Abstract
In Streptococcus mutans, ComX, an alternative sigma factor, drives the transcription of the “late-competence genes” required for genetic transformation. ComX activity is modulated by inputs from two signaling pathways, ComDE and ComRS, that respond to the competence stimulating peptide (CSP) and the SigX-inducing peptide (XIP), respectively. In particular, the comRS, encoding the ComR regulatory protein and the ComS precursor to XIP, functions as the proximal regulatory system for ComX activation. Here, we investigated the individual and combinatorial effects of CSP and XIP on genetic transformation and cell killing of S. mutans. Our transformation results confirm the recent reports by Mashburn-Warren et al. and Desai et al. that XIP functions optimally in a chemically defined medium (CDM), whereas its activity is inhibited when cells are grown in complex medium. Using tandem mass spectrometry (MS/MS) fragmentation, a drastic reduction in XIP levels in ComX-deficient cultures were observed, suggesting a ComX-mediated positive feedback mechanism for XIP synthesis. Our evaluation of cell viability in the presence of 10μM XIP resulted in the killing nearly 82% of the population. The killing activity was shown to be dependent on the presence of comR/S and comX. These results suggest a novel role for XIP as a compelling effector of cell death. This is the first report that demonstrates a role for XIP in cell killing.
INTRODUCTION
The acquisition of novel, heritable DNA by genetically competent bacteria not only propagates antibiotic resistance and virulence determinants, but also shapes bacterial genomes contributing to rapid evolutionary changes (Cody et al., 2003; Didelot & Maiden, 2010; Feil et al., 1999; Kroll et al., 1988; Seifert et al., 1988). In Streptococcus mutans, an oral resident associated with dental caries, competence development relies on multiple input systems that relay environmental signals to ultimately modulate the transcription of comX, encoding an alternate sigma factor, ComX (SigX) (Li et al., 2001; Aspiras et al., 2004). In S. mutans, competence does not develop in the absence of ComX, as it is critical for the expression of genes involved in DNA uptake and recombination (Aspiras et al., 2004). Expression of comX was first shown to be regulated by the ComDE two component signaling system comprising of a sensor kinase and a response regulator, respectively, which responds to accumulation of the competence stimulating peptide (CSP) (Li et al., 2001; Aspiras et al., 2004). Recently, Mashburn-Warren et al. (2010) identified the ComR regulatory protein of the ComRS signaling pathway as the proximal regulator necessary for comX expression. ComR, in conjunction with its cognate signal peptide, XIP (SigX inducing peptide), modulates comX transcription in S. mutans (Mashburn-Warren et al., 2010). The XIP precursor encoded by comS is consequently exported, processed to its mature form, and then internalized via the Opp/Ami transporter to interact with ComR for comX regulation (Mashburn-Warren et al., 2010; Desai et al., 2012). The loss of ComR abolishes comX expression and competence development, which cannot be restored by the addition of CSP. Furthermore, XIP does not require a functional comE gene to induce the expression of comX (Mashburn-Warren et al., 2010). These observations highlight the central role of ComRS in the regulation of comX.
Previously, it has been demonstrated that S. mutans cultures exposed to high CSP concentrations (2–4 μM) cause growth arrest and eventually undergo cell death by lysis (Perry et al. 2009, Qi et al. 2005). In this work, we asked whether synthetic XIP can elicit a similar response to cause cell death of S. mutans. Our viability assays revealed that supplementing 10μM XIP killed approximately 82% of the population. We further report that in addition to the comR/S, the presence of comX is vital for optimal killing. Moreover, we also report the effects of XIP on genetic transformation, which support findings by Mashburn Warren et al. (2011) and Desai et al., (2012). Further, using tandem mass spectrometry, we successfully detected the 7 amino acid XIP peptide (GLDWWSL) in the wild-type UA159 supernatant, but not in that of the ComS-deficient mutant. While these results concur with those recently reported by Khan et al. (2012), we further show that supernatant XIP levels are drastically reduced in ComX-deficient cultures, suggesting a positive role for ComX in ComS/XIP production, export or processing. Taken together, in addition to its widely discussed role in competence, our work reveals a novel role for XIP as a potent effector of cell death in S. mutans, which may be potentially used for the development of therapeutic strategies to prevent dental caries.
MATERIALS AND METHODS
Bacterial strains and growth conditions
S. mutans UA159 (Ajdic et al., 2002) and its mutant strains in comC (ΔSMcomC), comD (ΔSMcomD), comE (ΔSMcomE) (Li et al., 2001a) and comX (ΔSMcomX) (Li et al., 2002) were used in this study. The comS (ΔSMcomS) and the comR (ΔSMcomR) mutants were constructed using a non-polar ligation PCR mutagenesis method described previously (Lau et al., 2002). S. mutans strains were grown at 37°C with 5% CO2 in either Todd-Hewitt broth (Becton Dickinson, MD) containing 0.3% yeast extract (Difco Laboratories) (THYE), or chemically defined medium (CDM) described previously (Mashburn-Warren et al., 2010). Erythromycin and spectinomycin were used as needed at concentrations of 10μg/mL and 1mg/mL, respectively. Synthetic XIP (sXIP) and synthetic CSP (sCSP) peptides were synthesized using F-MOC chemistry (Advanced Protein Technology Centre, Hospital for Sick Kids, Toronto, Canada). Stock concentrations of 1μM of sXIP and 0.4mM sCSP were prepared in DMSO and water, respectively. Growth kinetics were monitored using an automated growth reader (Bioscreen C; Labsystems, Finland) as previously described (Senadheera et al., 2007).
Transformation frequency (TF) Assays
Overnight cells grown in THYE were pelleted, washed and resuspended in phosphate buffered saline (1x PBS). The resuspended culture was diluted 1:50 using prewarmed THYE or CDM and grown to an OD600 ~0.1. Next, 1μg/mL of the donor plasmid DNA (pDL277; specR) (LeBlanc et al., 1992) was added to 1mL aliquots of the culture in the presence or absence of CSP (0.4μM) or XIP (10μM) and samples were incubated for 90 min. For XIP, control cultures containing 1% DMSO were utilized. After incubation, cultures were serially diluted and plated on THYE plates with and without antibiotics. TF was calculated as transformant colony forming units (CFUs) divided by the total number of viable CFUs, times one hundred.
Cell Viability Assays
Overnight cultures in THYE were pelleted, washed, resuspended in sterile 1X PBS, and diluted 1:50 using warm THYE or CDM. Each suspension was supplemented with either 2μM CSP or 10μM XIP. Cultures without peptides or containing 1%DMSO were used as controls. All cultures were grown to an OD600~0.8, at which point, the cells were gently sonicated on ice and used for viability assays. Cell were serially diluted, plated on THYE agar and CFUs counted. Results standardized using cellular dry weight. These standardized values were then used to calculate the percentage survival by dividing the standardized number of viable cells after treatment by the standardized total number of cells without peptide, times 100.
Time-course Killing Analyses
Overnight cultures of UA159 in THYE were pelleted, washed, resuspended in sterile 1X PBS, and diluted 1:20 using warm CDM. The subcultures were allowed to grow to an OD600 of 0.4, after which they were split into two where one was exposed to 1% DMSO and the other to 10μM XIP. Cultures were further incubated and samples were taken at varying time points (0h, 1h, 2h, 3h, 4h and 5h) after exposure to XIP, gently sonicated, serially diluted and plated on THYE plates for CFU determination. Results were standardized using cellular dry weight. Percentage viability was calculated as the number of viable cells after treatment divided by the total number of cells without peptide, times 100.
Biofilm Formation Assays
Overnight cultures in THYE were pelleted, washed, resuspended in sterile 1X PBS, and diluted 1:100 using warm CDM. Each suspension was supplemented with either 1% DMSO or 10μM XIP and used to inoculate polystyrene plates. After 24h incubation, the biofilms were dried and strained with 0.1% Safranin Red.
Quantitative real-time PCR (qRT-PCR) analyses
Overnight cultures of UA159 and its derivatives were diluted 20X in fresh THYE or CDM and grown to an OD600 of 0.4–0.5 in the presence or absence of 0.4μM CSP or 10μM XIP, respectively. For growth in CDM, overnight cells were washed, resuspended in 1xPBS prior to inoculation and harvesting. Controls included, THYE without added peptide, as well as CDM with 1%DMSO. RNA isolation, DNAse treatment, cDNA synthesis, qRT-PCR and expression analyses were carried out as previously described (Senadheera et al., 2005). Primers used for qRT-PCR: comR (For: CGTTTAGGAGTGACGCTTGG, Rev: TGTTGGTCGCCATAGGTTG), comS (For: TTTTGATGGGTCTTGACTGG, Rev: TTTATTACTGTGCCGTGTTAGC) and comX (For: ACTGTTTGTCAAGTCGCGG Rev: TGCTCTCCTGCTACCAAGCG). Expression was normalized to that of 16SrRNA, and statistical analyses were performed on four independent experiments using Student’s t-test (P <0.05).
XIP detection and quantification
Overnight cultures in CDM were diluted 100-fold and grown for 48h at 37°C in 5% CO2 air mixture. Cell-free supernatants were obtained by centrifugation and filter sterilized using a 0.45μm syringe filter. Samples were lyophilized and, once dry, reconstituted in 2 mL of 5% MeOH/H2O (v/v) prior to analysis by HPLC-ESI-MS/MS (Dionex UltiMate 3000 HPLC system with variable UV detection in line to a Bruker amaZon X ion-trap mass spectrometer operating in positive ionization mode with auto MS/MS enabled). Analytical scale analysis was performed on a 250x 4.60 mm Phenomenex Luna 5μ C18(2) 100Å column (Serial no. 516161-20) with a flow rate of 1 ml min−1 and the following program consisting of solvents A (water + 0.1% formic acid) and B (acetonitrile + 0.1% formic acid): 0–2 min, equilibration at 5% B; 2–18 min, linear gradient to 100% B; 18–20 min, constant 100% B, 20–20.5 min, linear decrease to 5% B; 20.5–23 min re-equilibration at 5% B. The identity of XIP in culture supernatants was confirmed by comparison to the retention time and MS/MS fragmentation of sXIP. To quantify XIP levels a directed LC-MS/MS experiment was performed using selected-reaction monitoring (SRM) tandem mass spectrometry. The SRM m/z transition 876.4 → 658.4 was monitored, corresponding to a −SL loss from the GLDWWSL parent ion, generating a GLDWW daughter ion. Resulting peak areas were integrated and final concentrations calculated from a linear calibration curve created using CDM spiked with sXIP and processed in an identical way to CFSs. Peak areas from each sample were standardized to optical densities of 48-h culture samples prior to centrifugation for HPLC-MS analysis. Results were obtained for four independent experiments and statistics were conducted using the Student’s T-test.
RESULTS
Transformation frequencies (TF) with CSP and/or XIP in CDM and complex media
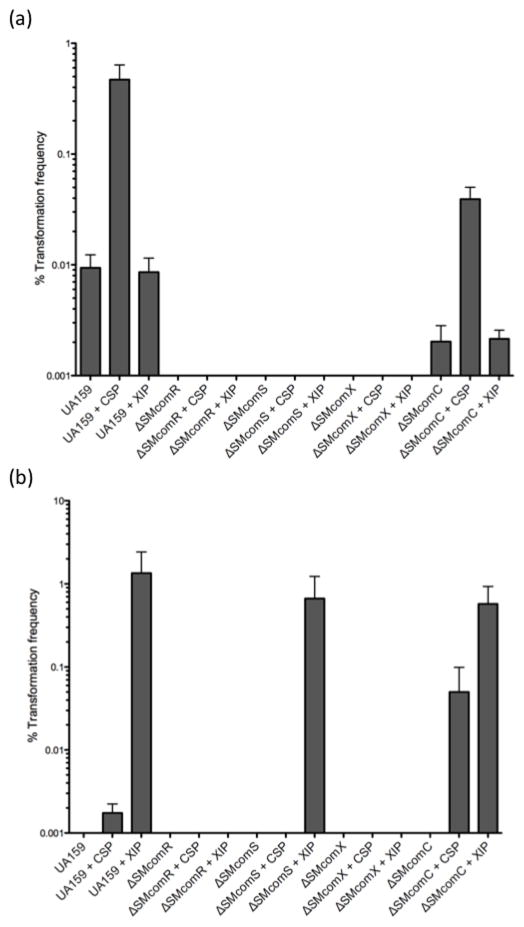
The initial finding that XIP induces genetic transformation via ComX was reported by Mashburn-Warren et al. (2010) using cells grown in CDM. Recent work by Desai et al., (2012) reported that the induction of comX by XIP was largely inhibited when grown in rich nutrient THB, a medium commonly used to study CSP-induced competence. In accordance with these reports, our TF assays show that XIP is optimally functional in CDM in eliciting transformation and its activity is inhibited when cells are grown in complex medium (i.e. THYE) (Fig. 1). In contrast, we observed that CSP was largely ineffective at inducing competence in CDM, and that it was optimally functional in complex medium (Fig. 1). Since CSP and XIP were shown not to function optimally in the same growth medium, we did not obtain significant combinatorial effects in either THYE or CDM (data not shown).
Fig. 1. Transformation frequency (TF) of S. mutans strains in THYE and CDM growth media.
S. mutans UA159 and mutant strains were subcultured to an OD600 of 0.1 in either THYE (a) or CDM (b). Plasmid DNA pDL277 (specr) was added alone or with 0.4μM sCSP or 10μM sXIP for transformation experiments. Results show the mean TF of three independent experiments ± standard error.
Measurement of XIP in the culture supernatants of S. mutans UA159, comR/S, comE and comX knockout mutants
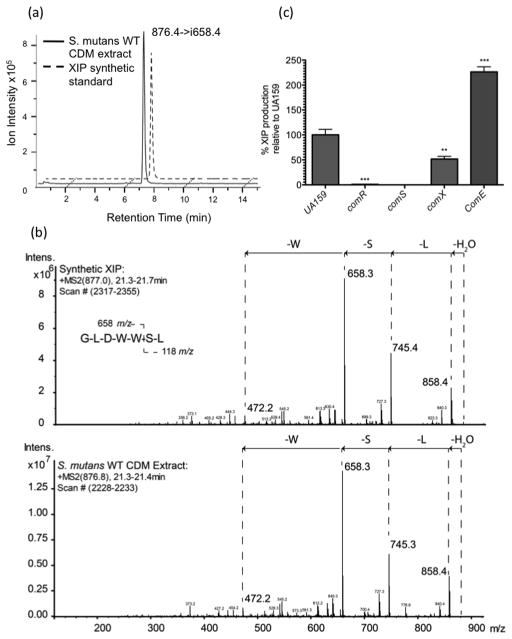
To elucidate the role of known S. mutans competence genes in the regulation of XIP production, its processing and/or secretion, we used HPLC-ESI-MS/MS to monitor extracellular XIP levels in comR/S, comE and comX-deficient mutants. We were able to successfully identify the presence of XIP in the wild-type supernatant by comparison of the retention time and of the fragmentation patterns to the sXIP standard (Fig 2(a) and (b)). We were able to detect XIP at concentrations ranging from 95 ng/mL to 750 ng/mL (or 109nM to 857nM), and consistent with the loss of transformability ΔSMcomS, XIP was absent in their cell free supernatants (Fig. 2(c)). These results are in accordance with that of Khan et al. (2012) who also reported their inability to detect mature XIP in culture supernatants of the ComS mutant. As expected of a positive regulator of comS expression, ΔSMcomR also displayed highly reduced levels of XIP. Our further quantification of XIP in the ComX and ComE mutants suggested a significant decrease (p<0.05) of this peptide in the ΔSMcomX supernatant, whereas it was significantly increased in the ΔSMcomE supernatant (Fig. 2(c)). These results suggested that while ComX positively influenced the production, processing and/or secretion of XIP, the ComDE two component system negatively affected one or more of these processes in S. mutans.
Fig. 2. Secretion of XIP by S. mutans UA159 and comX-, comR- and comS-mutant strains.
(a) Chromatograms resulting from selected-reaction monitoring (SRM) LC-MS experiments used for extraction and quantification of XIP. A transition corresponding to the loss of S and L residues from the XIP peptide parent ion at 876.4m/z, generating a daughter ion at 658.4 m/z was monitored. (b) A comparison of MS2 fragmentation pattern of authentic standard XIP peptide (top panel) and wild-type S. mutans sample (bottom panel). Specific amino acid and water losses are annotated. (c). XIP levels in the supernatant of each strain in stationary phase were quantified using HPLC-ESI-MS/MS. Results shown represent those from at least 3 separate experiments, error bars indicate standard deviation. Statistical analyses were performed using a Student’s T-test: *p<0.05.
Effect of XIP on cell viability of S. mutans UA159 grown in CDM
While investigating the effects of sXIP on genetic transformation, we noted that growth of UA159 was drastically impaired by the addition of 10μM XIP in CDM (Fig. 3a). Since this indicated a likely effect on cell death, we performed cell viability assays to determine whether XIP could act as a death effector of S. mutans. In the presence of 10μM XIP in CDM, we observed only an 18% survival rate relative to the no-peptide control, suggesting that XIP can function as a potent killing peptide under these conditions (Fig. 3b). XIP was unable to induce killing in ΔSMcomR and was also largely ineffective in ΔSMcomX, demonstrating the importance of both comR and comX in the killing activity of XIP. Furthermore, in ΔSMcomS and ΔSMcomC grown in CDM exposed to XIP, we noted 80% and 89% killing, respectively (Fig. 3b). In contrast to CDM, XIP was not able to induce killing when S. mutans strains were grown in THYE. To confirm the effect of XIP on cell viability, time course killing analyses were performed, which demonstrated a negative effect of XIP on the CFU counts of healthy cultures at varying time points (Fig. 3c). Furthermore, S. mutans was not able to form biofilms in the presence of XIP (Fig. 3d). This drastic effect on biofilm development may be attributed to XIP’s drastic effect on the viability of cells. These results suggest an important role for XIP as a novel killing peptide that can be targeted to kill S. mutans.
Fig. 3. Effect of CSP and XIP on cell survival of S. mutans.
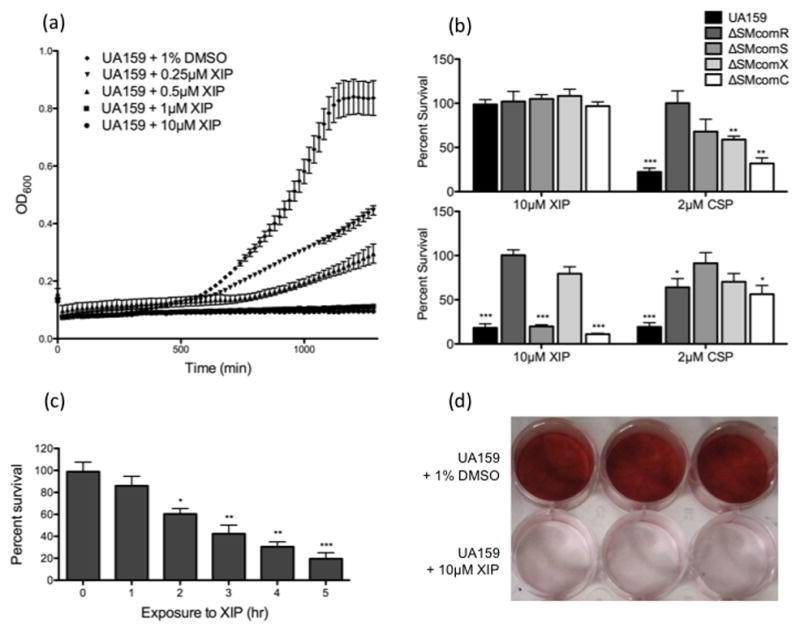
(a). The effect of XIP at varying concentrations (10μM, 1μM, 0.5μM, 0.25μM, 1% DMSO) on the growth of S. mutans UA159 was examined by monitoring OD600 using the Bioscreen C automated growth reader (b). S. mutans UA159, as well as comR, comS, comX and comC knockout mutant strains were grown in the presence or absence of either XIP (10μM) or CSP (2μM) to OD600~0.8 in THYE (top panel) or CDM (bottom panel). Viable CFUs were counted and results were standardized to the dry weight and presented as the percentage of the CFUs obtained in the absence of the peptide. Results shown here are the average of four independent experiments ± standard error. Statistical analyses were performed using Student’s T-test: *p<0.05, **p<0.01, ***p<0.001. (c). Actively growing cultures of UA159 were exposed to 10μM XIP for 0, 1, 2, 3, 4 and 5h. Following sonication, viable CFUs were counted and results were standardized to the dry weight and presented as the percentage of the CFUs obtained in the absence of the peptide. Results shown here are the average of four independent experiments ± standard error. Statistical analyses were performed using Student’s T-test: *p<0.05, **p<0.01, ***p<0.001. (d). S. mutans biofilms were grown on polystyrene plates overnight in the presence or absence of 10μM XIP, and stained with 0.1% Safranin Red. The image presented here is a representative of four independent experiments.
Similar to lysis by XIP, CSP-induced cell death was also largely diminished in the absence of comR/S or comX (Fig. 3), suggesting that the CSP-induced killing pathway previously described requires the presence of comR/S and comX for optimal killing.
Regulation of comR/S, and comX expression by XIP, CSP and the ComDE system
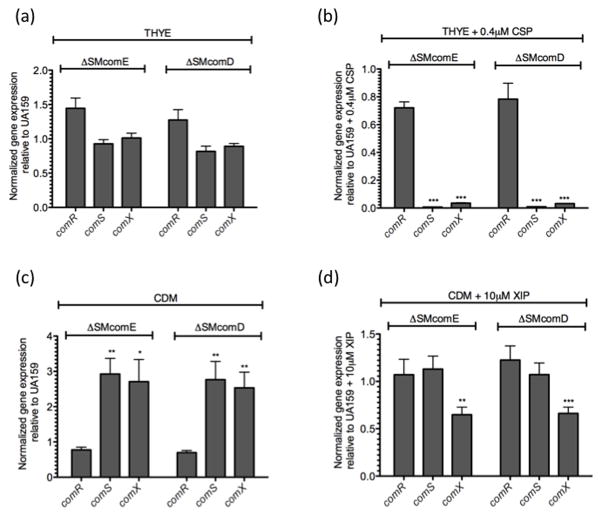
Our transformation and viability results as well as that obtained by Mashburn-Warren et al., (2010) and Desai et al., (2012), strongly suggest that the ComCDE system may regulate comX transcription through ComRS, although this was not directly tested. Hence, we examined comR/S and comX transcription in UA159, ΔSMcomD and ΔSMcomE strains grown with and without CSP or XIP. Due to the poor activity of CSP in CDM and no activity of XIP in THYE, experiments with CSP were performed in THYE, whereas those with XIP were conducted from cells grown in CDM. Supporting a hierarchal position of the ComCDE system upstream of ComRS, we observed that addition of CSP increased comS and comX expression by 73.9-fold and 2.3-fold, respectively (Fig. 4(a)). In THYE without added CSP, comR/S, and comX expression was not significantly affected by loss of comD/E relative to wild-type (Fig. 5(a)). However, with CSP, expression of comS was significantly decreased over 100-fold in both mutants (p<0.001), relative to wild-type (Fig. 5(b)). Addition of CSP also decreased comX expression by nearly 30-fold in ΔSMcomD and ΔSMcomE strains, respectively, compared with the parent (Fig. 5(b)). These results suggested that in complex medium, comS expression can be modulated by adding CSP, and that comS induction by the CSP is ComDE-dependent.
Fig. 4. Gene expression of comR, comS and comX in response to CSP and XIP.
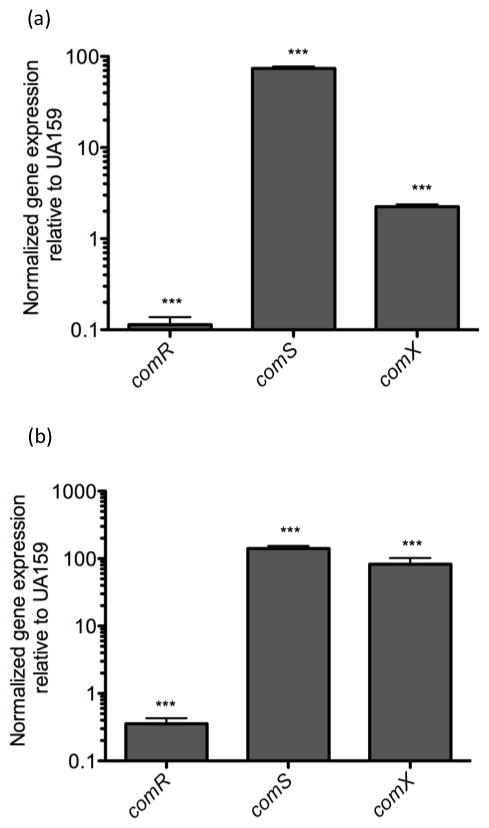
Real-time analysis of gene expression of comR, comS and comX were preformed using mid-logarithmic UA159 cells grown in THYE (a) and CDM (b) in the presence or absence of either 0.4μM CSP (a) or 10μM XIP (b). Gene expression was normalized to UA159 with no peptide added. Results shown here are the average of at least three independent experiments ± standard error. Statistical analyses were performed using Student’s T-test: ***p<0.001
Fig. 5. Differential regulation of comRS in THYE and CDM media grown in the presence or absence of CSP and XIP, respectively.
Real-time analysis of comR-, comS- and comX-specific expression in comD- and comE-knockout mutants grown in THYE (a) and CDM (c) media, as well as in the presence of either 0.4μM CSP (b) or 10μM XIP (d). Gene expression was normalized with 16SrRNA and fold-expression was calculated relative to that of UA159 held at a user-defined value of 1.0. Results shown are the mean expression values of at least three independent experiments ± standard error. Statistical analyses were performed using Student’s T-test: *p<0.05, **p<0.01, ***p<0.001.
In wild type, addition of sXIP increased expression of comX and comS by 83-fold and 141-fold, respectively (Fig. 5(b)), thus confirming the autoregulatory loop described by Mashburn-Warren et al., 2010. In ΔSMcomD and ΔSMcomE grown in CDM, comS and comX genes were upregulated almost 3-fold without added peptide, likely suggesting that ComDE may repress their expression in CDM medium (Fig. 5c). This finding was also supported by the high levels of XIP detected in the ΔSMcomE culture supernatant. Further, upon addition of sXIP to ΔSMcomD and ΔSMcomE mutants no change in comR and comS expression was observed (Fig. 5d), suggesting that the ComDE system does not affect XIP signaling, once the ComRS system is activated.
DISCUSSION
Competence has been observed in a number of bacteria to occur in conjunction with lysis of a subpopulation of cells (Claverys et al., 2007; Steinmoen et al., 2002; Perry et al, 2009; Lemme et al., 2011). The lysed subpopulation is thought to contribute to the genetic pool used for DNA uptake by the competent cells. Herein, we have demonstrated a role for the XIP competence peptide as potent modulator of cell death in S. mutans. Our viability assays show XIP can kill nearly 82% of the population when supplied at a concentration of 10μM. To our knowledge, this is the first report that demonstrates a function for XIP as an effector of cell death.
We further report that XIP-mediated killing works via the ComR/S system and ComX, which positions the ComR/S and ComX in a more centralized position in the killing pathway of S. mutans. Although previous reports have attributed CSP-induced lysis to an imbalance between the ComE-regulated mutacin V and its immunity protein ImmB (Lemme et al., 2011; Perry et al., 2009; Dufour et al., 2011), here we argue that competence-associated cell death in S. mutans, is instead, largely due to activity downstream of ComX. This is also supported by the fact that nlmC (synonyms: cipB, bsmA) encoding mutacin V also modulates comX activity, which in turn, may contribute to its killing activity (Dufour et al., 2011). We are currently examining genes downstream of ComX stimulated by XIP that may function as killing effectors using global transcriptome analysis.
Although the killing activity of CSP harbors specificity towards its parent strain (Qi et al., 2005), the spectrum of activity of XIP has yet to be determined. XIP contains a double-tryptophan (WW) motif conserved among short hydrophobic peptides of the pyogenic and bovis groups of Streptococci, located within a conserved genomic context (Mashburn-Warren et al., 2010). Similar peptides specific for Streptococcus agalactiae, Streptococcus porcinus, and Streptococcus parauberis have been shown to bear no effect on competence or growth of S. mutans, suggesting that these peptides may be specific to their parental strain (Desai et al., 2012). XIP therefore may be exploited for targeted killing of S. mutans.
Our transformation and cell viability results with CSP and/or XIP in both THYE and CDM media showed that these peptides do not function optimally under the same conditions. Our transformation results are in agreement with Desai et al., (2012) who reported that titration of THB into UA159 cultures in CDM inhibited XIP-induced transformability. While they demonstrated some level of activity of XIP in 100% THB, our results showed complete inhibition of XIP in THYE. It is likely that the yeast extract in THYE is largely responsible for the inhibition observed.
Despite the observation that XIP is inactive in THYE-grown cells, the comS gene is still required for transformation despite growth in a complex medium. Hence, it is possible that the ComS peptide may also function intracellularly without its export and subsequent import into the cell. We have also taken into consideration that conditions tested in complex medium may not be optimal for the expression of the XIP exporter, which can likely result in the accumulation of ComS inside the cell, making it vulnerable to intracellular cleavage.
Our expression analysis combined with LC-MS/MS in CDM demonstrates a negative-regulatory role for the ComDE system in XIP production. Kreth et al. (2007) reported that ComDE repressed comC expression prior to CSP stimulation. It is possible that ComDE may prevent premature expression of comS, thereby delaying competence induction in CDM to the latter stages of growth. As observed by Desai et al., (2012), competence in CDM is first observed in mid-logarithmic cells of S. mutans and continues well into the stationary phase.
We further observe that the amount of XIP was significantly reduced in ΔSMcomX, suggesting a ComX-mediated positive feedback mechanism for XIP synthesis. Putative ComX binding sites were located within the comR gene, upstream of comS, suggesting that ComX may directly regulate comS expression (Fig. 6a). This positive autoregulation of XIP production may contribute to the persistence of the competent state in CDM. Based on previous works and our findings presented here, we propose a growth condition-dependent model for genetic competence in S. mutans (Figure 6b).
Fig. 6. Model for competence and cell killing pathways of S. mutans in THYE versus CDM.
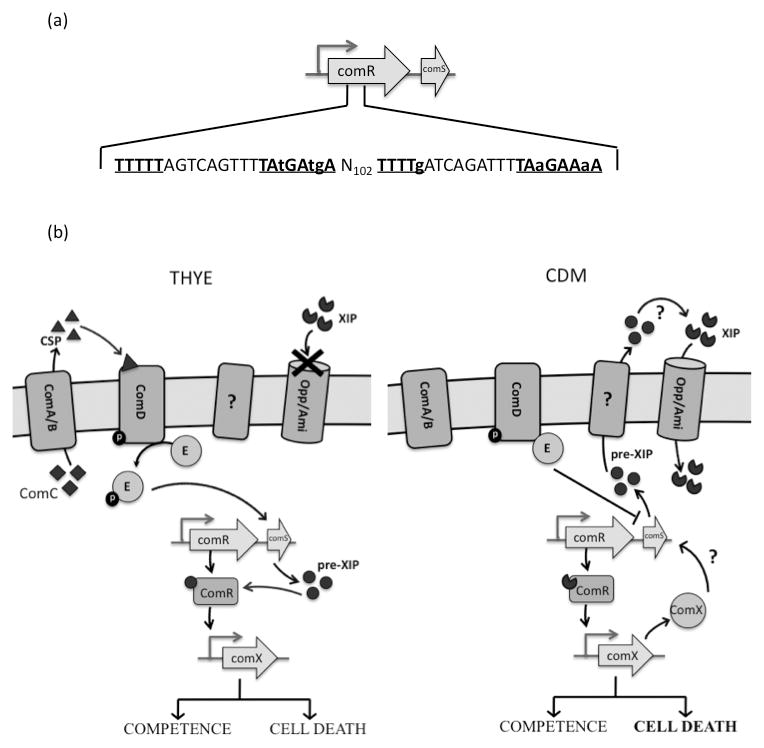
(a). Two putative ComX binding sites within the comR gene, located 102bp apart are underlined and shown in bold font (b). In complex medium (THYE), CSP is sensed by the ComDE system, which stimulates the expression of comS, leading to an intracellular accumulation of the ComS peptide or its cleavage products, which in turn, can activate ComR to induce comX expression. In CDM, the ComDE system represses comS expression. This repression is released by an unknown mechanism to upregulate comS, whose product is then secreted, processed and imported by the Opp/Ami transporter to activate comX transcription, as first described by Mashburn-Warren et al (2010). ComX positively regulates XIP production, which may contribute to the persistence of the competent state in CDM, as described by Desai et al (2012).
Acknowledgments
We thank Kirsten Krastel for technical assistance. We are thankful to Dr. Donald Morrison for his review of our manuscript and helpful suggestions provided along with Dr. Lauren Mashburn-Warren and Dr. Mike Federle. D.G.C. is a recipient of the NIH grant R01DE013230-03 and CIHR-MT15431.
References
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras MB, Ellen RP, Cvitkovitch DG. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- Cody AJ, Field D, Feil EJ, Stringer S, Deadman ME, Tsolaki AG, Gratz B, Bouchet V, Goldstein R, et al. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect Genet Evol. 2003;3:57–66. doi: 10.1016/s1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. Development of competence for genetic transformation by Streptococcus mutans in a chemically defined medium. Journal of Bacteriology. 2012 doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Cordova M, Cvitkovitch DG, CML Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol. 2011;193:6552–6559. doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Maiden MC, Achtman M, Spratt BG. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- Khan R, Rukke HV, Filho APR, Fimland G, Arntzen MO, Thiede B, Petersen FC. Extracellular identification of a processed type II ComR/ComS phermone of Streptococcus mutans. J Bacteriol. 2012 doi: 10.1128/JB.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, Cvitkovitch DG, Shi W, Qi F. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology. 2007;153:1799–1807. doi: 10.1099/mic.0.2007/005975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll JS, Hopkins I, Moxon ER. Capsule loss in Haemophilus influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell. 1988;53:347–356. doi: 10.1016/0092-8674(88)90155-9. [DOI] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 1992;28:130–145. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. Subpopulation-Specific Transcriptome Analysis of Competence-Stimulating-Peptide-Induced Streptococcus mutans. J Bacteriol. 2011;193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001a;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol. 2001b;183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002;184:6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Cvitkovitch DG, Levesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Kreth J, Levesque CM, Kay O, Mair RW, Shi W, Cvitkovitch DG, Goodman SD. Peptide pheromone induced cell death of Streptococcus mutans. FEMS Microbiol Lett. 2005;251:321–326. doi: 10.1016/j.femsle.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Seifert HS, Ajioka RS, Marchal C, Sparling PF, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336:392–395. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- Senadheera MD, Lee AWC, Hung DCI, Spatafora GA, Goodman SD, Cvitkovitch DG. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol. 2007;189:1451–1458. doi: 10.1128/JB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmoen H, Knutsen E, Havarstein LS. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc Natl Acad Sci U S A. 2002;99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]