Abstract
Testing of tumor tissue remains the recommended method for detecting the presence of somatic mutations in human malignancies. V600E is the most frequent somatic point mutation in metastatic melanoma, providing a unique molecular marker for this malignancy. In addition, tumors carrying this mutation are primary candidates for BRAF-targeted therapy. Although metastatic melanoma patients usually have sufficient tumor tissue available for genetic analyses, the detection of V600E in blood can have prognostic and predictive value. In addition, patients are rarely re-biopsied and genetic testing in blood can be useful for monitoring response to therapy. Cell-free DNA (cfDNA) and cell-free RNA (cfRNA), RNA associated to platelets and circulating tumor cells (CTCs) are some of the materials that can be derived from the blood of cancer patients. cfDNA can be easily purified from serum and plasma and contains DNA fragments of tumor origin. For this reason, it is the most widely used material for the detection of somatic mutations in blood. Several methodologies have been used to determine V600E status in the cfDNA of metastatic melanoma and some studies have demonstrated that the identification and follow-up of V600E in cfDNA can have prognostic and predictive value.
Keywords: BRAF, plasma, serum, circulating-free RNA (cfDNA), melanoma
“Liquid biopsies” and cell-free DNA (cfDNA)
Tumour biopsy tissue, either from the primary tumour, cytological samples or metastases, is the standard material to determine somatic mutations and other genetic alterations before the start of treatment. The exact definition of a biopsy is “the removal for diagnostic study of a piece of tissue from a living body”. However, the term “liquid biopsy”, originally introduced for the analysis of circulating tumor cells (CTCs) is currently used for analysis of all materials derived from blood.
Tumor biopsies sometimes carry risks for patients, are painful, costly and time consuming; while liquid biopsies do not have any of these disadvantages. Also, a single tissue biopsy may not represent the complexities of tumour heterogeneity, both within a tumour and between a primary tumour and metastases. In contrast, liquid biopsies can capture the entire heterogeneity of the disease and offer additional, valuable prognostic and predictive information. Finally, tumour genotypes are unstable and actually change under selection pressure; for instance, genetic alterations leading to drug resistance (secondary mutations, gene amplifications and others) often appear in tumors treated with targeted drugs. Tumor rebiopsies are extremely infrequent and liquid biopsies offer the opportunity to take serial samples in order to monitor the course of the disease and, very particularly, tumor genomic changes in real time (1-3). Instead of waiting for scans or other imaging results, liquid biopsies can identify at an earlier stage if a treatment is not working, sparing the patient unnecessary toxicities, and identify new targetable alterations that might emerge during the course of the disease, helping to provide patients with the right treatment without delay.
In short, liquid biopsies can provide a non-invasive, ongoing picture of a patient’s malignancy, providing valuable insight into how best to fight it. In addition to offering clues about stage and spread, they can be used to monitor the effects of treatment, giving an early warning about recurrence and also about the reasons for treatment resistance (4).
CTCs have been the most studied material in liquid biopsies. Although they can provide information at both the genetic and cellular level, they are relatively scarce (particularly in some malignancies) and require sophisticated technologies for collection and analysis. For this reason, cfDNA is emerging as an effective alternative to CTCs in liquid biopsy, being much easier to collect, purify and analyze. In cancer patients, it contains DNA fragments of tumor origin (often referred as “circulating tumor DNA”, ctDNA) and can be readily used for the detection of somatic mutations and other tumor molecular alterations (5-8).
BRAF V600E mutation in melanoma
The BRAF gene is located in the 7q34 chromosomal region and encodes a serine/threonine kinase responsible for activating the MEK-ERK cell signaling pathway downstream of RAS (9). Somatic BRAF mutations occur in tumors of the colon (10), thyroid (11), lung (12) and, particularly, in melanomas (13). Mutations in the BRAF gene have been reported in 30% of primary and 50-60% of metastatic melanoma patients (14), a percentage that raises to >80% in melanomas located in intermittently exposed body sites, such as the trunk, arms and legs (15). The substitution of valine by glutamic acid in codon 600 (V600E), caused by a transversion T→A at nucleotide 1799 (T1799A), accounts for over 90% of BRAF mutations in melanoma and represents the most prominent molecular marker of the disease. Other mutations, such as V600K, V600M, V600R, V600D and V600G, are less common. The V600E mutated protein shows constitutive kinase activity and triggers downstream signaling in the Ras-Raf-MEK pathway, which results in increased cell proliferation, resistance to apoptosis and tumor progression.
Prognosis has been traditionally very poor for patients with cutaneous metastatic melanoma with response rates to conventional chemotherapy around 10% (16,17). However, the treatment perspectives of BRAF melanoma patients are changing. Based on the results of phase III trials, targeted agents that inhibit key effector kinases of the MAPK signaling pathway (including BRAF and MEK inhibitors) have been recently approved by the US Food and Drug Administration in the metastatic setting (18-20). BRAF inhibitors, such as vemurafenib or dabrafenib, achieve high response rates and significantly improve the survival of melanoma patients carrying BRAF activating mutations. However, response duration is usually limited to less than one year and 20% of patients are intrinsically resistant, highlighting the need for further advances in therapy. Testing BRAF mutational status is required in order to select patients for treatment with these inhibitors.
Detection of V600E mutation in cell-free DNA (cfDNA) from melanoma patients
The BRAF V600E mutation is the most relevant alteration in metastatic melanoma, and most efforts in the field of “liquid biopsy” for this malignancy have been focused to develop methodologies to detect V600E in cfDNA.
The percentage of cfDNA that corresponds to ctDNA depends on several factors, such as tumor burden, but can also reflect more complex mechanisms of the tumor biology, but it usually represents less than 1% (21). For this reason, the detection of somatic mutations (such as V600E) in cfDNA requires highly sensitive techniques. Standard approaches in tumor tissue samples, such as Sanger sequencing or pyrosequencing, are not sufficiently sensitive. In consequence, other methodologies have been tested during the last decade to assess BRAF in cfDNA, such as ARMS, BEAMing or RT-PCR, that can detect one copy of V600E allele in a background of 50 to 10,000 copies of wt allele (analytical sensitivity 2-0.01%) (22-26) (Table 1). In our laboratory we have also developed a rapid, sensitive methodology for the detection and quantification of V600E mutation in cfDNA isolated from plasma and serum based on a quantitative 5’-nuclease PCR (Taqman®) in presence of a peptide-nucleic acid (PNA) designed to inhibit the amplification of the wt allele (27). Our technique compares favorably to other methodologies, being able to determine BRAF mutations at a level as low as 1 copy of the mutant allele against a background of 20,000 copies of wt alleles (0.005% analytical sensitivity). In addition, it is a quantitative technique, allowing the calculation of (I) the absolute amount of V600E in cfDNA (expressed as pg of V600E genomes/µL) and (II) the percentage of V600E mutated alleles in cfDNA (expressed as % of V600E allele).
Table 1. Comparison of methods used for the detection of the BRAFV600E mutation in the cfDNA of metastatic melanoma patients.
| Author | Nº patients [Adv stage] | BRAF | Stage | Treatment | Sample | Method | Limit of detection | Diagnostic sensitivity |
|---|---|---|---|---|---|---|---|---|
| Daniotti (22) | 41 [13] | Mut, wt | I, II, III, IV | – | Serum/plasma | Allele specific PCR ASPCR-1 | 0.25% | 38% |
| Shinozaki (23) | 48 [29] | Mut/wt | I, II, III, IV | Biochemo | Serum | Real-time quantitative PCR + PNA clamp and FRET LNA probe | 0.10% | 39% |
| Board (24) | 45 | Mut | IV | MEK1/2 inh | Serum | ARMS | 0.01% | 56% |
| Aung (25) | 221 | Mut/wt | IV | MEK1/2 inh | Serum/plasma | ARMS | 2.00% | 44-52% |
| Ascierto (26) | 72 | Mut | IV | BRAF inh | – | BEAMing | – | 79% |
| Gonzalez-Cao (27) | 22 | Mut | IV | BRAF inh | Serum/plasma | Taqman assay + PNA clamp | 0.01% | 58% |
Key issues when applying a test to determine mutations in “liquid biopsies” are its diagnostic sensitivity and specificity. Sensitivity is the percentage of actual positives which are correctly identified as such; in our case, the percentage of patients mutated in tissue where the mutation is also detected in blood. The sensitivity of a test, also called the true positive rate, is complementary to the false negative rate. Specificity is defined as the proportion of negatives which are correctly identified as such; in our case, the percentage of wild-type patients in tissue where the mutation is not detected in blood. The specificity of a test, also known as the true negative rate, is complementary to the false positive rate. In order to be applied in the clinical setting, the specificity of a test to determine mutations in blood must be close to 100%, in order words, the test must not offer false positive results.
The diagnostic sensitivity of the methods described in the literature to determine the V600E mutation in the cfDNA of melanoma patients range from 38% to 79% (22-27). In the case of ARMS, 44-56% has been reported while the sensitivity of RT-PCR derived techniques is 38-39%; specificity ranging from 85% to 94% in both cases. The assay we have developed (Q-RT-PCR in presence of PNA) compares very favorably with these two other techniques, having 70% sensitivity and 100% specificity (Table 1). It must be noted that, in metastatic melanoma, there is usually enough tumor tissue available for genetic analyses. Therefore, the objective of V600E testing in the cfDNA of melanoma patients is not to be a surrogate of solid biopsy but rather to offer clinically relevant information. In consequence, a very high sensitivity is not as relevant as in other malignancies, such as advanced non-small lung cancer, where tumor samples appropriate for genetic testing are often more difficult to obtain.
There is some controversy about the optimal source of cfDNA for mutation analysis, with a majority of investigators selecting only plasma but others using serum or both. The sensitivity of V600E detection in cfDNA purified from plasma using our test was higher than in cfDNA from serum, similarly to what has been described in other cfDNA-based mutation assays. We found that purification from serum yielded higher amounts of total cfDNA, as estimated by the Ct of the BRAF wt allele. In contrast, the absolute and relative abundances of V600E in positive bloods were significantly higher in plasma. Mean abundance in serum was 1.13 pg/µL (range, 0-6.01 pg/µL) vs. 3.15 pg/µL in plasma (range, 0-26.93 pg/µL) and relative abundance (allelic fraction) in serum was 1.15% (range, 0-9.18%) vs. 7.94% (range, 0-55.52%) in plasma. The V600E allelic fractions in serum or plasma cfDNA and the V600E allelic fraction in tumor tissue did not correlate.
The differences in cfDNA concentration and relative abundance of V600E, which have also been reported previously (25), probably explain the different sensitivities of mutation detection between serum and plasma. We developed our assay for both fluids, which can easily be obtained at the same time, allowing us to detect the mutation also in those few patients where it is only present in cfDNA derived from serum.
Prognostic value of V600E mutation in cell-free DNA (cfDNA)
Several studies have demonstrated a prognostic value of mutation analysis in cfDNA in different types of malignancies (5,28). In the case of metastatic melanoma, a first study using a RT-PCR derived technique in patients treated with chemotherapy with unknown BRAF status in tumor found no significant association between the V600E mutation in cfDNA and response to therapy. However, the patients where the mutation persisted post-treatment had a significantly worse overall survival (OS) (23). Another study using ARMS reported that patient positive for the V600E mutation in both pretreatment serum and tumor did not have a shorter PFS after chemotherapy treatment than those positive only in tumor tissue (24). Post-treatment samples were not analyzed in these cases. Finally, a recent report using BEAMing technology found a significant correlation between the baseline tumor burden and the levels of V600E mutation in cfDNA and significant associations of baseline cfDNA V600E allelic fraction with overall response rate and PFS, although the hazard ratio was very modest (HR =1.09) (26).
In the case of the PNA Q-RT-PCR test we have developed, we investigated its possible clinical application by analyzing blood samples at presentation in a cohort of 22 stage IV melanoma patients harboring the V600E mutation in tumor tissue. All patients were treated with BRAF inhibitors (vemurafenib or dabrafenib). We observed that positivity for V600E in pretreatment cfDNA identified those patients with a very poor prognosis in spite of BRAF inhibitor treatment. Patients with the V600E mutation detected in cfDNA had a median PFS of only 3.6 months, compared to 13.4 months in negative patients and a median OS of 7 vs. 21.8 months, respectively. We are currently testing our methodology and validating its clinical utility in two prospective studies conducted by the Spanish Melanoma Group that are enrolling a significantly higher number of BRAF mutated patients (29).
In our study, clinical stage classification showed a significant association with the cfDNA results. The frequency of detection of the V600E mutation was higher in stage M1b or M1c than inM1a patients (P=0.018). The quantification of the V600E mutation in the cfDNA of positive patients also was higher in patients with a high number of metastatic sites (Figure 1) and showed a trend to increase with more advanced tumor stage (Figure 2). A previous study has also shown that the levels of V600E mutation in pretreatment cfDNA correlate with tumor burden (26). It is reasonable to think that a more aggressive genotype correlates with higher tumor burden at time of diagnosis, higher amount of tumor DNA released to the blood and a shorter duration of clinical response.
Figure 1.
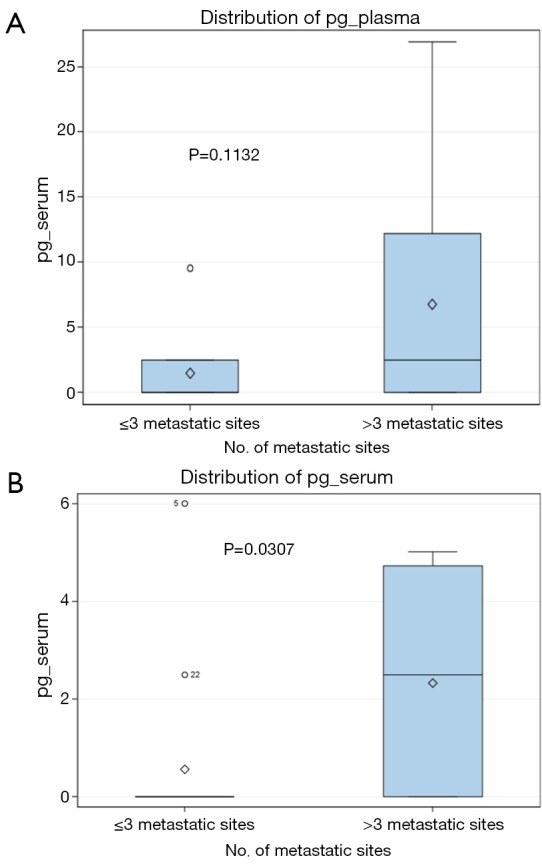
Quantification of the BRAFV600E mutation in the plasma (A) and serum (B) of advanced stage melanoma patients, expressed in pg of mutated genome/µL of fluid. Patients are classified according to the number of metastatic sites.
Figure 2.
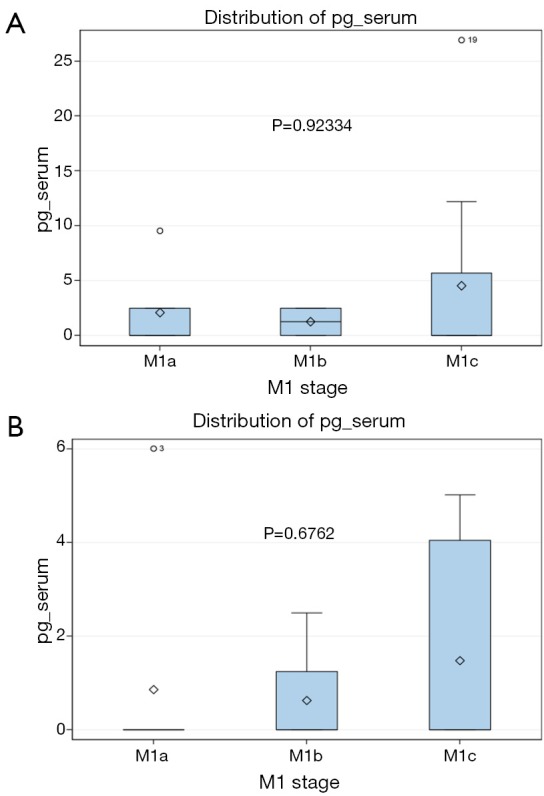
Quantification of the BRAFV600E mutation in the plasma (A) and serum (B) of advanced stage melanoma patients, expressed in pg of mutated genome/µL of fluid. Patients are classified according to clinical stage.
Serial monitoring of V600E status in cell-free DNA (cfDNA)
In the first published analysis of cfDNA performed along BRAF inhibitor treatment (30) in melanoma patients, a good correlation was found between clinical evolution and the amount of mutation in cfDNA along treatment. Also, it was found that in some case there was an increased in BRAF cfDNA amount 42 to 112 days before the clinical progression was detected (5).
In our case, serial blood samples from three melanoma patients positive for the V600E mutation in tissue were collected. Although this number is very limited, our results confirm that testing for BRAF in cfDNA can provide early information about outcome. In particular, we have observed that early negativization after treatment with BRAF inhibitors might predict a good clinical response. The short half-life of circulating mutant DNA is known to be approximately 2 h, allowing evaluation of rapid tumor changes (15). Also, we have observed the possibility of a false increased in BRAF cfDNA in the first hours of treatment. In the follow-up of one patient responding to BRAF inhibitors, we observed an increase in the absolute quantity of V600E mutation at 48 h after starting treatment, while the ratio of the mutant vs. wt allele did not change. We can hypothesize that this was due to an early release of DNA from melanoma cells undergoing apoptosis in response to treatment. Subsequently, the V600E allele in cfDNA decreased as early as 7 days post-treatment and was undetectable after 30 days. In another long responder patient, the V600E mutation was detected in plasma, disappeared after 30 days and remained undetectable until disease progression, when it reappeared in serum. Finally, a third patient had a markedly increased 60 days post-treatment in BRAFV600E ctDNA, he experienced a rapid progression and exitus. These observations suggest that close, periodic monitoring of BRAFV600E mutation in cfDNA could help to select the patients who should continue on BRAF inhibitors and to avoid inactive treatment for resistant patients.
Conclusions
Several methodologies have been tested for mutation analysis of V600E in serum/plasma cfDNA. Some of them have excellent specificity, good sensitivity and are likely to be implemented in the clinical setting for the management of melanoma patients. Although the concordance between “liquid biopsy” and “solid biopsy” (tumor tissue) is not perfect, mutation assessment in cfDNA has prognostic value and allows to monitor the tumor in real time offering valuable information about the course of the disease.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.De Mattos-Arruda L, Cortes J, Santarpia L, et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol 2013;10:377-89. [DOI] [PubMed] [Google Scholar]
- 2.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [DOI] [PubMed] [Google Scholar]
- 3.Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med 2013;5:207ps14. [DOI] [PubMed]
- 4.Karachaliou N, Mayo-de-Las-Casas C, Molina-Vila MA, et al. Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med 2015;3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [DOI] [PubMed] [Google Scholar]
- 6.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [DOI] [PubMed] [Google Scholar]
- 7.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [DOI] [PubMed]
- 8.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [DOI] [PubMed] [Google Scholar]
- 9.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855-67. [DOI] [PubMed] [Google Scholar]
- 10.French AJ, Sargent DJ, Burgart LJ, et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res 2008;14:3408-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojciechowska K, Lewinski A. BRAF mutations in papillary thyroid carcinoma. Endocr Regul 2006;40:129-38. [PubMed] [Google Scholar]
- 12.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [DOI] [PubMed] [Google Scholar]
- 13.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [DOI] [PubMed] [Google Scholar]
- 14.Shinozaki M, Fujimoto A, Morton DL, et al. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res 2004;10:1753-7. [DOI] [PubMed] [Google Scholar]
- 15.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135-47. [DOI] [PubMed] [Google Scholar]
- 16.Chapman PB, Einhorn LH, Meyers ML, et al. Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J Clin Oncol 1999;17:2745-51. [DOI] [PubMed] [Google Scholar]
- 17.Patel PM, Suciu S, Mortier L, et al. Extended schedule, escalated dose temozolomide versus dacarbazine in stage IV melanoma: final results of a randomised phase III study (EORTC 18032). Eur J Cancer 2011;47:1476-83. [DOI] [PubMed] [Google Scholar]
- 18.Flaherty KT, Robert C, Hersey P, et al. Improved Survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107-14. [DOI] [PubMed] [Google Scholar]
- 19.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012;380:358-65. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer 2013;49:1297-304. [DOI] [PubMed] [Google Scholar]
- 22.Daniotti M, Vallacchi V, Rivoltini L, et al. Detection of mutated BRAFV600E variant in circulating DNA of stage III-IV melanoma patients. Int J Cancer 2007;120:2439-44. [DOI] [PubMed] [Google Scholar]
- 23.Shinozaki M, O'Day SJ, Kitago M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res 2007;13:2068-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Board RE, Ellison G, Orr MC, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer 2009;101:1724-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aung KL, Donald E, Ellison G, et al. Analytical validation of BRAF mutation testing from circulating free DNA using the amplification refractory mutation testing system. J Mol Diagn 2014;16:343-9. [DOI] [PubMed] [Google Scholar]
- 26.Ascierto PA, Minor D, Ribas A, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 2013;31:3205-11. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Cao M, Mayo-de-las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res 2015;25:486-95. [DOI] [PubMed] [Google Scholar]
- 28.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Cao M, Soriano V, Rodriguez D, et al. BRAF mutation analysis in tumoral cell free DNA (cfDNA) of melanoma patients: preliminary results from the Spanish Melanoma Group prospective study GEM1304. Annals Oncol 2014;25:iv384. [Google Scholar]
- 30.Panka DJ, Buchbinder E, Giobbie-Hurder A, et al. Clinical utility of a blood-based BRAF(V600E) mutation assay in melanoma. Mol Cancer Ther 2014;13:3210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


