofer S. R., Bradley R. D. 2006. Intron 2 of the alcohol dehydrogenase gene (ADH1-I2): a nuclear DNA marker for mammalian systematics. Occasional Papers, Museum of Texas Tech University 256:1–16. [Google Scholar]
- Behrensmeyer A. K., Turner A. 2013. Taxonomic occurrences of Peromyscus recorded in the Paleobiology Database. Fossilworks http://fossilworks.org Accessed 3 April 2014.
- Bradley R. D., Baker R. J. 2001. A test of the genetic species concept: cytochrome-b sequences and mammals. Journal of Mammalogy 82:960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R. D., Carroll D. S., Haynie M. L., MuÑiz Martínez R., Hamilton M. J., Kilpatrick C. W. 2004a. A new species of Peromyscus from western Mexico. Journal of Mammalogy 85:1184–1193. [Google Scholar]
- Bradley R. D., Durish N. D., Rogers D. S., Miller J. R., Engstrom M. D., Kilpatrick C. W. 2007. Toward a molecular phylogeny for Peromyscus: evidence from mitochondrial cytochrome-b sequences. Journal of Mammalogy 88:1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R. D., Edwards C. W., Carroll D. S., Kilpatrick C. W. 2004b. Phylogenetic relationships of Neotomine-Peromyscine rodents: based on DNA sequences from the mitochondrial cytochrome-b gene. Journal of Mammalogy 85:389–395. [Google Scholar]
- Bradley R. D., et al. 2014. Morphometric, karyotypic, and molecular evidence for a new species of Peromyscus (Cricetidae: Neotominae) from Nayarit, Mexico. Journal of Mammalogy 95:176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell V., Lapointe F.-J. 2009. The use and validity of composite taxa in phylogenetic analysis. Systematic Biology 58:560–572. [DOI] [PubMed] [Google Scholar]
- Carleton M. D. 1980. Phylogenetic relationships in neotomine-peromyscine rodents (Muroidea) and a reappraisal of the dichotomy within New World Cricetinae. Miscellaneous Publications of the Museum of Zoology 157:146. [Google Scholar]
- Carleton M. D. 1989. Sytematics and evolution. Pp. 7–141 in Advances in the study of Peromyscus (Rodentia) (G. L. Kirkland and J. N. Layne, eds.). Texas Tech University Press, Lubbock. [Google Scholar]
- Carroll D. S., Bradley R. D., Edwards C. W. 2005. Systematics of the genus Sigmodon: DNA sequences from beta-fibrinogen and Cytochrome b . Southwestern Naturalist 50:342–349. [Google Scholar]
- Cassiliano M. L. 1999. Biostratigraphy of Blancan and Irvingtonian mammals in the Fish Creek-Vallecito section, southern California, and a review of the Blancan–Irvingtonian boundary. Journal of Vertebrate Paleontology 19:169–186. [Google Scholar]
- Chambers R. R., Sudman P. D., Bradley R. D. 2009. A phylogenetic assessment of pocket gophers (Geomys): evidence from nuclear and mitochondrial genes. Journal of Mammalogy 90:537–547. [Google Scholar]
- Dalquest W. W. 1962. The Good Creek Formation, Pleistocene of Texas, and its fauna. Journal of Paleontology 36:568–582. [Google Scholar]
- DeBry R. W., Sagel R. M. 2001. Phylogeny of Rodentia (Mammalia) inferred from the nuclear-encoded gene IRBP. Molecular Phylogenetics and Evolution 19:290–301. [DOI] [PubMed] [Google Scholar]
- Douady C. J., Delsuc F., Boucher Y., Doolittle W. F., Douzery E. J. P. 2003. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Molecular Biology and Evolution 20:248–254. [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. 2008. Tracer v1.5 http://tree.bio.ed.ac.uk/software/tracer/ Accessed 27 October 2014.
- Drummond A. J., Suchard M. A., Xie D., Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durish N. D., Halcomb K. E., Kilpatrick C. W., Bradley R. D. 2004. Molecular systematics of the Peromyscus truei species group. Journal of Mammalogy 85:1160–1169. [Google Scholar]
- Edwards C. W., Fulhorst C. F., Bradley R. D. 2001. Molecular phylogenetics of the Neotoma albigula species group: further evidence of a paraphyletic assemblage. Journal of Mammalogy 82:267–279. [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Gene Codes Corporation. 2013. Sequencher version 5.0 sequence analysis software. Gene Codes Corporation, Ann Arbor, Michigan: http://www.genecodes.com. Accessed 1 September 2013. [Google Scholar]
- Haddrath O., Baker A. J. 2012. Multiple nuclear genes and retroposons support vicariance and dispersal of the palaeognaths, and an Early Cretaceous origin of modern birds. Proceedings of the Royal Society of London, B. Biological Sciences 279:4617–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J. D., Bradley R. D. 2008. Molecular diversity within Melanomys caliginosus (Rodentia: Oryzomyini): evidence for multiple species. Occasional Papers, Museum of Texas Tech University 275:1–11. [PMC free article] [PubMed] [Google Scholar]
- Helgen K. M., Cole F. R., Helgen L. E., Wilson D. E. 2009. Generic revision in the holarctic ground squirrel genus Spermophilus . Journal of Mammalogy 90:270–305. [Google Scholar]
- Hooper E. T. 1958. The male phallus in mice of the genus Peromyscus . Miscellaneous Publications of the Museum of Zoology, University of Michigan 105:1–24. [Google Scholar]
- Hooper E. T. 1968. Classification. Pp. 27–74 in Biology of Peromyscus (Rodentia) (J. A. King, ed.). Special Publication, American Society of Mammalogists, Stillwater, Oklahoma. [Google Scholar]
- Hooper E. T., Musser G. G. 1964. The glans penis in neotropical Cricetines (Family Muridae) with comments on classification of muroid rodents. Miscellaneous Publications of the Museum of Zoology, University of Michigan 123:1–57. [Google Scholar]
- Huelsenbeck J. P., Larget B., Miller R. E., Ronquist F. 2002. Potential applications and pitfalls of Bayesian inference of phylogeny. Systematic Biology 51:673–688. [DOI] [PubMed] [Google Scholar]
- Irwin D., Kocher T., Wilson A. 1991. Evolution of the cytochrome b gene of mammals. Journal of Molecular Evolution 32:128–144. [DOI] [PubMed] [Google Scholar]
- Jansa S., Voss R. 2000. Phylogenetic studies on didelphid marsupials I. Introduction and preliminary results from nuclear IRBP gene sequences. Journal of Mammalian Evolution 7:43–77. [Google Scholar]
- Karow P., Morgan G., Portell R., Simmons E., Auffenberg K. 1996. Middle Pleistocene (early Rancholabrean) vertebrates and associated marine and non-marine invertebrates from Oldsmar, Pinellas County, Florida. Pp. 97–133 in Palaeoecology and Palaeoenvironments of Late Cenozoic mammals tributes to the career of C S (Rufus) Churcher (K. Stewart and K. Seymour, eds.). University of Toronto Press, Toronto, Canada. [Google Scholar]
- Kass R. E., Raftery A. E. 1995. Bayes factors. Journal of the American Statistical Association 90:773–795. [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–120. [DOI] [PubMed] [Google Scholar]
- León-Paniagua L., Navarro-Sigüenza A. G., Hernández-Baños B. E., Morales J. C. 2007. Diversification of the arboreal mice of the genus Habromys (Rodentia: Cricetidae: Neotominae) in the Mesoamerican highlands. Molecular Phylogenetics and Evolution 42:653–664. [DOI] [PubMed] [Google Scholar]
- Longhofer L. K., Bradley R. D. 2006. Molecular systematics of the genus Neotoma based on DNA sequences from intron 2 of the alcohol dehydrogenase gene. Journal of Mammalogy 87:961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matocq M. D., Shurtliff Q. R., Feldman C. R. 2007. Phylogenetics of the woodrat genus Neotoma (Rodentia: Muridae): implications for the evolution of phenotypic variation in male external genitalia. Molecular Phylogenetics and Evolution 42:637–652. [DOI] [PubMed] [Google Scholar]
- McKenna M. C., Bell S. K. 1997. Classification of mammals above the species level. Columbia University Press, New York. [Google Scholar]
- Miller J. R., Engstrom M. D. 2008. The relationships of major lineages within Peromyscine rodents: a molecular phylogenetic hypothesis and systematic reappraisal. Journal of Mammalogy 89:1279–1295. [Google Scholar]
- Musser G. G., Carleton M. D. 2005. Superfamily Muroidea. Pp. 501–775 in Mammal species of the world: a taxonomic and geographic reference (D. E. Wilson and D. M. Reeder, eds.). John Hopkins University Press, Baltimore, Maryland. [Google Scholar]
- Nylander J. A. A. 2004. MrModeltest v2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden. [Google Scholar]
- Osgood W. H. 1909. Revisions of the mice of the American genus Peromyscus . North American Fauna 28:1–285. [Google Scholar]
- Peppers L. L., Bradley R. D. 2000. Cryptic species in Sigmodon hispidus: evidence from DNA sequences. Journal of Mammalogy 81:332–343. [Google Scholar]
- Reeder S. A., Bradley R. D. 2004. Molecular systematics of Neotomine-Peromyscine rodents based on the dentin matrix protein 1 gene. Journal of Mammalogy 85:1194–1200. [Google Scholar]
- Reeder S. A., Bradley R. D. 2007. Phylogenetic relationships of Neotomine-Peromyscine rodents using DNA sequences from beta fibrinogen and cytochrome b . Pp. 883–900 in The quintessential naturalist: honoring the life and legacy of Oliver P. Pearson (D. A. Kelt, E. P. Lessa, J. A. Salazar-Bravo, and J. L. Patton, eds.). University of California, Publications in Zoology, Berkeley. [Google Scholar]
- Reeder S. A., Carroll D. S., Edwards C. W., Kilpatrick C. W., Bradley R. D. 2006. Neotomine–Peromyscine rodent systematics based on combined analyses of nuclear and mitochondrial DNA sequences. Molecular Phylogenetics and Evolution 40:251–258. [DOI] [PubMed] [Google Scholar]
- Rennert P. D., Kilpatrick C. W. 1986. Biochemical systematics of populations of Peromyscus boylii. I. Populations from east-central Mexico with low fundamental numbers. Journal of Mammalogy 67:481–488. [Google Scholar]
- Rennert P. D., Kilpatrick C. W. 1987. Biochemical systematics of populations of Peromyscus boylii. II. Chromosomally variable populations from eastern and southern Mexico. Journal of Mammalogy 68:799–811. [Google Scholar]
- Riddle B. R., Hafner D. J., Alexander L. F. 2000. Phylogeography and systematics of the Peromyscus eremicus species group and the historical biogeography of North American warm regional deserts. Molecular Phylogenetics and Evolution 17:145–160. [DOI] [PubMed] [Google Scholar]
- Rogers D. S., Engstrom M. D., Arellano E. 2005. Phylo- genetic relationships among Peromyscine rodents: allozyme evidence. Pp. 427–440 in Contribuciones mastozoológicas en Homenaje a Bernardo Villa (V. Sanchez-Cordero and R. A. Medellin, eds.). Instituto de Biología e Instituto de Ecología, Universidad Nacional Autónoma de México, Mexico. [Google Scholar]
- Ronquist F., et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylo- genetic tree selection. Systematic Biology 51:492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. [DOI] [PubMed] [Google Scholar]
- Smith M. F., Patton J. L. 1993. The diversification of South American murid rodents: evidence from mitochondrial DNA sequence data for the Akodontine tribe. Biological Journal of the Linnean Society 50:149–177. [Google Scholar]
- Smith M., Patton J. 1999. Phylogenetic relationships and the radiation of Sigmodontine rodents in South America: evidence from cytochrome b . Journal of Mammalian Evolution 6:89–128. [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. [DOI] [PubMed] [Google Scholar]
- Stanhope M. J., Czelusniak J., Si J.-S., Nickerson J., Goodman M. 1992. A molecular perspective on mammalian evolution from the gene encoding interphotoreceptor retinoid binding protein, with convincing evidence for bat monophyly. Molecular Phylogenetics and Evolution 1:148–160. [DOI] [PubMed] [Google Scholar]
- Suchard M. A., Weiss R. E., Sinsheimer J. S. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Molecular Biology and Evolution 18:1001–1013. [DOI] [PubMed] [Google Scholar]
- Sullivan J., Kilpatrick C. W. 1991. Biochemical systematics of the Peromyscus aztecus assemblage. Journal of Mammalogy 72:681–691. [Google Scholar]
- Sullivan J. M., Kilpatrick C. W., Rennert P. D. 1991. Biochemical systematics of the Peromyscus boylii species group. Journal of Mammalogy 72:669–680. [Google Scholar]
- Sullivan J., Markert J. A., Kilpatrick C. W. 1997. Phylogeography and molecular systematics of the Peromyscus aztecus species group (Rodentia: Muridae) inferred using parsimony and likelihood. Systematic Biology 46:426–440. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I., Kilpatrick C. W., Schmidly D. J., Bradley R. D. 2000. Molecular phylogenetics of the Peromyscus boylii species group (Rodentia: Muridae) based on mitochondrial cytochrome b sequences. Molecular Phylogenetics and Evolution 16:366–378. [DOI] [PubMed] [Google Scholar]
- Townsend T. M., et al. 2011. Phylogeny of Iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species-tree approaches for an ancient, rapid radiation. Molecular Phylogenetics and Evolution 61:363–380. [DOI] [PubMed] [Google Scholar]
- Weksler M. 2003. Phylogeny of Neotropical Oryzomyine rodents (Muridae: Sigmodontinae) based on the nuclear IRBP exon. Molecular Phylogenetics and Evolution 29:331–349. [DOI] [PubMed] [Google Scholar]
- Wickliffe J. K., Hoffmann F. G., Carroll D. S., Duninia-Barkovskaya Y., Bradley R. D. 2003. PCR and sequencing primers for intron 7 (Fgb-I7) of the fibrinogen, B beta polypeptide (Fgb) in mammals: a novel nuclear DNA phylogenetic marker. Occasional Papers, Museum of Texas Tech University 219:1–6. [Google Scholar]