Abstract
E3 ligases are genetically implicated in many human diseases, yet E3 enzyme mechanisms are not fully understood, and there is a strong need for pharmacological probes of E3s. We report the discovery that the HECT E3 Nedd4-1 is a processive enzyme and that disruption of its processivity by biochemical mutations or small molecules switches Nedd4-1 from a processive to a distributive mechanism of polyubiquitin chain synthesis. Furthermore, we discovered and structurally characterized the first covalent inhibitor of Nedd4-1, which switches Nedd4-1 from a processive to a distributive mechanism. To visualize the binding mode of the Nedd4-1 inhibitor, we used X-ray crystallography and solved the first structure of a Nedd4-1 family ligase bound to an inhibitor. Importantly, our study shows that processive Nedd4-1, but not the distributive Nedd4-1:inhibitor complex, is able to synthesize polyubiquitin chains on the substrate in the presence of the deubiquitinating enzyme USP8. Therefore, inhibition of E3 ligase processivity is a viable strategy to design E3 inhibitors. Our study provides fundamental insights into the HECT E3 mechanism and uncovers a novel class of HECT E3 inhibitors.
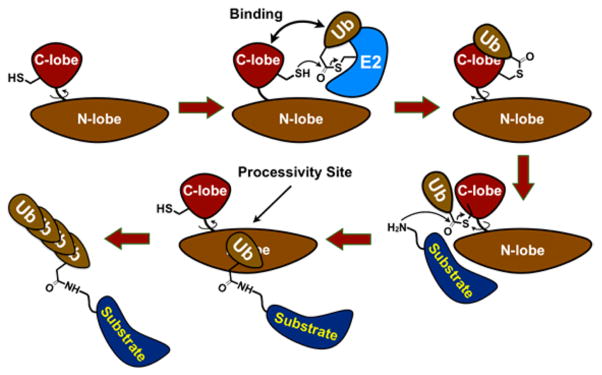
The E3 ligases regulate all aspects of biology, and there is a strong need for E3 ligase inhibitors and probes.1–3 Nedd4-1 is a HECT E3 ubiquitin (Ub) ligase (~28 known) and regulates mammalian metabolism, growth, and development. 4 Furthermore, it is a promising drug target to treat cancers,5 obesity,6 Parkinson’s disease,7 and viral infections.8 HECT E3s form an obligatory HECT E3 ~ Ub thioester during the catalytic cycle for the subsequent ligation of the Ub onto the substrate lysine. Current biochemical studies of HECT E3s suggest a mode of chain elongation which may occur by either a processive or a distributive mechanism (Figures 1, S1).9–13 In this model, the last Ub of the growing polyUb chain binds the N-lobe of the catalytic HECT domain proximal to the C-lobe, which positions this polyUb chain for the addition of another Ub molecule for polyUb chain growth. However, whether HECT E3 ligases are processive or distributive enzymes, and how this process might be targeted for inhibition, had not been completely investigated. Herein, we present the first rigorous proof that Nedd4-1 is a processive enzyme and describe the discovery of a first-in-class Nedd4-1 inhibitor. The discovered Nedd4-1 inhibitor is the first example of an E3 inhibitor that switches the enzyme from a processive to a distributive mechanism of polyUb chain synthesis. Furthermore, when Nedd4-1 becomes distributive, substrate ubiquitination can be efficiently antagonized by the deubiquitinating enzyme USP8 in vitro.
Figure 1.
Simplified model of HECT E3-mediated protein ubiquitination. E2 ~ Ub thioester binds HECT E3 domain, followed by transthiolation reaction producing HECT E3 ~ Ub thioester. Subsequently, HECT E3 ~ Ub ligates Ub onto the lysine of the substrate, followed by polyUb chain growth. C- and N-lobes of HECT domain rotate relative to each other during the catalytic cycle.
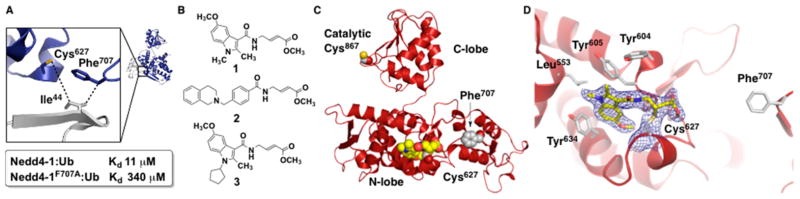
The crystal structure of the Nedd4-1 HECT domain with Ub bound to the N-lobe10,11 has two solvent-exposed Cys residues: the catalytic Cys867 and the noncatalytic Cys627. Cys627 is located in the N-lobe of Nedd4-1 near Phe707 of Nedd4-1 and Ile44 of Ub, two residues that form critical hydrophobic contacts (Figure 2A). Mutation of Phe707 to Ala in Nedd4-1 disrupts Nedd4-1:Ub binding and affects the kinetics of polyUb chain growth.11 The equivalent F618A mutation in Rsp5, the Saccharomyces cerevisiae homologue of Nedd4-1, also disrupts its binding to Ub and results in temperature-sensitive growth defects, suggesting an essential function of this site in vivo.12 Therefore, inhibitors that target these cysteines would either inhibit Nedd4-1 completely (Cys867) or inhibit the ability of Nedd4-1 to elongate polyUb chains (Cys627). To search for Nedd4-1 inhibitors, we used our previously reported covalent tethering method.14 The HECT domain of Nedd4-1 was treated with a mixture of electrophilic fragments, and compounds 1 and 2 were identified as weak covalent modifiers of cysteine residues in Nedd4-1 using mass spectrometry (Figures 2B, S2). Point mutation studies revealed that compounds 1 and 2 selectively reacted with the noncatalytic Cys627 of Nedd4-1 (Figure S3) in a time- and concentration-dependent manner (Figure S4). The catalytic Cys867 of Nedd4- 1 was more reactive than Cys627 with the N-acetyl electrophile 4 that has only CH3 as a directing group (Figure S5), suggesting that compounds 1 and 2 are specific hits, since they modify the less reactive noncatalytic cysteine in the presence of the more reactive catalytic cysteine. Furthermore, compound 1 demonstrated an initial structure–activity relationship, a further indication that covalent labeling of Nedd4-1 by 1 was specific (Figure S6).
Figure 2.
(A) The hotspot interface of Nedd4-1 (blue) and Ub (gray) in the Nedd4-1:Ub complex (PDB ID 2XBB) responsible for polyUb chain elongation, with the key side chains depicted as sticks. (B) Electrophilic compounds 1–3 used in this work. Compounds 1 and 2 are specific hits, and compound 3 is an optimized covalent inhibitor of Nedd4-1. (C) Cartoon depiction of the crystal structure of Nedd4-1 bound to fragment hit 1 with key side chains and the fragment shown as spheres. (D) Close-up view of the small molecule-binding site with the key side chains depicted as sticks and colored by atom type. The 2Fo – Fc electron density map (blue mesh, contoured at 1.0 σ) is presented for Cys627 and 1.
To visualize the binding mode of compound 1, we crystallized the Nedd4-1:1 complex and solved the structure to 2.44 Å resolution (PDB ID: 5C91) (Figures 2C, S7). Notably, this structure is the first of a HECT E3 Ub ligase bound to a small molecule. The overall conformation is virtually identical to the previously reported structure of Nedd4-110,11 (root-mean-square deviation (rmsd): 0.316 Å; Figure S7). Our structure confirms that 1 forms a stable covalent bond with Cys627 and reveals that the hydrophobic indole core of 1 is oriented toward a pocket of the N-lobe formed by residues Leu553, Glu554, Asn602, Tyr604, Tyr605, Leu607, and Tyr634 (Figures 2D, S7). This ligand orientation explains why compound 6, which contains an EtO group on the indole core, did not label Nedd4-1, since the ligand binding pocket cannot accommodate this sterically bulkier group (Figure S6). The aromatic edge-to-face interactions of Tyr605 and Tyr634 with the indole moiety of 1 provide further stabilization of the ligand conformation, while a hydrogen bond between the backbone carbonyl oxygen of Tyr605 and the amide NH of 1 positions the connecting region between Cys627 and the indole group.
The ester methoxy group points inward and toward the cavity formed by Gly606O, Asn621N, Asn623O, and Asn623Cβ, which are within 3.4–4.4 Å of the methyl group. However, since the methyl ester group is freely rotatable around the C–C bond, our crystallographic data cannot exclude a partial conformation in which the methoxy and the carbonyl groups are exchanged. Interestingly, it was shown that Tyr605 of Nedd4-1 is also important for noncovalent Ub binding and polyUb chain elongation by Nedd4-1.11 Since 1 forms an edgeto- face binding interaction with Tyr605, the ligand should block the interaction between Leu73 of Ub and Tyr605 of Nedd4-1 observed in the Nedd4-1 HECT:Ub complex structure.
Our initial experiments showed that labeling of Nedd4-1 with 1 could be completely inhibited by 60 μM of Ub (Figure S8), which is the approximate concentration of Ub in cells.15 Therefore, further improvements in the potency of 1 were necessary. Since the N-methyl group in 1 was shown to tolerate substitutions, we prepared a series of N-substituted analogues 3 and 11–13 (Figure S9). Of these, the N-cyclopentyl analogue 3 was the most potent. Indicative of its selectivity, compound 3 did not label the HECT domains of the related ligases WWP1 or E6-AP that do not have cysteine at this position (Figures S10, S11B), but it was effective at labeling the highly homologous HECT ligase Nedd4-2, which has an almost identical noncovalent Ub-binding site and a cysteine at that position (Figure S11). Furthermore, compound 3 did not react with the deubiquitinase USP8, Human Rhinovirus 3C protease, the E1 enzyme Ube1, or the E2 enzyme UbcH5a, all of which have reactive catalytic cysteines (Figure S12).
Fluorescence polarization (FP) experiments using fluorescein- Ub were used to confirm that 3 inhibits the Nedd4-1:Ub interaction. Compound 3 disrupted Nedd4-1:Ub binding with second-order inhibition kinetics (KI 29.3 μM, kinact 5.8 × 10−5 s−1; kinact/KI = 1.98 M−1 s−1) and was 22.2-fold more potent than compound 1. (Figure S13). Notably, the FP assay requires a high concentration of Nedd4-1 (8 μM); so with a KI of 29.3 μM we achieve half-maximal covalent inhibition at only a 3.7-fold excess of inhibitor relative to Nedd4-1. Furthermore, in contrast to indole 1, compound 3 was able to label Nedd4-1 even in the presence of 60 μM Ub, indicative of its increased potency (Figure S14). To investigate if compound 3 affects the ability of Nedd4-1 to elongate polyUb chains we used Wbp2-C-K222 as a substrate.13 Wbp2-C-K222 has only one acceptor lysine residue (Lys222) and a single cysteine residue (Cys73), which we modified with 5-iodo-acetamidofluorescein for quantification purposes (abbreviated Flu-Wbp2; Figure S15). Remarkably, we observed that the catalytic HECT domain of Nedd4-1 covalently modified with compound 3 was not able to build long polyUb chains on Flu-Wbp2, as compared to Nedd4-1 treated with DMSO or the negative control electrophile 14 (Figure S16). Importantly, this inhibition does not occur because 3 inhibits the formation of the Nedd4-1 ~ Ub thioester (Figure S17). However, since the Nedd4-1:3 complex could still build short polyUb chains on Flu-Wbp2, we asked if these chains are built with a processive or a distributive mechanism.
We hypothesized that since compound 3 disrupts Ub binding to the noncovalent Ub-binding site of Nedd4-1, the ubiquitinated substrate would be more likely to dissociate from the Nedd4-1:inhibitor complex in between rounds of addition of Ub to the growing chain. Therefore, inhibitor-bound Nedd4-1 should assemble polyUb chains via a distributive mechanism. To distinguish between processive and distributive mechanisms, we used an assay wherein full length Nedd4-1 with or without inhibitor pretreatment is allowed to ubiquitinate Flu-Wbp2 for 1 min, followed by addition of a 200-fold excess of nonfluorescent Wbp2. If Nedd4-1 is processive, it should remain bound to Flu-Wbp2-Ubx and continue to elongate the polyUb chain on Flu-Wbp2 even in the presence of the large excess of nonfluorescent Wbp2. If Nedd4-1 is distributive, it should dissociate from Flu-Wbp2-Ubx between rounds of ubiquitination. In this case, Flu-Wbp2-Ubx will be outcompeted by nonfluorescent Wbp2 and polyUb chain growth on Flu-Wbp2 will be inhibited.
For these experiments, we used full-length Nedd4-1 with the activating E554A mutation, which disrupts the autoinhibitory conformation of wild-type full length Nedd4-1.16 We found that E554A Nedd4-1 was processive and efficiently converted Flu-Wbp2 into ≥Ub4-modified Flu-Wbp2 even after addition of a 200-fold excess of nonfluorescent Wbp2 (Figure 3A,B). However, in the case of the Nedd4-1:3 complex (Figure S18), we found that ubiquitination of Flu-Wbp2 was significantly inhibited upon addition of a 200-fold excess of nonfluorescent Wbp2 (Figure 3C,D). Furthermore, consumption of monoubiquitinated Flu-Wbp2 and the formation of Ub2/Ub3 and ≥Ub4-modified Flu-Wbp2 were also inhibited (Figure 3C,D). This observation indicates that inhibitor-bound Nedd4-1 dissociates from Flu-Wbp2-Ubx before adding Ubx+1 and is therefore distributive. Similar results were observed for the Nedd4-1 E554A F707A mutant (Figure S19). These experiments prove for the first time that Nedd4-1 is processive, and when the noncovalent interaction between the N-lobe and Ub is disrupted by compound 3 or the F707A mutation, the enzyme becomes distributive. Previously, it was assumed, but not rigorously proven, that HECT E3s are processive and not distributive enzymes.
Figure 3.
Covalent inhibitor 3 switches Nedd4-1 from a processive to a distributive enzymatic mechanism. (A) Full length Nedd4-1 with the activating E554A mutation (150 nM) was incubated with fluorescent Flu-Wbp2 substrate (100 nM) in the presence of ATP, Ub, E1, and E2 enzymes. After 1 min, a 200-fold excess of nonfluorescent Wbp2 substrate or empty buffer was added to the reaction mixture, and further ubiquitination of Flu-Wbp2 was monitored. (B) The amount of ubiquitinated Flu-Wbp2 was plotted as a function of time. (C) The same experiment was conducted with full-length Nedd4-1 E554A covalently modified with compound 3. (D) The amount of ubiquitinated Flu-Wbp2 in (C) was plotted as a function of time.
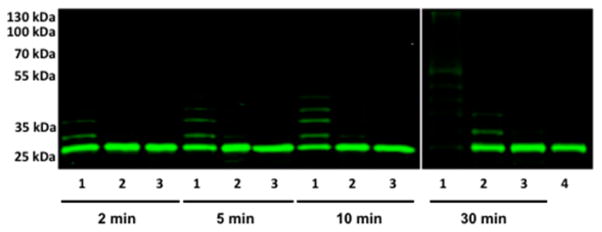
Since endogenous intracellular deubiquitinating enzymes (DUBs) reverse protein ubiquitination, we hypothesized that distributive Nedd4-1 would be more susceptible to antagonism by DUBs than processive Nedd4-1. To test this hypothesis, full length Nedd4-1 E554A with or without compound 3 bound and the Nedd4-1 E554A F707A mutant were allowed to ubiquitinate Flu-Wbp2 in the presence of the DUB USP8. In vitro, USP8 can disassemble K48- and K63-linked polyUb chains,17 while Nedd4-1 predominantly makes K63-linked chains.11 Remarkably, we observed that untreated Nedd4-1 robustly polyubiquitinated Flu-Wbp2 in the presence of USP8 after 30 min. However, neither compound 3-treated Nedd4-1 nor the F707A mutant were able to consume Flu-Wbp2 or build ≥Ub4 chains on Flu-Wbp2 in the presence of USP8, even though they consumed Flu-Wbp2 in the absence of USP8 (Figures 4, S20). Since distributive Nedd4-1 dissociates from the substrate in between rounds of Ub addition to the growing polyUb chain, this provides an opportunity for the DUB to hydrolyze the Ub chain before another Ub can be added to it by Nedd4-1. Processive catalysis appears to be a necessary condition for the formation of ≥Ub4 chains on the substrate in the presence of DUB.
Figure 4.
Ubiquitination of Flu-Wbp2 (100 nM) by a full length Nedd4-1 (150 nM) at different time points in the presence of the deubiquitinase USP8 (200 nM) shows that distributive Nedd4-1 is inhibited by USP8, but processive Nedd4-1 is not. Lane 1: DMSO treated Nedd4-1 full length E554A; lane 2: compound 3 treated Nedd4-1 full length E554A; lane 3: Nedd4-1 full length E554A F707A; and lane 4: no ATP control.
In summary, we report the first rigorous proof that the HECT E3 Nedd4-1 is a processive enzyme and that disrupting noncovalent binding of Ub to the N-lobe of Nedd4-1 switches Nedd4-1 to a distributive enzyme. Furthermore, we report the discovery and structural characterization of a covalent Nedd4-1 inhibitor that targets this processive site and switches Nedd4-1 from a processive to a distributive mechanism. We also show that introducing a DUB antagonist into the assay augments the inhibitory effect of compound 3 on Nedd4-1, while the untreated enzyme is still able to build long polyUb chains. E3 ligase inhibitors with this mechanism of action were not previously known, and this work outlines a conceptually new design strategy for E3 ligase inhibitors. Taken together, our studies provide fundamental insights into the HECT E3 enzymatic mechanism and represent an important case study in the emerging area of E3 ligase inhibitor discovery.18 Further studies of Nedd4-1 inhibitors will be reported in due course.
Supplementary Material
Acknowledgments
Funding was provided from Northwestern University (A.V.S.), Pew Charitable Trusts (A.V.S.), National Institutes of Health Grants R01GM115632 (A.V.S.), GM58518 (A.C.R.), T32GM105538 (S.G.K.), and F32GM105339 (A.T.S.), Deutsche Forschungsgemeinschaft (DFG) Sp 1476/1-1 (I.S.), Chemistry of Life Processes Institute Lambert Fellowship (Z.X.), and the ACS Medicinal Chemistry Fellowship (S.G.K.). We thank Simona Polo (IFOM, Milan, Italy) and Jon Huibregtse (University of Texas) for Nedd4-1 and Wbp2-C-K222 constructs. We thank Rama Mishra and the Center for Molecular Innovation and Drug Discovery for assisting with the initial design of the library of electrophilic fragments.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b06839.
Experimental procedures, analytical data, supporting figures, and schemes (PDF)
References
- 1.Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Buckley DL, Crews CM. Angew Chem, Int Ed. 2014;53:2312–30. doi: 10.1002/anie.201307761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rotin D, Kumar S. Nat Rev Mol Cell Biol. 2009;10:398. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 4.Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, Daly RJ, Kumar S, Yang B. Sci Signaling. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi YJ, Wang JR, Chandarlapaty S, Cross J, Thompson C, Rosen N, Jiang XJ. Nat Struct Mol Biol. 2014;21:522. doi: 10.1038/nsmb.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JJ, Ferry RJ, Diao SY, Xue BZ, Bahouth SW, Liao FF. Endocrinology. 2015;156:1283. doi: 10.1210/en.2014-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, Caldwell GA, Rochet JC, Buchwald SL, Lindquist S. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. J Virol. 2014;88:7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamadurai HB, Qiu Y, Deng A, Harrison JS, MacDonald C, Actis M, Rodrigues P, Miller DJ, Souphron J, Lewis SM, Kurinov I, Fujii N, Hammel M, Piper R, Kuhlman B, Schulman BA. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maspero E, Valentini E, Mari S, Cecatiello V, Soffientini P, Pasqualato S, Polo S. Nat Struct Mol Biol. 2013;20:696. doi: 10.1038/nsmb.2566. [DOI] [PubMed] [Google Scholar]
- 11.Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, Polo S. EMBO Rep. 2011;12:342. doi: 10.1038/embor.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French ME, Kretzmann BR, Hicke L. J Biol Chem. 2009;284:12071. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HC, Steffen AM, Oldham ML, Chen J, Huibregtse JM. EMBO Rep. 2011;12:334–341. doi: 10.1038/embor.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathman SG, Xu Z, Statsyuk AV. J Med Chem. 2014;57:4969. doi: 10.1021/jm500345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Nat Methods. 2011;8:691. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mari S, Ruetalo N, Maspero E, Stoffregen MC, Pasqualato S, Polo S, Wiesner S. Structure. 2014;22:1639. doi: 10.1016/j.str.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Reyes-Turcu FE, Wilkinson KD. Chem Rev. 2009;109:1495. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mund T, Lewis MJ, Maslen S, Pelham HR. Proc Natl Acad Sci U S A. 2014;111:16736. doi: 10.1073/pnas.1412152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.