Abstract
Background
The zoonotic bacterium Campylobacter jejuni has a broad host range but is especially associated with birds, both domestic and wild. Earlier studies have indicated thrushes of the genus Turdus in Europe to be frequently colonized with C. jejuni, and predominately with host-associated specific genotypes. The European Blackbird Turdus merula has a large distribution in Europe, including some oceanic islands, and was also introduced to Australia by European immigrants in the 1850s.
Methods
The host specificity and temporal stability of European Blackbird C. jejuni was investigated with multilocus sequence typing in a set of isolates collected from Sweden, Australia, and The Azores.
Results
Remarkably, we found that the Swedish, Australian, and Azorean isolates were genetically highly similar, despite extensive spatial and temporal isolation. This indicates adaptation, exquisite specificity, and stability in time for European Blackbirds, which is in sharp contrast with the high levels of recombination and mutation found in poultry-related C. jejuni genotypes.
Conclusion
The maintenance of host-specific signals in spatially and temporally separated C. jejuni populations suggests the existence of strong purifying selection for this bacterium in European Blackbirds.
Keywords: gastrointestinal pathogen, Campylobacter jejuni, Blackbird, MLST, population structure
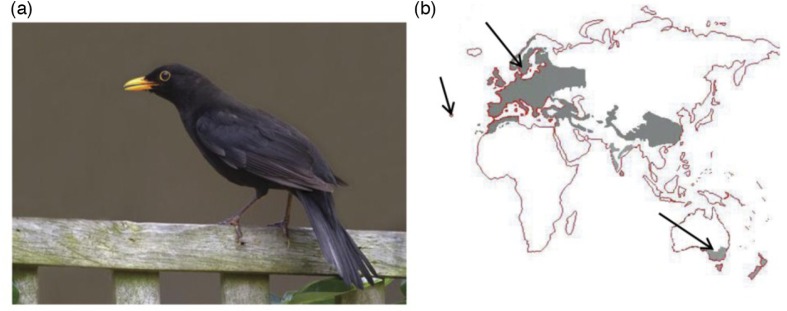
When immigrants from Europe colonized Australia, the new settlers faced a very different environment. Many were homesick and missed the birds and animals that were familiar to them. Acclimatization societies were established to help the new Australians adjust to their foreign surroundings, which led to the introduction of English birds (1, 2). European Blackbirds (Fig. 1a) have a beautiful and well-known song and it was perceived that their presence would help the immigrants settle in to their new environment (1). Therefore, European Blackbirds were released between 1850 and 1870 in Australia, with the first releases in Melbourne and Sydney. The introduction was successful, and Blackbirds are now established in south-eastern Australia, with the most recent population estimates being 1–6 birds/ha around Melbourne. The birds are sedentary and do not migrate in Australia, whereas in Europe they are partial migrants (2) (Fig. 1b). In both regions, Blackbirds feed on terrestrial invertebrates, particularly earthworms, snails, and insects as well as on seeds and fruit, but the relative importance of food items differs between localities and the time of year (2, 3). The native distribution of the Blackbird includes most of mainland Europe, parts of Russia, and North Africa. There are isolated populations on some oceanic islands, including The Azores where the endemic T. merula azorensis is abundant (3) (Fig. 1b). Blackbird populations have been separated from Europe for 150 generations in Australia and around 10,000 generations on The Azores with essentially no interchange due to migration.
Fig. 1.
(a) European Blackbird (Turdus merula), NSW, Australia. Photo: Grant Brosie. (b) The global distribution of Blackbird populations shown in grey. Arrows indicate the sampling locations.
The significance of Campylobacter in human disease was first realized in the early 1980s (4). Campylobacter jejuni infection is now recognized as one of the most common causes of bacterial gastroenteritis (campylobacteriosis) in humans in large parts of the world. It is typically acquired through the consumption of contaminated food, particularly commercial poultry, or the environment and usually involves self-limiting symptoms such as diarrhea, vomiting, and dehydration and is seldom spread from person to person (5). Certain wild bird species can also carry C. jejuni, but it is still not fully understood whether wild birds play a role in the spread of C. jejuni to other hosts, or whether they can act as a natural reservoir for the genotypes involved in human disease (6). C. jejuni does not normally produce disease symptoms in birds, and has been regarded as commensals of the gastrointestinal tract of both domestic and wild birds (3). However, a study with wild European Robins (Erithacus rubecula) showed that infection with human-associated C. jejuni induced weight loss (7). New strains are continuously emerging, as the result of mutations and high recombination frequency (8). Wild bird genotypes seem to exhibit stronger patterns of host specificity and higher genetic diversity than human-/farm-related C. jejuni, and also less similarity to human-associated C. jejuni (6, 9, 10). Nevertheless, despite intensive efforts, the survival properties of C. jejuni population structures, as well as the species-specific, temporal, and geographic stability of C. jejuni are not fully understood. Due to the history of the Blackbird and the isolated population, these factors can be investigated.
Materials and methods
In total, 109 C. jejuni isolates (103 isolates from Sweden, 4 isolates from Australia, and 2 isolates from The Azores) were characterized by multilocus sequence typing (MLST) and incorporated in the analysis (Table 1). All samples except those from The Azores have previously been published as a part in a large data set. MLST is commonly used for genetic studies of C. jejuni (8, 11). The samples from Sweden and Australia have previously been used in a study investigating C. jejuni epidemiology in numerous species of wild birds, chickens, and humans (10). Cultivation, morphology, and phenotypic characterization of C. jejuni followed standard protocols (12). Polymerase chain reaction amplification and nucleotide sequencing of each isolate was performed using established C. jejuni MLST techniques (12) and the use of the Campylobacter database (www.pubmlst.org/campylobacter/) to establish sequence types (ST) (8, 11) Table 1.
Table 1.
C. jejuni genotypes isolated from Blackbird samples collected in Sweden, Australia, and The Azores
| Country | Year | aspAa | glnAa | gltAa | glyAa | pgma | tkta | uncAa | STb | CCc | Frequencyd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sweden | 2001 | 48 | 7 | 10 | 4 | 1 | 7 | 1 | 11 | 45 | 5 |
| Sweden | 2001 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | 45 | 45 | 1 |
| Sweden | 2000 | 4 | 7 | 10 | 4 | 42 | 7 | 1 | 137 | 45 | 1 |
| Sweden | 2001 | 17 | 2 | 8 | 5 | 8 | 2 | 4 | 177 | 177 | 1 |
| Sweden | 2001 | 4 | 7 | 40 | 4 | 42 | 51 | 1 | 267 | 283 | 1 |
| Sweden | 2000/2001 | 10 | 81 | 50 | 99 | 120 | 76 | 52 | 677 | 677 | 4 |
| Sweden | 2001 | 10 | 2 | 107 | 137 | 120 | 76 | 1 | 1080 | – | 1 |
| Sweden | 2000/2001 | 97 | 121 | 97 | 125 | 174 | 152 | 100 | 1229 | – | 8 |
| Sweden | 2001 | 99 | 127 | 91 | 136 | 184 | 148 | 109 | 1249 | – | 2 |
| Sweden | 2001 | 64 | 85 | 71 | 100 | 134 | 187 | 55 | 1252 | – | 1 |
| Sweden | 2001 | 64 | 93 | 100 | 3 | 175 | 143 | 16 | 1254 | – | 1 |
| Sweden | 2000/2001 | 99 | 136 | 92 | 128 | 173 | 150 | 113 | 1260 | – | 9 |
| Sweden | 2000/2001 | 99 | 127 | 91 | 125 | 170 | 148 | 109 | 1266 | – | 6 |
| Sweden | 2000 | 110 | 120 | 101 | 145 | 196 | 141 | 84 | 1271 | 1325 | 1 |
| Sweden | 2000/2001 | 96 | 135 | 99 | 136 | 183 | 140 | 113 | 1277 | – | 4 |
| Sweden | 2001 | 95 | 2 | 94 | 127 | 172 | 144 | 114 | 1286 | – | 3 |
| Sweden | 2000/2001 | 101 | 122 | 99 | 125 | 182 | 151 | 88 | 1290 | – | 3 |
| Sweden | 2001 | 99 | 127 | 91 | 125 | 170 | 148 | 115 | 1297 | – | 1 |
| Sweden | 2000/2001 | 99 | 118 | 91 | 125 | 170 | 142 | 111 | 1303 | – | 10 |
| Sweden | 2000/2001 | 100 | 142 | 93 | 135 | 190 | 145 | 81 | 1304 | 1304 | 5 |
| Sweden | 2001 | 95 | 2 | 94 | 127 | 181 | 144 | 110 | 1316 | – | 6 |
| Sweden | 2001 | 99 | 128 | 91 | 125 | 170 | 146 | 111 | 1324 | – | 5 |
| Sweden | 2000/2001 | 94 | 120 | 101 | 145 | 196 | 141 | 84 | 1325 | 1325 | 2 |
| Sweden | 2001 | 94 | 120 | 101 | 125 | 196 | 141 | 84 | 1331 | 1325 | 1 |
| Sweden | 2001 | 99 | 129 | 92 | 128 | 173 | 150 | 113 | 1337 | – | 1 |
| Sweden | 2000/2001 | 98 | 122 | 98 | 125 | 180 | 150 | 113 | 1342 | – | 12 |
| Sweden | 2001 | 94 | 117 | 90 | 125 | 174 | 147 | 100 | 1351 | – | 3 |
| Sweden | 2001 | 10 | 119 | 50 | 126 | 171 | 194 | 112 | 1367 | – | 5 |
| Australia | 2006 | 99 | 128 | 91 | 125 | 170 | 146 | 111 | 1324 | – | 1 |
| Australia | 2006 | 98 | 122 | 98 | 125 | 180 | 150 | 113 | 1342 | – | 1 |
| Australia | 2006 | 98 | 122 | 98 | 125 | 74 | 150 | 113 | 3067 | – | 1 |
| Austra lia | 2006 | 35 | 2 | 9 | 51 | 8 | 46 | 21 | 3068 | 682 | 1 |
| Azores | 2007 | 99 | 127 | 91 | 125 | 184 | 148 | 109 | 3576 | – | 2 |
Allele nucleotide sequence number in the MLST database (www.pubmlst.org/campylobacter/).
Sequence type numbers that are unique making up the allelic profile of the seven loci.
Clonal complex.
Frequency of each isolate with the same ST.
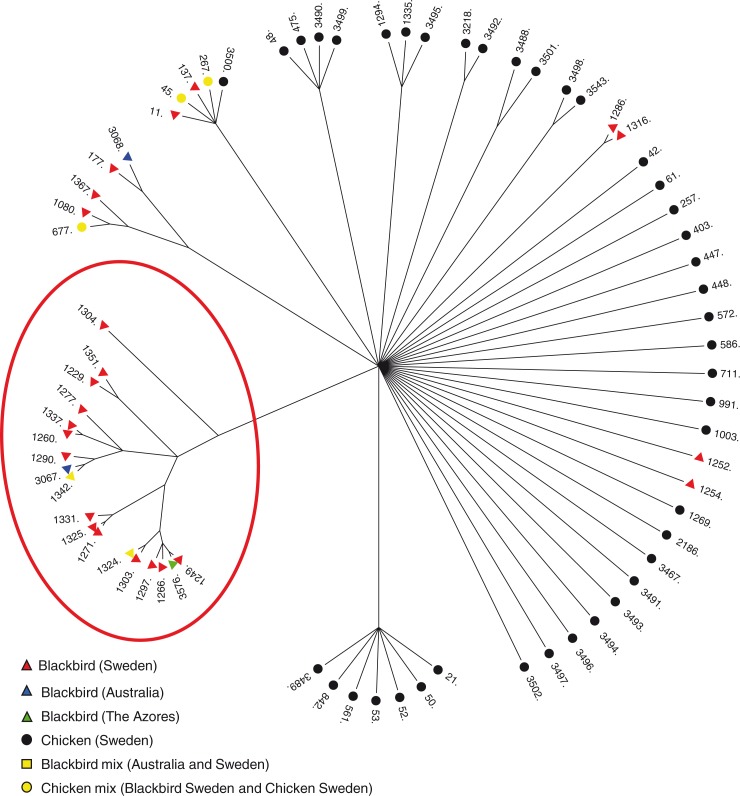
The genetic relationship between the STs isolated from Blackbirds and published STs from chicken samples (12) was analyzed using ClonalFrame software (Fig. 2) (9, 11, 13). The model determines and takes into account the fact that bacterial microevolution distinguishes point mutations from imported chromosomal recombination events, which allows a more accurate calculation of clonal relationships in C. jejuni. The tree was constructed using 50,000 burn-in cycles and 100,000 further iterations with a 75% consensus tree that is representative of three independent ClonalFrame analyses (13).
Fig. 2.
The genetic relationship between the C. jejuni STs isolated from blackbirds and chickens is visualized with a ClonalFrame analysis which demonstrates that the majority of Blackbirds cluster together (within the red circle). ST-1324 and ST-1342 were found in Blackbirds from both Australia and Sweden and are indicated as a Blackbird mixture. ST-677, ST45, and ST267 were found in Blackbirds from both Sweden and Chicken isolates and are indicated as a chicken mix. The chicken data set represents the human- and food-related C. jejuni.
Results and discussion
Blackbirds commonly carry C. jejuni and the establishment of separate Blackbird populations, such as those in Australia and The Azores, enables us to study the population dynamics of C. jejuni and the impact of migration, different food sources, and the geographic associations, as part of a huge natural experiment. To investigate differences in C. jejuni between different Blackbird populations MLST data from migratory Blackbirds in Sweden (10) as well as from isolated, non-migratory populations in Australia and The Azores were incorporated in the analysis (Table 1). Examinations of all genotypes found in C. jejuni MLST databases show that farm-/human-related genotypes are not widely genetically separated and often even share genotypes. Therefore, in this study, chicken genotypes (10, 12) were included in the analysis to represent the cluster of farm-/human-related genotypes. ClonalFrame (13 was used to estimate genealogy, and showed that the majority of Blackbird isolates formed separate clusters from chicken isolates (Fig. 2). Frequently identified STs found within the Blackbird cluster were ST-1229, -1260, -1303, and -1342, which accounted for almost 40% of the data set (Table 1). Some Blackbird C. jejuni genotypes (ST-11, 267, 137, and 1080) were identified (7%), which cluster with chicken genotypes, suggesting a transfer of bacteria in proximity to human/farm surroundings (10). Remarkably, two of the four genotypes from Australian samples (ST-1324 and -1342) were also detected in Swedish samples, and the remaining two (ST-3067 and -3068), although not identical, were genetically very similar (10). Furthermore, the C. jejuni isolates from The Azores (ST-3576) were identical to each other and were closely related to ST-1249 identified in Sweden (Table 1, Fig. 2).
Assessments of ClonalFrame analysis showed a high degree of similarity between the STs from Blackbird populations of different isolated localities, indicating a low rate of differentiation among the geographically separated C. jejuni, and that C. jejuni populations of Blackbirds from Sweden, Australia, and The Azores are either identical or genetically very similar. The genotypes detected are frequently found in Blackbirds, and these results demonstrate that geography, migration, different food sources, and in particular large periods of time have minor effects on the structure of C. jejuni populations in Blackbirds.
It has previously been shown that certain C. jejuni clones can persist over long periods of time in different environments. For instance, a strain from a human outbreak in England in 1981 was found 20 years later in a chicken outbreak, this in spite of the frequent recombination events that can occur in this bacterium (9). However, the genotype stability in Blackbirds in Australia, which were separated 150 years ago and have been maintained over 150 generations, indicates much greater temporal stability. The isolates from The Azores further indicate that genetic stability is independent of both time and geography. It is not possible to date when and how the bacterium colonized the birds on The Azores, but it should be noted that the avifauna is isolated, and that many species that occur there have evolved into clearly distinctive taxa or even new species such as Monteiro's Storm-Petrel (Oceanodroma monteiroi) and The Azores Bullfinch (Pyrrhula murina).
Our study demonstrates that even though C. jejuni is known to be a highly diverse and a rapidly evolving bacterium, a remarkably low level of genetic differentiation and high species fidelity was found in C. jejuni in Blackbirds even after thousands of years of isolation. This indicates that there exists an extraordinarily strong evolutionary adaptation and association of specific C. jejuni genotypes with particular bird species. To investigate further, the next step would be sampling longitudinally in Blackbird populations, their food sources, in other potential hosts, and also in the surrounding environment in a bigger scale. Additionally, it would be highly interesting to make a deeper genetic characterization of the isolates to assess whether the striking host-specific signal observed at the MLST loci is maintained at the genomic level.
Conflict of interest and funding
The authors declare no conflicts of interests.
References
- 1.Campell AJ, Kendall H, editors. Birds of a feather. Vol. 5. Melbourne: Australasian Ornithologists’ Union; 1906. Union, AOURAO. The emu; pp. 1905–1906. [Google Scholar]
- 2.Higgins PJ, Peter JM, Cowling SJ, editors. Boatbill to starlings. Vol. 7. Melbourne: Oxford University Press; 2006. Handbook of Australian, New Zealand & Antarctic birds. [Google Scholar]
- 3.Brooks JD. Handbook of the birds of Europe, the Middle East and North Africa. In: Cramp S, editor. The birds of the Western Palearctic. Vol. 5. Oxford: Oxford University Press; 1988. [Google Scholar]
- 4.Luechtefeld NA, Blaser MJ, Reller LB, Wang WL. Isolation of Campylobacter fetus subsp. jejuni from migratory waterfowl. J Clin Microbiol. 1980;12:406–8. doi: 10.1128/jcm.12.3.406-408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skirrow MB, Blaser MJ. Clinical aspects of Campylobacter infection. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ed. Washington, DC: ASM Press; 2000. pp. 69–82. [Google Scholar]
- 6.Colles FM, McCarthy ND, Howe JC, Devereux CL, Gosler AG, Maiden MC. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling) Environ Microbiol. 2009;11:258–67. doi: 10.1111/j.1462-2920.2008.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldenström J, Axelsson-Olsson D, Olsen B, Hasselquist D, Griekspoor P, Jansson L, et al. Campylobacter jejuni colonization in wild birds: results from an infection experiment. PLoS One. 2010;5:e9082. doi: 10.1371/journal.pone.0009082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, et al. Multilocus sequence typing system for Campylobacter jejuni . J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colles FM, Dingle KE, Cody AJ, Maiden MC. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl Environ Microbiol. 2008;74:3583–90. doi: 10.1128/AEM.02491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, et al. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol. 2013;22:1463–72. doi: 10.1111/mec.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingle KE, Colles FM, Ure R, Wagenaar JA, Duim B, Bolton FJ, et al. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg Infect Dis. 2002;8:949–55. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griekspoor P, Engvall EO, Olsen B, Waldenström J. Multilocus sequence typing of Campylobacter jejuni from broilers. Vet Microbiol. 2010;140:180–5. doi: 10.1016/j.vetmic.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–66. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]