Abstract
Radiolabels can be used to detect small biomolecules with high sensitivity and specificity, and without interfering with the biochemical activity of the labeled molecule. For instance, the radiolabeled glucose analogue, [18F]fluorodeoxyglucose (FDG), is routinely used in positron emission tomography (PET) scans for cancer diagnosis, staging and monitoring. However, despite their widespread usage, conventional radionuclide techniques are unable to measure the variability and modulation of FDG uptake in single cells. We present here a novel microfluidic technique, dubbed droplet radiofluidics, that can measure radiotracer uptake for single cells encapsulated into an array of microdroplets. The advantages of this approach are multiple. First, droplets can be quickly and easily positioned in a predetermined pattern for optimal imaging throughput. Second, droplet encapsulation reduces cell efflux as a confounding factor, because any effluxed radionuclide is trapped in the droplet. Last, multiplexed measurements can be performed using fluorescent labels. In this new approach, intracellular radiotracers are imaged on a conventional fluorescence microscope by capturing individual flashes of visible light that are produced as individual positrons, emitted during radioactive decay, traverse a scintillator plate placed below the cells. This method is used to measure the cell-to-cell heterogeneity in the uptake of tracers such as FDG in cell lines and cultured primary cells. The capacity of the platform to perform multiplexed measurements was demonstrated by measuring differential FDG uptake in single cells subjected to different incubation conditions and expressing different types of glucose transporters. This method opens many new avenues of research in basic cell biology and human disease by capturing the full range of stochastic variations in highly heterogeneous cell populations in a repeatable and high-throughput manner.
Introduction
Methods that can analyze the heterogeneous states and phenotypes of single cells have been garnering increased research attention in recent years.1–5 Fluorescence methods such as flow cytometry and microscopy have long been used for this purpose, but their limitation is that most small molecules lack intrinsic fluorescence and cannot be fluorescently labeled without greatly interfering with their biochemical activity.6 Radionuclide labeling has the advantage that it can be used to detect a small molecule with high sensitivity, both in vitro and in vivo, without altering the structure of the labeled molecule. The imaging of radiotracer distributions is routinely performed using positron emission tomography (PET) for cancer diagnosis, staging and monitoring.7 The radiotracer most commonly used in PET is the glucose analog [18F]fluorodeoxyglucose (FDG). Retention of FDG in tissues is correlated with high glucose metabolism and is used to image malignant tumors.8,9 However, despite its widespread usage in research and hospitals, many questions and contradictory observations arise about the variability and modulation of FDG uptake in cancer cells and even within cells from the same cell line.10–17 Conventional radionuclide detection methods, such are liquid scintillation counting, PET, and autoradiography, are limited to the measurement of radiotracer uptake in bulk samples. We present here a novel droplet microfluidic method that allows the measurement of radiotracer uptake in single cells and use this technique to measure the distribution of FDG uptake in single colon and breast cancer cells.
Microfluidics has been previously explored in nuclear medicine, primarily for the synthesis of radiopharmaceuticals such as PET tracers18–20 and for in vitro assays. For instance, Sweedler et al. analyzed the content of a single neuron cell by combining capillary electrophoresis with analysis by matrix-assisted laser desorption/ionization mass spectroscopy (MALDI) and radionuclide detection.21,22 Microfluidic chambers have been developed to probe kinase enzyme activity by measuring substrate phosphorylation using radioactive 32P, and to measure the fast metabolic response of live cancer cells to drugs.23,24 However, this imaging method has low throughput due to the small number of cell imaging compartments within the microfluidics chip and the poor spatial resolution of the radiation detector, and in general they do not allow large numbers of single cells to be measured.
We have recently shown that by detecting positron emission through a scintillator substrate, radionuclide detection can be extended to the single-cell level using an imaging technique called radioluminescence microscopy.25,26 Radioluminescence microscopy holds much promise for the study of the behavior of small molecules that cannot be easily labeled with a fluorescent dye, such as ions, amino acids, metabolites, and drugs. For instance, radioluminescence microscopy could be used to investigate the influence of epigenetics alterations on glucose metabolism27,28 and therapeutic radioiodine uptake in thyroid cancer29,30.
However, there are still constraints with the technique in its current state with regards to throughput and accuracy. The full potential of the technique is not achieved as cells tend to gather in clusters that make them undistinguishable from one another on the radioluminescence image due to the current resolution of radioluminescence microscopy of about 40 micrometers. To maximize throughput, cells should be spatially arranged in patterns amenable to image analysis. Increasing throughput is necessary for longitudinal studies of cells and would help capture the full range of variations in a heterogeneous population of cells. Another factor that degrades the accuracy of single-cell radionuclide measurements is radiotracer efflux. In a bulk measurement, radiotracer that is effluxed from the cell remain within the sample and therefore does not affect quantitative accuracy. However, in a single-cell assay, effluxed radiotracer is no longer tied to a specific cell and can (1) lower the perceived radioactivity of individual cells, and (2) increase the background signal. The use of droplet microfluidics—i.e. micrometer-sized aqueous droplets made and transported in inert oil—can overcome these experimental limitations in throughput and sensitivity.
Droplet microfluidics has rapidly advanced in recent years to a broad range of chemical31 and biological assays32. This progression has been driven by the large number of independent measurements made possible by picoliter droplets. Droplets can encapsulate cells for sequential or parallel large-scale biological assays.33–36 The parallel analysis of confined single cells is of particular interest for radiolabel studies and we present here a new platform, dubbed “droplet radiofluidics”, where single cells are encapsulated into a picoliter droplet array and imaged using radioluminescence microscopy.
This new approach has a number of advantages: First, because the cells are encapsulated in a hermetic droplet, cellular efflux does not affect the measurement because the radioactivity taken up by the cell remains trapped in the droplet and can thus be attributed to that cell; Second, using a technique called “Rails and Anchors”,37 droplets can be quickly and reliably positioned using passive forces to form an array, thus increasing throughput by optimizing cell-to-cell spacing for radioluminescence assays. Third, droplets loaded with different types of cells can be mixed and imaged simultaneously, using fluorescent dyes within the droplets to keep track of the various cell types. The advantage of such multiplexed measurements is that they are less subject to experimental biases since multiple cell populations are measured using the exact same parameters. Last, droplet microfluidics is suitable for reliably manipulating cells in suspension, including small samples such as rare circulating tumor cells or stem cells. This is an improvement over radioluminescence microscopy, which requires cell to be adherent.
We present below the droplet radiofluidics technique and its application to the profiling of single-cell glucose metabolism using FDG, a metabolic substrate of clinical relevance for its widespread usage in PET scans. We also use the multiplex capacities of the technique to show that, at the single-cell level, FDG uptake is differentially modulated by incubation conditions and the activity of the glucose transporter 1.
Materials and Methods
[18F]Fluorodeoxyglucose(FDG)
The glucose analog FDG was prepared from mannose triflate precursor via nucleophilic 18F-fluorination and hydrolysis. The 18F was produced in a GE PETtrace cyclotron. The production was proceeded on a cassette based automated synthetic module (FASTlab, GE healthcare). Quality control criteria were set and the tests were performed according to USP823. Due to its short lifetime, FDG was used within 8 hours after it was produced and dosed at the levels described below for experiments. The radioactivity was measured with a dose calibrator.
Cells
MDA-MB-231 human breast cancer cells and RKO human colon carcinoma cells were grown at 37 °C and 5% CO2 in RPMI medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Life Technologies, Carlsbad, CA). Cells were trypsinized and counted before being used in microscopy studies. For standard radioluminescence imaging, glass-bottom imaging dishes were coated with 5μg/mL human fibronectin (BD bioscience) and cells were plated two days prior to imaging. For droplet radioluminescence imaging, cells were harvested just prior to experiments. Where indicated, cells were stained with 5μM CellTracker Red CMTPX Dye (Invitrogen) using the manufacturer’s protocol. RKO cells in which the glucose transporter 1 (GLUT-1)38 was knocked out were produced using standard techniques.
Prior to imaging, cells were fasted in glucose-free RPMI medium supplemented with 10% fetal bovine serum for 45 minutes at 37 °C and 5% CO2. Subsequently, FDG ~500 microcurie (μCi) per mL ) was introduced in the media and allowed to be taken up by the cells for 45 min at 37°C and 5% CO2. Cells were then rinsed three times with RPMI medium supplemented with 10% fetal bovine serum to remove residual FDG. The cells were then trypsinized and concentrated to approximately 107 cells/mL for droplet radiofluidics experiments.
Microfluidic Device
Polydimethylsiloxane (PDMS) microfluidic chips with channel depth modulations were fabricated using the dry-film photoresist soft lithography technique described by Stephan et. al.39 since it enables rapid prototyping of multi-level structures. The PDMS chips were plasma-bonded to square 1 cm2 cadmium tungstate (CdWO4) scintillator of 0.5 mm thickness. To render the internal channel surface hydrophobic, Novec™ 1720 Electronic Grade Coating (3M) was flowed into the microchannel and the chip was heated for 30 minutes at 150 °C. The surface treatment prevented wetting and contact of the aqueous droplets with the channel walls.
Droplet Radiofluidics
Droplets were formed using two separate flow focusers and droplets flowed into a 2 mm wide channel containing an array of anchors (Figure 1b). The channel height was 25 μm. The circular anchors had a diameter of 50 μm, a depth of 25 μm and were spaced 150 μm apart. The oil phase used was fluorinated oil containing surfactant, QX100 (Biorad). The aqueous droplets contained 0.1 % m/v pluronic F-68 (Affymetrix) for all experiments. Pluronic F-68 was found to reduce undesired droplet fusion. For this combination of channel depth, anchor depth and fluids, droplets would remain in the anchors for external oil flows of less than about 100 μL/min. The array could be entirely cleared of all droplets at higher external oil flows (about 200 μL/min). Fluid flow was controlled using computer-controlled syringe pumps (Nemesys, Cetoni). All images were taken with an inverted bioluminescence microscope (Olympus LV200) equipped with a light-tight enclosure and multicolor fluorescence imaging capacity.
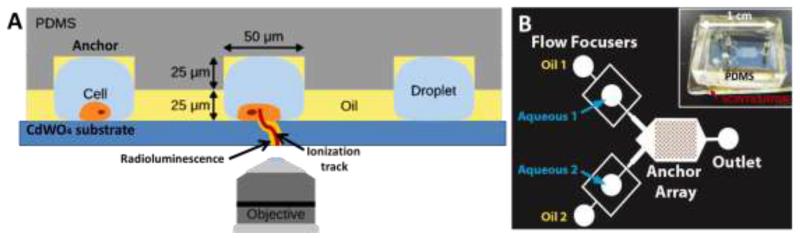
Figure 1.
a) The principal of droplet radiofluidics is presented by a cross-sectional diagram of the platform. Droplets encapsulating cells are anchored in locations of higher channel height. The radioactive decay of intracellular FDG leads to a beta particle producing an ionization track and to the emission of visible light in the CdWO4 scintillator, which is collected by a high numerical aperture objective. b) Microfluidic channel design used for droplet radioluminescence. The chip contains two flow focusers to make droplets. The wide part of the channel contains an anchor array shown in dark grey with wells that are 25 μm higher than the rest of the channel. Inset: Picture of device.
Image Analysis and Quantitation
The radioluminescence image was obtained by our methodology called “optical reconstruction of the beta-ionization track” (ORBIT) as described in detail in Pratx et al.26 Briefly, serial images of individual ionization tracks were acquired at a high acquisition rate (50-200 ms integration time) using an EM-CCD (Hamamatsu ImageEM C9100-14) at maximum gain. The acquisition time was chosen to average about 10 decay events per frame (typically 50-200 ms). Each frame was first filtered with a Gaussian kernel to reduce spatially uncorrelated shot noise. The processed image was later segmented using a constant threshold set above the noise floor. The final ORBIT image was reconstructed by aggregating the center of mass of each detected track for every image. The resulting image was then filtered with a Gaussian kernel to account for the localization uncertainty.
In the following results, the number of FDG molecules refers to the number molecules present at the beginning of the acquisition, inferred from the number of observed decays. It can be obtained from the standard decay equation for radioisotopes A(t) = A(0) exp(-λt), where A is the time-varying activity and λ is the rate constant of radioactive decay, which is related to the half-life, t1/2, by the expression λ = ln(2) /t1/2. When both sides are integrated over the total observation time T, the left side of the equation yields the number of observed decays N during this time. Solving the expression for the initial radioactivity A(0) and converting it to the initial number of molecules N0 via N0=A0/λ gives an expression for the initial number of observed FDG molecules, which we use throughout this work:
| (1) |
The yield factor compensates for the fact that the decay of 18F yields a positron only 97% of the time. The remaining decays occur by electron capture, which is not observable by radioluminescence microscopy. N0 excludes the FDG molecules in the cell that have decayed prior to the experiment.
Safety considerations
Radioactive compounds, such as [18F]fluorodeoxyglucose (FDG), pose particular health risks. Institutional protocols should be followed for the handling and disposal of all radioactive materials.
Results and Discussion
Droplets are ideal vessels for carrying cells; however, techniques are needed to control the position of picoliter droplets. Two-dimensional arrays simplify the observation, manipulation, and analysis of large sets of separate measurements. We use a technique called “Rails and Anchors”37 to produce a droplet array. The droplets in the microchannels are not spheres but rather “pancake” shaped, squeezed between the top and bottom surfaces of the channel. These flattened droplets can be positioned using a reduction in surface energy due to the expansion into a well that is microfabricated in the PDMS chip. This method allows the quick and passive formation of an array of droplets.
Figure 1a shows a cross-sectional view of the technique to demonstrate the general principal of droplet radiofluidics. The PDMS chip is directly bonded to the optically transparent CdWO4 scintillator. Following the incubation of the cells with the radionuclide probe FDG, cells are encapsulated in the picoliter droplets. The chip uses flow focusers40 to produce droplets (Figure 1b). There are two different flow focusers, to allow the production of two distinct populations of droplets. These droplets flow into a wide channel containing an anchor array consisting of microfabricated circular wells in the top of the channel. Droplets expand into the anchors and are therefore anchored in place despite the external flow of oil forming a droplet array. The droplets remain anchored for the duration of the radioluminescence acquisition (approximately 45 minutes). Figure 2a and b show a top view of the droplet array. Once the droplets are immobilized, the cells sediment in the droplet ensuring their close proximity to the scintillator below. The transparency of the scintillator plate in the visible range allows for conventional microscopy of the same sample.
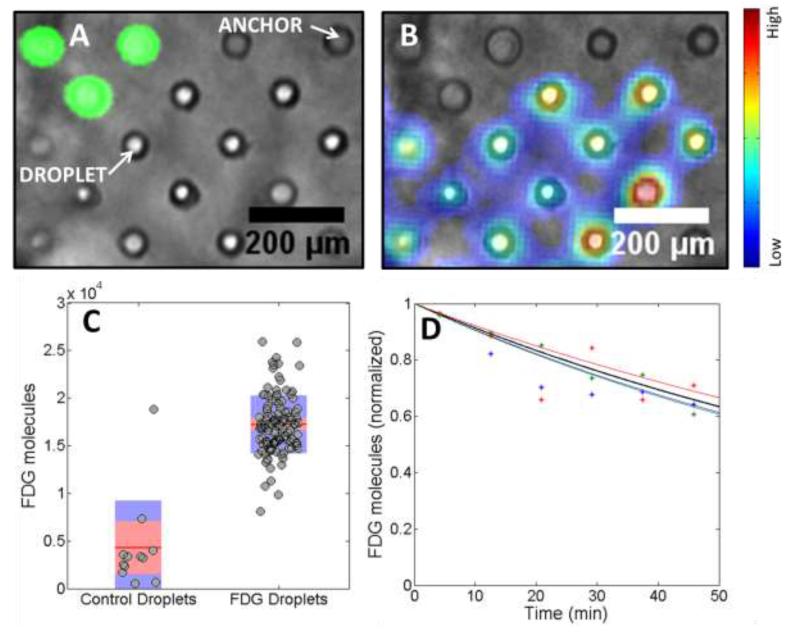
Figure 2.
Comparison of droplets containing a green fluorophore and a radioactive glucose analog, FDG. a) Fluorescence image of a subsection of the droplet array superimposed on a bright-field image. The green droplets contain fluorescein while clear droplets contain FDG b) Radioluminescence image superimposed on a bright-field image. Heat map intensity ranges from blue for low counts to red for high radiolabel count. c) Analysis of the number of observed FDG molecules for single droplets of control and FDG droplets. The average value is indicated by a red line. The standard deviation and the 95% confidence interval are indicated by a blue and red bar, respectively. d) Detected decays over time. The theoretical decay curve of 18F is shown in black. Data for three single droplets are shown in red, blue and green along with their fitted decay curves (solid lines).
The radioactive decay of probes inside the cells or droplet produces a beta particle (a positron in the case of 18F). Positrons can travel up to hundreds of micrometers through matter at relativistic speed, leaving in their wake an ionization track.41 Some of the positrons will propagate downwards through the scintillator, emitting visible-range photons on their path, in a process called radioluminescence (also known as scintillation).42,43 Decay events occurring further from the scintillator would have lower detection efficiency due to geometric factors and the beta particle’s finite travel distance. The small height of the confined droplet (50 μm) ensures that effluxed radiotracers also remain close to the scintillator surface. The visible light produced as the positron travels in the scintillator is collected by a high numerical aperture objective. The best spatial resolution is obtained by moving the microscope focal plane to the top surface of the scintillator near the point of entrance of the positron. After the acquisition, high external oil flow rates are used to eject the droplets from the anchors for subsequent experiments. A picture of the radiofluidic device is shown in the inset of Figure 1b.
To validate the technique and to highlight the differences with conventional fluorescence microscopy, two types of droplets, one containing radioactive [18F]fluorodeoxyglucose (FDG, 700 μCi/mL at the start of the 50 minute acquisition) and a smaller number of control droplets containing 5μM fluorescein were flowed into the anchor array and populated the anchors in a random manner. FDG is a radioactive glucose analogue and is widely used as a PET imaging agent, with a radioactive half-life of 109.8 minutes. Figure 2a shows a fluorescence image with blue excitation of a subsection of the array superimposed on a brightfield image showing droplets and anchors. The FDG droplets are approximately the same size as the anchors and a few anchors remain empty. In the fluorescence image, only the droplets containing fluorescein are bright and therefore the fluorescence image serves to unequivocally identify droplet type in the array. The light produced by beta decay into the scintillator is several orders of magnitude dimmer and therefore not visible in the fluorescence image (Figure 2a).
In contrast, when the excitation light is turned off, the only source of light is produced by radioluminescence. Thousands of short frames are acquired in rapid succession with short exposure time and high electron-multiplication gain.26 Because beta decay occurs stochastically, each short-exposure image contains only a few ionization tracks, which vary in their shape and intensity (supplemental video S-1). The locations of individual decay events are reconstructed to produce an image of the estimated distribution of beta decay events, superimposed on the bright field image (Figure 2b). The distribution reconstructed by the algorithm is a quantitative representation of the observed FDG probe quantity in the droplet at the start of the acquisition.
Comparison of Figure 2a and b confirms that droplets light up either in the fluorescence or in the radioluminescence image, but not in both. By defining regions of interest, each droplet is attributed a radioluminescence intensity as summarized for the two types of droplets in Fig 2c. This result confirms that radioluminescence is only produced in droplets containing FDG (Fig. 2c). The analysis measured on average 17,200±300 molecules per FDG-containing droplet (N=118) in contrast to 4300±1500 FDG molecules per control droplets (N=12). The variability of FDG signal is partly due to droplet size heterogeneity. Indeed, by producing droplets with a wider range of sizes, FDG signal was shown to correlate to the size of the droplet (supplemental figure S-1a). The sensitivity of the imaging system was also found to be constant across the field of view (supplemental figure S-1b). The low-level radioluminescence observed for control droplets is due to optical crosstalk from neighboring FDG-containing droplets and from background signal from long-range gamma rays. While crosstalk can be reduced by increasing the spacing of the wells, in general a small amount of crosstalk can be tolerated to allow for denser arrays and higher cell throughput. Fig. 2c also showed that one particular control droplet had unusually high FDG signal, an indication that it was displaced by an FDG droplet during the radioluminescence acquisition.
Finally, we plotted the number of detected decay events for three single droplets over time and fitted the resulting curve using an exponential decay model (Fig. 2d). The three fits show an average half-life of 112±9 min and follow closely the theoretical decay curve of 18F (half-life of 109.8 min). This indicates that the developed droplet system prevents FDG molecules from leaving a droplet and effectively preserves the initial concentration of radiolabeled small molecules. This experiment validates the suitability and sensitivity of the droplet microfluidic device for radioluminescence experiments.
The radiofluidics platform was used to observe two MDA-MB-231 cell populations that were exposed to FDG for a short or long incubation (60 minutes incubation versus 15 minutes incubation and 45 minutes efflux). After exposure, the cells were washed three times with buffer to remove residual FDG. Using a dose calibrator, the bulk radioactivity of the population of cells exposed to FDG for a long incubation was found to be 41±2 μCi/mL compared to 5±2 μCi/mL for the short incubation. However, as a bulk cell measurement, the dose calibrator does not give information on the cell-to-cell variability on the uptake of FDG and therefore lacks potentially critical information that lies in the uptake distribution of individual cells. The two cells populations were encapsulated separately into droplets and anchored to the droplet array. Fluorescein was again used to distinguish the two cell populations and a red fluorescent membrane label was used to identify cells in the droplets.
Figure 3a shows a fluorescence image superimposed on a bright-field image of a subsection of the array, which contains a mixture of the two populations of cells. Cells that underwent a long exposure to FDG are marked green while those subjected to a short exposure have been marked blue. Single cells from both cell populations are visible as red dots inside the droplets and are indicated by an arrow in the image. Some droplets are empty while others contain one or two cells, as expected from a Poisson distribution of cell occupancy.44 A few droplets appear to be immobilized next to an anchor, likely due to localized wetting interaction with the channel surface. Figure 3b shows the radioluminescence image superimposed on the bright-field image. The strongest signal corresponds to the cells that were incubated longer with FDG and to droplets containing multiple cells. The radioluminescence signal attributed to each cell is summarized in Figure 3c with long incubation giving an average of 500±80 (average ± error on mean) (N=12) FDG molecules per cell and the short incubation giving an average of 160±30 (N=20) FDG molecules per cell. For droplets containing multiple cells, the radioluminescence signal was normalized by the number of cells. This quantitative cell analysis confirms bulk measurements that show an increase in radioactivity for longer FDG incubation.
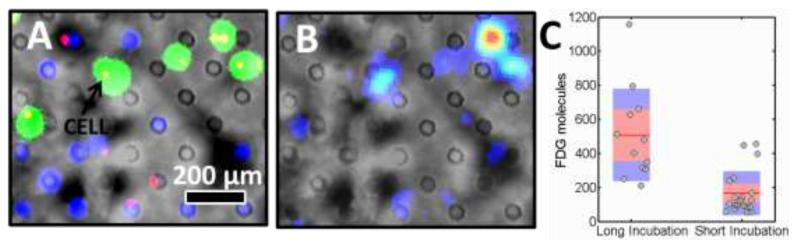
Figure 3.
Analysis of a breast cancer cell line (MDA-MB-231) for short (15 minute) and long (60 minutes) incubations with FDG. a) Fluorescence image of a subsection of the droplet array superimposed on a bright-field image. The green droplets were incubated with FDG for 60 minutes while blue droplets were incubated for 15 minutes and allowed to efflux for 45 minutes. Individual cells are marked in red. b) Radioluminescence image superimposed on a bright-field image. Intensity ranges from blue for low counts to red for high counts of radiolabels and primarily coincides with the position of long incubation cells. c) Analysis of the number of observed FDG molecules for single cells with long and short incubation. The average value is indicated by a red line. The standard deviation and the 95% confidence interval are indicated by a blue and red bar, respectively.
However, even within the same cell population, there is substantial cell-to-cell variability in the FDG uptake, for example spanning 200 to 1180 observed FDG molecules per cell for the long incubation with FDG. This variability is not visible in the bulk samples. Our system shows a spread in single-cell uptake values, which is reported here by the coefficient of variation—i.e. standard deviation divided by the mean uptake—of 50% for the long incubation and 80% for the short incubation. Both standard deviations are well above the shot noise, which would yield a 4% and 8% spread for the long and short incubation, respectively. Furthermore, the large coefficient of variation is not due to noise generated by the imaging field as the coefficient of variation of uniform size FDG droplets shown in Fig. 2 is only 16%. Consequentially, biological variations between cells are the primary cause for the large standard deviation. The larger spread for the short incubation can be explained by noting that variations in cellular processes that govern influx and efflux of FDG are non-linear in time. Indeed, uptake of FDG is known to reach equilibrium after a long-enough exposure to the tracer, which could potentially explain the difference in cell-to-cell variability between the two incubation conditions.
We also measured differential FDG uptake in an isogenic cancer model, composed of two genetically matched cell lines in which the glucose transporter 1 (GLUT-1) was either wild-type (RKO-WT) or knocked-out (RKO-KO). The knockout of GLUT-1 decreases by a factor of 3 the uptake of FDG both in bulk cell radioactivity measurements and on individual cells measured using standard radioluminescence microscopy measurements (supplemental figure S-2). The two cell lines were encapsulated in droplets and imaged as previously described. Droplets containing the knockout RKO cells were tagged with fluorescein and labeled green, and cells were marked in red (Figure 4a). The RKO cell line has a tendency to form small cell clumps and some droplets in the image contained multiple cells. The matching radioluminescence image shows that most of the signal can be attributed to droplets containing WT cells (Figure 4b). The analysis of single cells, normalized to the number of cells per droplet, shows a significantly lower FDG signal for the RKO KO cells (60±20, N=12) compared to the RKO WT cells (200±20, N=26). This result shows that, at the single-cell level, the knockout significantly reduces, but does not entirely block, the uptake of FDG. The uptake of the FDG into the knockout cells is driven by other glucose transporters of the family, mainly GLUT-3. However, these alternative pathways do not entirely compensate for the loss of GLUT-1, resulting in lower FDG uptake in the knockout cell line. A similar plot and conclusion is obtained when droplets containing multiple cells are excluded from the analysis.
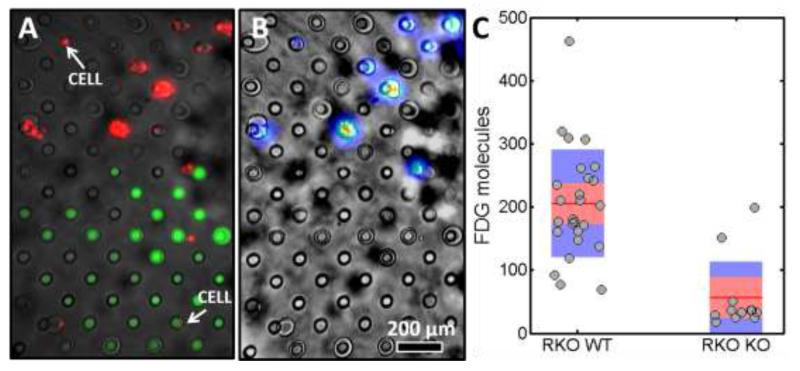
Figure 4.
Analysis of a colon cancer cells with wild-type (RKO WT) and glucose transporter 1 knockout (RKO KO). a) Fluorescence image of a subsection of the droplet array superimposed on a bright-field image. The clear droplets contain control cells while green droplets contain cells with the knockout. Individual cells are marked in red. b) Radioluminescence image superimposed on a bright-field image. Intensity ranges from blue for low counts to red for high counts of radiolabels and primarily coincides with the position of WT cells. c) Analysis of the number of observed FDG molecules for single cells for the control and knockout. The average value is indicated by a red line. The standard deviation and the 95% confidence interval are indicated by a blue and red bar, respectively.
Within both the WT and KO cells, a large spread is observed in individual cell’s uptake of FDG. The spread, which is reported here by the coefficient of variation, is 40% for the RKO WT cells and 100% for the RKO KO cells. The relative shot noise for the two samples is 7% and 13% for the RKO WT and RKO KO cells, respectively. This indicates that the alternative FDG uptake mechanism, driven mainly by GLUT-3, yields higher biological variability between single cells. This result is partly expected due to the fact that, in the WT cell line, variations in GLUT-1 and GLUT-3 expression would average out. A further possibility is that GLUT-1 expression may be also more tightly regulated than GLUT-3, thus reducing the overall variability.
Single-cell analyses using droplet microfluidics can be utilized to predict the therapeutic response on populations of heterogeneous cancer cells. For instance, bulk data suggest that inhibition of GLUT-1 would dramatically starve cancer of glucose. However, single-cell data paint a more nuanced picture: a small fraction of the RKO-KO cells have FDG uptake equivalent to an average RKO-WT cell. This suggests that anti-cancer drugs that inhibit GLUT-1 may be only selecting a subpopulation of cancer cells that have strong GLUT-3 expression and may still possess the ability to take up as much glucose as required for continued proliferation.
Conclusions
In summary, we have presented a novel droplet radiofluidics platform for analyzing the uptake of radiolabeled molecules by single cells encapsulated into a microdroplet array. This technique was used to measure the single-cell uptake of the glucose analogue FDG, used widely in PET scans to detect and monitor tumors. The experiments revealed large cell-to-cell heterogeneity in FDG uptake within the same cell line. The single cell uptake of the radiotracer was modulated by incubation conditions. The knockout of the glucose transporter 1 was shown to reduce (but not exclude) uptake of FDG except for a small fraction of cells that still exhibited significant glucose transport. Future studies, using this high-throughput platform, will seek to correlate FDG intake to cell cycle and gene expression profiles. Currently, there are conflicting results on the factors that modulate FDG uptake, 10–17 although the uptake value is a clinical predictor used by physicians.
As shown here, droplet radiofluidics is suitable for the study of cells in suspension and does not require cells to be adherent. It is therefore ideal for studying immune cells, circulating tumor cells, or primary cells derived from patients’ tumors. Droplet encapsulation facilitates the arraying of cells in two dimensions and can increase the number of cells that can be analyzed at one time. This can be further improved by incorporating microfluidic techniques that ensure single cell occupancy in droplets45,46 and by using thinner scintillators and smaller droplets to obtain better spatial resolution and lower optical crosstalk from neighboring droplets. Furthermore, different populations of cells can be imaged and compared simultaneously by using fluorescent dyes to label the content of the droplets, which allows for less systematic bias in differential measurements. Last, by trapping the radionuclide into the droplet at the time of encapsulation, the platform circumvents cell efflux as a confounding factor in the analysis.
In the future, droplets can also be recovered off-chip and coupled to techniques that have been developed for droplet sorting and selection.47 Furthermore, with the addition of a destabilizing surfactant,34 droplets can be broken to recover cells which can then be further analyzed (genomics, proteomics, metabolomics, etc.).
By including existing radiopharmaceuticals in the repertoire of dyes that can be used to label live cells, droplet radiofluidics will dramatically increase the ability to probe biochemical processes in single cells. Specific examples that can only be studied at the single cell level using radiolabels include most small-molecule drugs, the uptake of radioiodine, amino acid metabolism, glucose metabolism, and targeted radionuclide therapy.27–30,48 These measurements can be correlated with standard measurements of cellular states (e.g. cell cycle, redox status, proliferation, ROS production, etc.) obtained using the fluorescence channel. The study of single cells using existing small-molecule radiotracers will reveal new behaviors that cannot be observed with existing imaging techniques and thus contribute to our knowledge and understanding of basic cell biology and human disease.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Stanford small-animal imaging facility (SCI3); the Olympus Corporation for providing the LV200; Stanford Cyclotron Radiochemistry Facility Staffs (Murugesan Subbarayan, George Montoya and Shawn Scatliffe) for producing/packing 18F-FDG. We would also like to thank International Electronic Components Inc. for their generous donation of dry photoresist films and Rob Campell for the notBoxPlot matlab function. This work was supported in part by a grant from the National Institutes of Health (R01CA186275) and by a Prostate Cancer Research Program fellowship (W81XWH-14-1-0288). The radiotracer production was supported in part by a Stanford University, Department of Radiology Startup Funding (FTC), and a NCI ICMIC P50 CA114747 grant.
Footnotes
Supporting Information Available
This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Cai L, Friedman N, Xie XS. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- (2).Fritzsch FSO, Dusny C, Frick O, Schmid A. Annu. Rev. Chem. Biomol. Eng. 2012;3:129–155. doi: 10.1146/annurev-chembioeng-062011-081056. [DOI] [PubMed] [Google Scholar]
- (3).Spudich J., Jr Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- (4).Spiller DG, Wood CD, Rand DA, White MRH. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- (5).Willingham MC, Cornwell MM, Cardarelli CO, Gottesman MM, Pastan I. Cancer Res. 1986;46:5941–5946. [PubMed] [Google Scholar]
- (6).Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin C-C, Wang MC, Min W. Nat. Methods. 2014;11:410–412. doi: 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Surasi DS, Bhambhvani P, Baldwin JA, Almodovar SE, O’Malley JP. J. Nucl. Med. Technol. 2014;42:5–13. doi: 10.2967/jnmt.113.132621. [DOI] [PubMed] [Google Scholar]
- (8).Phelps ME. J. Nucl. Med. 1999;41:661–681. [PubMed] [Google Scholar]
- (9).Cairns RA, Harris IS, Mak TW. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- (10).Aloj L, Caraco C, Jagoda E, Eckelman WC, Neumann RD. Cancer Res. 1999;3:4709–4714. [PubMed] [Google Scholar]
- (11).Reske S, Grillenberger K, Glatting G, Port M, Hildebrandt M, Gansauge F, Beger H-G. J. Nucl. Med. 1997;38:1344–1348. [PubMed] [Google Scholar]
- (12).Brown RS, Wahl RL. Cancer. 1993;72:2979–2985. doi: 10.1002/1097-0142(19931115)72:10<2979::aid-cncr2820721020>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- (13).Rempel A, Mathupala SP, Griffin CA, Hawkins AL, Pedersen PL. Cancer Res. 1996;56:2468–2471. [PubMed] [Google Scholar]
- (14).Caracó C, Aloj L, Chen LY, Chou JY, Eckelman WC. J. Biol. Chem. 2000;275:18489–18494. doi: 10.1074/jbc.M908096199. [DOI] [PubMed] [Google Scholar]
- (15).Torizuka T, Zasadny KR, Recker B, Wahl RL. Radiology. 1998;207:767–774. doi: 10.1148/radiology.207.3.9609902. [DOI] [PubMed] [Google Scholar]
- (16).Zhao S, Kuge Y, Mochizuki T, Takahashi T, Nakada K, Sato M, Takei T, Tamaki N. J. Nucl. Med. 2005;46:675–682. [PubMed] [Google Scholar]
- (17).Avril N. J. Nucl. Med. 2004;45:930–932. [PubMed] [Google Scholar]
- (18).Elizarov AM. Lab Chip. 2009;9:1326–1333. doi: 10.1039/b820299k. [DOI] [PubMed] [Google Scholar]
- (19).Audrain H. Angew. Chem. Int. Ed. Engl. 2007;46:1772–1775. doi: 10.1002/anie.200603509. [DOI] [PubMed] [Google Scholar]
- (20).Arima V, Pascali G, Lade O, Kretschmer HR, Bernsdorf I, Hammond V, Watts P, De Leonardis F, Tarn MD, Pamme N, Cvetkovic BZ, Dittrich PS, Vasovic N, Duane R, Jaksic A, Zacheo A, Zizzari A, Marra L, Perrone E, Salvadori PA, Rinaldi R. Lab Chip. 2013;13:2328–2336. doi: 10.1039/c3lc00055a. [DOI] [PubMed] [Google Scholar]
- (21).Page JS, Rubakhin SS, Sweedler JV. Anal. Chem. 2002;74:497–503. doi: 10.1021/ac0156621. [DOI] [PubMed] [Google Scholar]
- (22).Miao H, Rubakhin SS, Scanlan CR, Wang L, Sweedler JV. J. Neurochem. 2006;97:595–606. doi: 10.1111/j.1471-4159.2006.03791.x. [DOI] [PubMed] [Google Scholar]
- (23).Fang C, Wang Y, Vu NT, Lin W-Y, Hsieh Y-T, Rubbi L, Phelps ME, Müschen M, Kim Y-M, Chatziioannou AF, Tseng H-R, Graeber TG. Cancer Res. 2010;70:8299–8308. doi: 10.1158/0008-5472.CAN-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang J, Hwang K, Braas D, Dooraghi A, Nathanson D, Campbell DO, Gu Y, Sandberg T, Mischel P, Radu C, Chatziioannou AF, Phelps ME, Christofk H, Heath JR. J. Nucl. Med. 2013;54:1820–1824. doi: 10.2967/jnumed.112.118497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pratx G, Chen K, Sun C, Martin L, Carpenter CM, Olcott PD, Xing L. PLoS One. 2012;7:e46285. doi: 10.1371/journal.pone.0046285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pratx G, Chen K, Sun C, Axente M, Sasportas L, Carpenter C, Xing L. J. Nucl. Med. 2013;54:1841–1846. doi: 10.2967/jnumed.112.113365. [DOI] [PubMed] [Google Scholar]
- (27).Gerhäuser C. Biomed. Res. 2012;23:69–89. [Google Scholar]
- (28).Yun J, Johnson JL, Hanigan CL, Locasale JW. Front. Oncol. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Brent GA, Kogai T. Nat. Rev. Endocrinol. 2013;9:508–509. doi: 10.1038/nrendo.2013.132. [DOI] [PubMed] [Google Scholar]
- (30).Derwahl M. J. Clin. Endocrinol. Metab. 2011;96:610–613. doi: 10.1210/jc.2010-2826. [DOI] [PubMed] [Google Scholar]
- (31).Song H, Chen DL, Ismagilov RF. Angew. Chem. Int. Ed. Engl. 2006;45:7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Guo MT, Rotem A, Heyman JA, Weitz DA. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- (33).Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Proc. Natl. Acad. Sci. 2009;106:14195. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Debs B. El, Utharala R, Balyasnikova IV, Griffiths AD, Merten CA. Proc. Natl. Acad. Sci. 2012;190:11570–11575. doi: 10.1073/pnas.1204514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret J-C, Marquez M, Klibanov AM, Griffiths AD, Weitz DA. Proc. Natl. Acad. Sci. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Huebner A, Bratton D, Whyte G, Yang M, Demello AJ, Abell C, Hollfelder F. Lab Chip. 2009;9:692. doi: 10.1039/b813709a. [DOI] [PubMed] [Google Scholar]
- (37).Abbyad P, Dangla R, Alexandrou A, Baroud CN. Lab Chip. 2011;11:813–821. doi: 10.1039/c0lc00104j. [DOI] [PubMed] [Google Scholar]
- (38).Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JKV, Markowitz S, Zhou S, Diaz LA, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Stephan K, Pittet P, Renaud L, Kleimann P, Morin P, Ouaini N, Ferrigno R. J. Micromech. Microeng. 2007;17:N69. [Google Scholar]
- (40).Anna SL, Bontoux N, Stone HA. Appl. Phys. Lett. 2003;82:364–366. [Google Scholar]
- (41).Levin CS, Hoffman EJ. Phys. Med. Biol. 1999;44:781–799. doi: 10.1088/0031-9155/44/3/019. [DOI] [PubMed] [Google Scholar]
- (42).Melcher CL. Nucl. Instruments Methods Phys. Res. Sect. A. 2005;537:6–14. [Google Scholar]
- (43).Moses WW. Nucl. Instruments Methods Phys. Res. Sect. A. 2002;487:123–128. [Google Scholar]
- (44).Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Köster S, Duan H, Holze C, Weitz DA, Griffiths AD, Merten CA. Chem. Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- (45).Chabert M, Viovy J-LL. Proc. Natl. Acad. Sci. 2008;105:3191. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Edd JF, Di Carlo D, Humphry KJ, Köster S, Irimia D, Weitz DA, Toner M. Lab Chip. 2008;8:1262–1264. doi: 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Baret J-C, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD. Lab Chip. 2009;9:1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- (48).Tseng J-C, Wang Y, Banerjee P, Kung AL. Mol. Imaging Biol. 2012;14:553–560. doi: 10.1007/s11307-012-0545-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.