Abstract
Krüppel-like factor (KLF)-10 is an important transcriptional regulator of TGF-β1 signaling in both CD8+ and CD4+ T lymphocytes. In the present study, we demonstrate a novel role for KLF10 in the regulation of TGFβRII expression with functional relevance in macrophage differentiation and activation. We first show that transfer of KLF10−/− bone marrow-derived macrophages into wild-type (WT) mice leads to exacerbation of experimental colitis. At the cell biological level, using two phenotypic strategies, we show that KLF10-deficient mice have an altered colonic macrophage phenotype with higher frequency of proinflammatory LyC6+MHCII+ cells and a reciprocal decrease of the anti-inflammatory LyC6−MHCII+ subset. Additionally, the anti-inflammatory CD11b+CX3CR1hi subset of colonic macrophages is significantly decreased in KLF10−/− compared with WT mice under inflammatory conditions. Molecularly, CD11b+ colonic macrophages from KLF10−/− mice exhibit a proinflammatory cytokine profile with increased production of TNF-α and lower production of IL-10 in response to LPS stimulation. Because KLF10 is a transcription factor, we explored how this protein may regulate macrophage function. Consequently, we analyzed the expression of TGFβRII expression in colonic macrophages and found that, in the absence of KLF10, macrophages express lower levels of TGFβRII and display an attenuated Smad-2 phosphorylation following TGF-β1 stimulation. We further show that KLF10 directly binds to the TGFβRII promoter in macrophages, leading to enhanced gene expression through histone H3 acetylation. Collectively, our data reveal a critical role for KLF10 in the epigenetic regulation of TGFβRII expression in macrophages and the acquisition of a “regulatory” phenotype that contributes to intestinal mucosal homeostasis.
Keywords: colitis, colonic macrophages, epigenetics, histone acetyl transferase, Krüppel-like factor
macrophages are the most abundant mononuclear phagocytes in the healthy intestinal lamina propria (LP) and are important regulators of immune responses during intestinal inflammation (26, 29, 42). In the murine system, intestinal macrophages rely on constant replenishment by Ly6C+ blood monocytes conditioned to acquire a noninflammatory profile and differentiate into chemokine receptor CX3CR1hi “regulatory” macrophages (1, 2, 20, 33, 42). During experimental colitis, however, the conditioning of Ly6C+ blood monocytes is impaired, giving rise to proinflammatory macrophages and migratory dendritic cells that perpetuate intestinal inflammation. IL-10/IL10R (27, 41) and TGF-β/TGFβR signaling in the mononuclear phagocyte system have emerged as important regulators of inflammation “anergy” characteristic of LP macrophages in mice and human (24, 25, 28). However, the molecular cues that imprint this inflammatory anergy downstream of either TGF-β or IL-10 signaling are poorly defined (28).
The significance of TGF-β signaling, in particular, in dendritic cells and macrophages has been recently highlighted by the observation that mice with a dendritic cell-specific deletion of TGF-β receptor II (TβRII) develop a multiorgan autoimmune inflammation, including colitis and gastritis (24). Moreover, CD68TGFβDNRII mice with macrophage-specific expression of a truncated TGF-βRII show enhanced colitis susceptibility and reduced recovery following intestinal injury with dextran sulfate sodium (DSS) (25). These data reveal an important contribution of TGF-β signaling in the suppression of intestinal inflammation, at least in part, through direct effects on macrophage function (25, 28). In fact, these considerations led us in this study to search for cellular and molecular mechanisms that acting downstream of TGF-β signaling may contribute to colitis. In this regard, recent studies have identified a role for TGF-β-inducible early gene 1 or KLF10, which belongs to the family of Sp1/Krüppel-like zinc finger transcription factors (31), in T lymphocytes and innate immune cells (5, 23, 34, 39).
KLF proteins constitute a family of transcription factors that regulate the expression of a large number of genes with established relevance to cell proliferation, apoptosis, differentiation, and transformation (6, 19, 36, 40). Given the established importance of KLF10 in TGF-β signaling in both T and innate immune cells and of TGF-β signaling in the suppression of intestinal inflammation, we hypothesized that KLF10 may contribute to intestinal mucosal homeostasis through regulation of colonic macrophage phenotype and function. Congruent with this hypothesis, we here report that indeed KLF10 plays an important role in the differentiation of intestinal macrophages toward an anti-inflammatory phenotype which contributes to the regulation of colitis in vivo (4). More specifically, we demonstrate that, through its effect on chromatin remodeling, KLF10 regulates TGFβRII expression in murine macrophages via histone H3 modification. In the absence of KLF10, colonic macrophages display a lower frequency of the P3 plus P4 anti-inflammatory subset (defined as Ly6C−MHCII+) and produce less IL-10 in response to LPS stimulation. Consistent with the anti-inflammatory role of KLF10 in macrophage regulation, transfer of KLF10−/− bone marrow-derived macrophages (BMDM) to wild-type (WT) mice leads to worsening experimental colitis following DSS administration.
Combined, these results advance our understanding on the KLF10-dependent molecular mechanisms underlying the function of TGF-β signaling in macrophages, while at the same time shedding light into the innate immune contributions to colitis. Therefore, this new knowledge has both cell biological and biomedical relevance and should be taken into consideration as a potential pathophysiological mechanism in inflammatory processes.
MATERIALS AND METHODS
Mouse strains.
C57BL/6 mice were purchased from the Jackson Laboratory. KLF10−/− mice were kindly provided by Thomas C. Spelsberg (Mayo Clinic, Rochester, MN) (30). The CAR transgenic mouse was obtained through the NIAID Exchange Program of the National Institutes of Health: Balb/cJ[Tg]CARdelta1-[Tg]DO11.10 mouse line 4285 (35). All of the mice used in experiments were 6–8 wk in age and have been maintained under specific pathogen-free conditions. The mice were age matched in experiments comparing WT with KLF10−/−. All animal experiments were performed per the recommendations outlined in the Guide for Care and Use of Laboratory Animals from the National Institutes of Health as required by Mayo Clinic. These guidelines were incorporated into the present study protocol (IACUC no. A13313), which was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Mayo Clinic, Rochester, MN.
Cell isolation.
Lamina propria mononuclear cells (LPMC) from mouse colons were isolated as previously described (20). LPMCs were further separated into CD11b+ cells using anti-CD11b Micro-Beads (Miltenyi Biotec, Auburn, CA). BMDM were obtained as previously described (27).
Flow cytometry.
Macrophage subsets in the LP of WT and KLF10−/− mice were analyzed by flow cytometry using the following surface markers: CD45, CD11b, CD64, CD103, Ly6C, MHCII, and CX3CR1 (eBioscience) (27, 33). Proinflammatory population was defined as Ly6C+MHCII+/− and anti-inflammatory as Ly6C+MHCII+/− (2). The percentage of CX3CR1hi or CX3CR1int macrophage subset between WT and KLF10−/− mice was also analyzed as previously described (20).
Western blot.
Colonic CD11b+ mononuclear cells were incubated with TGFβ1 for different time points. The cells were lysed in RIPA buffer (1× lysis buffer-150 with protease inhibitors) for p-SMAD2 measurement or Laemmli buffer for total SMAD2 measurements. Proteins were separated by 10% SDS/PAGE, transferred to membrane, blocked with 5% milk, and probed overnight with primary antibodies at the following dilutions: p-SMAD2 (1:5,000; Cell Signaling, Danvers, MA), SMAD2 (1:1,000; Cell Signaling), and β-actin (1:1,000). Blots were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; Santa Cruz Biotechnology, Dallas, TX) and developed with chemiluminescence.
Transfection and luciferase assays.
Two million THP-1 cells were transfected by using the Amaxa Cell Line Nucleofector Kit V from Lonza (Köln, Germany) according to the optimized protocol provided with the kit. Two micrograms of plasmid DNA for TβRIIluc promoter, KLF10, or empty vector were added to the cells. The nucleofection was done with a NucleofectorII device. Cells were divided into triplicate and allowed to sit for 24 h. Luciferase assays were done according to the manufacturer's recommendations (Promega, Madison, WI).
ChiP assays.
Chromatin immunoprecipitation (ChiP) assays were performed as previously described (36). Two million murine CAR-expressing BMDM were transfected with KLF10-His adenovirus or empty vector. After 48 h cells were treated with 1% formaldehyde to cross-link histones to DNA. Fixed cells were sonicated to yield chromatin fragments of 200-1,000 bp. Antibody used in the ChiP assays was Omni-probe (D-8) (catalog no. sc7270) from Santa Cruz Biotechnology (Santa Cruz, CA). DNA was recovered by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation with the addition of an inert carrier. Options for critically relevant control samples include total IgG or preenriched chromatin (input). We chose to control with preenriched chromatin because nonspecific IgG frequently does not control adequately for nonspecific cross-reactivity. Furthermore, the chromatin input generates a more accurate estimation of biases introduced through sonication of chromatin and subsequent PCR (36). The TGF-βRII ChiP primers have been previously described (38). The anti-acetyl-histone H3 antibody (no. 06-599, H3K9, H3K14, H3K18, H3K23) was purchased from EMD Millipore, (Billerica, MA) and the anti-KAT2B/PCAF antibody (no. 12188) was purchased from Abcam (Cambridge, MA).
DSS colitis model and BM macrophage transfer.
Induction of DSS colitis was performed as previously described (7). To analyze the effect of bone marrow (BM) macrophage transfer to DSS colitis we transferred WT or KLF10−/− BMDM 1 day prior and 1 day after the administration of DSS and followed the mice for disease activity and histological score (20, 37).
Measurement of cytokines by ELISA.
Culture supernatants of stimulated colonic macrophages and BMDM were analyzed by ELISA.
Statistical methodology.
Statistical analyses were performed with GraphPad Software version 4 (GraphPad Software, La Jolla, CA). Descriptive analyses including means and standard deviations were performed in normally distributed data. A t-test was used to compare means between two groups. Paired t-test was used to compare means between paired samples. A P value of <0.05 was considered as statistically significant.
RESULTS
Role of KLF10-deficient macrophages in worsening colitis in wild-type mice.
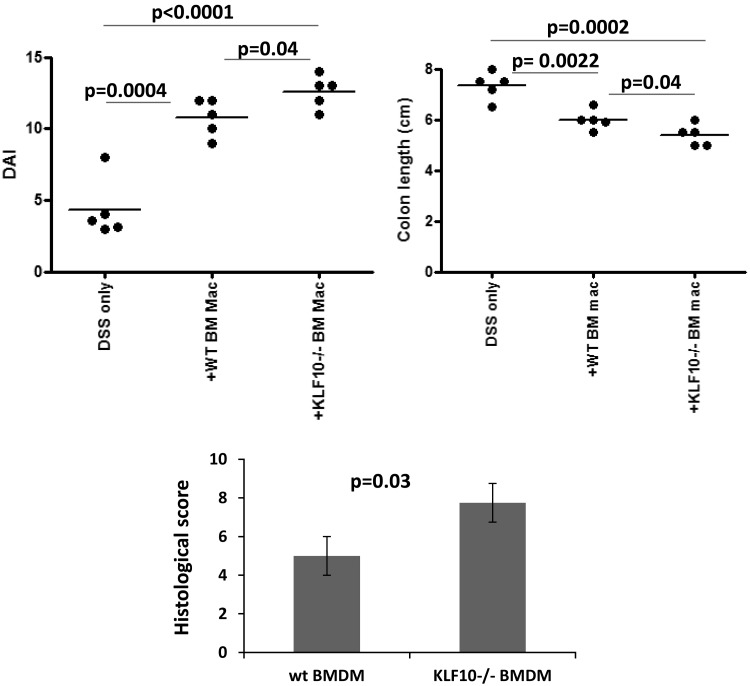
We have previously shown that KLF10−/− mice show enhanced susceptibility to colitis, partially through dysregulation of FOXP3 expression in Tregs (36, 37). However, the precise cellular and molecular pathways, regarding the participation of other cell types in this process, are not fully characterized. We have recently shown that the susceptibility to DSS colitis in KLF10-deficient mice is transferable with BM (37). Given the importance of mucosal macrophages in colitis susceptibility in mice and humans, we reasoned that KLF10 deficiency in innate immune cells may contribute to colitis susceptibility through dysregulation of macrophages. To address the contribution of KLF10 deficiency in macrophages to colitis susceptibility, we transferred BMDM from WT or KLF10−/− mice into WT mice and assessed their susceptibility to colitis. As shown in Fig. 1, DSS-treated WT mice that received KLF10−/− BMDM developed more severe colitis compared with the ones that received WT BMDM, as indicated by the disease activity index (DAI), colon length, and histological score (20). Interestingly, the mice that received either WT or KLF10−/− BMDM developed more severe colitis compared with DSS-treated mice that received no macrophages (Fig. 1). The DAI and colon length in mice that received WT BMDM was 10.8 ± 0.58 and 6 ± 0.17 cm vs. 12.6 ± 0.5 and 5.4 ± 0.18 cm in mice that received KLF10−/− BMDM, respectively (n = 5, P = 0.04 for both). Similarly the histological inflammatory score was 5 ± 0.6 in mice that received WT BMDM vs. 7.75 ± 0.8 in mice that received KLF10−/− BMDM (P = 0.03). The observed worsening of the colitis after transfer of either WT or KLF10−/− BMDM is likely the result of the proinflammatory differentiation of transferred monocytes/macrophages in the context of colitis (2, 26), and this appears to be more pronounced in the KLF10−/− transferred macrophages. These data indicate a novel pathophysiological role for the transferred KLF10−/− macrophages in enhancing colitis severity.
Fig. 1.
Transfer of KLF10−/− bone marrow-derived macrophages (BMDM) worsens dextran sulfate sodium (DSS) colitis. BMDM obtained from wild-type (WT) or KLF10−/− mice were administered intraperitoneally at days −1 and +1 to WT mice who were subjected to 3% DSS treatment at day 0. Disease activity index (DAI, top left), colon length (top right), and histological score (bottom) were compared between the different treatment groups.
Genetic inactivation of KLF10 in the germline leads to an altered colonic macrophage phenotype in vivo.
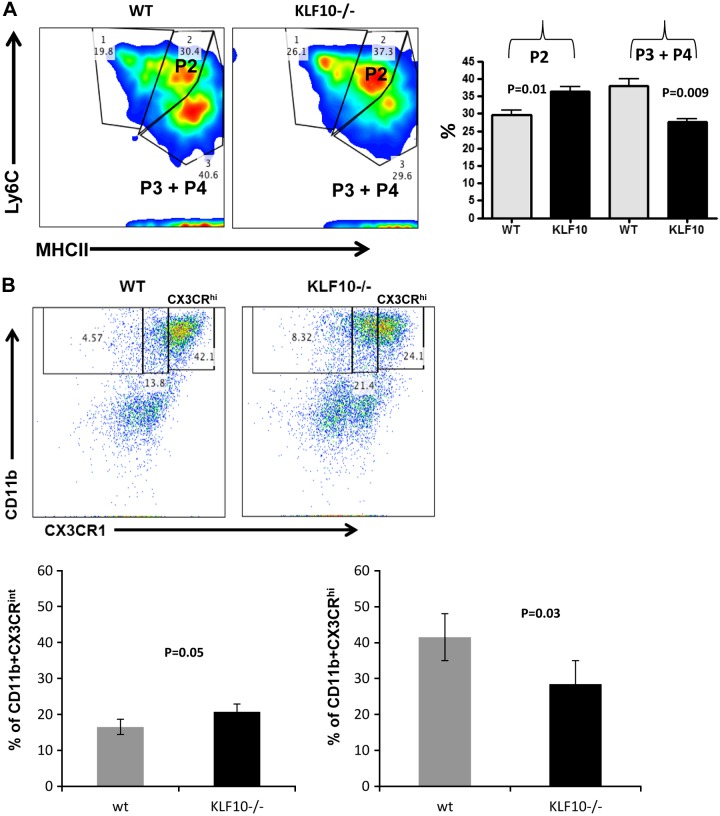
To start elucidating the mechanisms by which KLF10 deficiency in macrophages enhances colitis susceptibility, we analyzed the effect of KLF10 deficiency on colonic macrophage physiology, by first analyzing the phenotype of colonic macrophages isolated from KLF10−/− or WT mice. Given the complexity of the phenotype of the macrophage subsets that have been previously characterized from the small intestine and colon (1, 2, 26, 33, 42), we focused our attention on the phenotypic characterization of colonic macrophages defined as CD45+CD11b+CD64+CD103− cells (27, 42). Gating on these cells, we then analyzed the different macrophage subsets based on the expression of Ly6C and MHCII as proposed by Tamoutounour et al. (33). By using this gating strategy, colonic macrophages can be characterized as proinflammatory P2 stage (Ly6C+MHCII+) or anti-inflammatory P3 plus P4 stage (Ly6C−MHCII+) (27, 33). We found that, in the absence of KLF10, the frequency of proinflammatory P2 stage subset was significantly increased compared with WT colons (Fig. 2A). The percentage of the P2 subset was 29.6 ± 1.2% in WT mice compared with 36.4 ± 1.2% in KLF10−/− mice (n = 4, P = 0.01). Similarly, there was a reciprocal decrease in the frequency of anti-inflammatory P3 plus P4 stage in the colons of KLF10−/− mice (Fig. 2A). The percentage of the P3 plus P4 subset was 38 ± 2% in WT mice compared with 27.7 ± 1% (n = 4, P = 0.009).
Fig. 2.
Characterization of colonic macrophages in adult (6-to 8-wk-old) KLF10−/− mice. A: Lamina propria mononuclear cells (LPMC) were isolated from WT or KLF10−/− mice and stained for CD45, CD11b, CD64, CD103, LyC6, and MHCII and analyzed by flow cytometry. Cells were gated on CD45+ live CD11b+CD64+CD103− subset and analyzed for the expression of Ly6C and MHCII. The different macrophage subsets were then characterized as proinflammatory (Ly6C+MHCII+) P2 or anti-inflammatory (Ly6C−MHCII+) P3 and P4. Average percentages ± SE of the different colonic macrophage subsets P2, P3, and P4 in WT and KLF10−/− colon are shown (right, n = 4). B: representative dot-plot analysis of CX3CR1 expression among CD11b+ murine colonic mononuclear cells between WT and KLF10−/− mice at day 8 following DSS treatment. Bottom: average percentages of CX3CR1int or CX3CR1hi subset in WT vs. KLF10−/− mice (n = 4 mice/group).
Recruited Ly6C+ blood monocytes to the healthy murine colon acquire a noninflammatory gene-expression profile and differentiate into resident chemokine receptor CX3CR1hi anti-inflammatory macrophages (20, 43). Moreover, during DSS colitis there is a colonic recruitment of CX3CR1int cells that are proinflammatory (2, 43). To further define the effect of KLF10 deficiency in the expression of CX3CR1 by resident and newly recruited colonic macrophages, we also analyzed the expression of CX3CR1 among CD11b+ mononuclear cells between KLF10 and WT mice. We observed no significant differences in the proportion of CD11b+CX3CR1hi subset between WT and KLF10−/− mice isolated from the colon (28.6% ± 2.93 vs. 29 ± 2.6, n = 5, P = NS). However, we observed a significant increase of the colonic CX3CR1int subset in KLF10−/− compared with WT mice. The frequency of CD11b+CX3CR1int subset was 16.3 ± 1.8% in WT vs. 21.5 ± 2% in KLF10−/− mice (n = 5, P = 0.03). More importantly after the induction of DSS colitis, the colonic CD11b+CX3CR1hi subset increased significantly more in WT mice compared with KLF10−/− mice. For example, by day 8 of the experiment 41.5 ± 5.6% of WT CD11b+ cells acquired high levels of CX3CR1 vs. 28.4 ± 4.5% of KLF10−/− mice (n = 4, P = 0.03) (Fig. 2B). Although the difference was not as pronounced for the CD11b+CX3CR1int subset, we observed a higher frequency of CD11b+CX3CR1int cells in KLF10−/− compared with WT mice following DSS colitis (20.8 ± 0.45% vs. 16.5 ± 1.4%, n = 3; P = 0.05) (Fig. 2B). Collectively, these results implicate an important role for KLF10 in the differentiation of intestinal macrophages in health and during acute colitis.
KLF10-deficient macrophages display a dysregulation in the profile of proinflammatory cytokines.
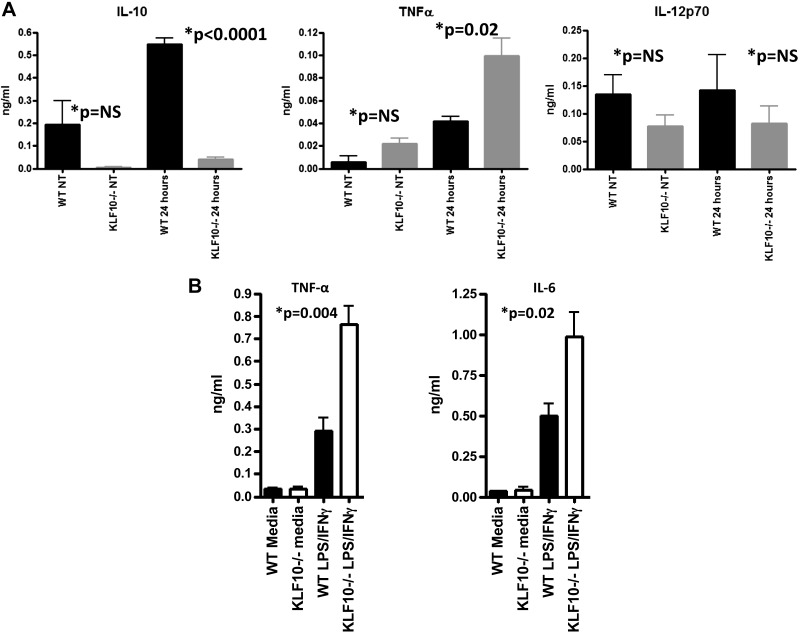
Since the CD11b+CX3CRint macrophage subset is being characterized as proinflammatory with the ability to produce high levels of TNF-α and the CD11b+CX3CRhi subset as anti-inflammatory through the production of IL-10 (8, 41), we next analyzed the cytokine profile of colonic macrophages isolated from the colons of KLF10−/− or WT mice following LPS stimulation in vitro. As shown in Fig. 3A, colonic macrophages from KLF10−/− mice produce significantly higher levels of TNF-α and lower levels of IL-10 compared with macrophages isolated from WT mice following LPS stimulation. The amount of IL-10 released by LPS-stimulated WT colonic macrophages was 0.55 ± 0.03 compared with 0.04 ± 0.009 ng/ml (n = 3; P < 0.0001) of similarly stimulated KLF10−/− colonic macrophages. The amount of TNF-α released by LPS-stimulated WT colonic macrophages was 0.042 ± 0.004 ng/ml compared with 0.1 ± 0.016 ng/ml released by KLF10−/− colonic macrophages (Fig. 3A). We found no significant differences between WT and KLF10−/− colonic macrophages in the release of IL-12 p70 following LPS stimulation (Fig. 3A). Similarly, stimulation of KLF10−/− BMDM with IFN-γ and LPS led to higher release of TNF-α and IL-6 compared with WT BMDM. Stimulated WT BMDM released 0.29 ± 0.05 ng/ml of TNF-α compared with 0.76 ± 0.08 ng/ml of KLF10−/− stimulated cells (n = 3, P = 0.004). Similarly the amount of IL-6 released by WT BMDM was 0.5 ± 0.07 vs. 0.98 ± 0.15 ng/ml of stimulated KLF10−/− cells (n = 3, P = 0.02) (Fig. 3B), indicating that the absence of KLF10 directly affects the proinflammatory activation of macrophages irrespective of their location or state of differentiation.
Fig. 3.
Cytokine dysregulation of KLF10−/− macrophages. A: cytokine release by CD11b+ colonic mononuclear cells isolated from WT or KLF10−/− mice. Cells were incubated in medium with or without LPS (100 ng/ml) for 24 h and supernatants were collected and analyzed for the expression of IL-10, TNF-α, and IL-12p70. The data represent means ± SD of 3 independent experiments from 3 different WT or KLF10−/− mice. NT, not treated; NS, not significant. B: cytokine release by WT or KLF10−/− BMDM. BMDM were stimulated with LPS plus IFN-γ for 24 h and supernatants were analyzed for the production of TNF-α and IL-6. The data represent means ± SD of 3 independent experiments from 3 different WT or KLF10−/− mice.
Genetic inactivation of KLF10 directly impairs TGF-βRII expression and TGF-β signaling in macrophages.
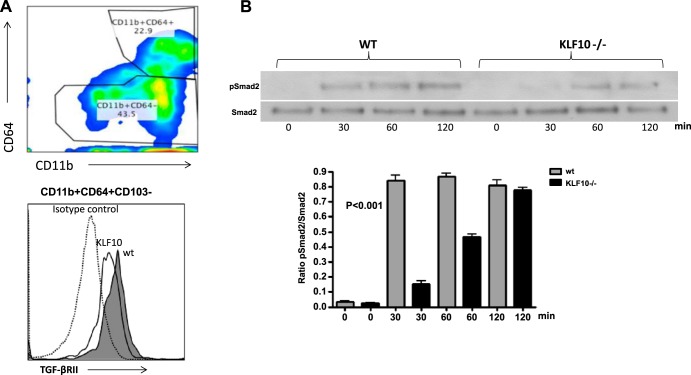
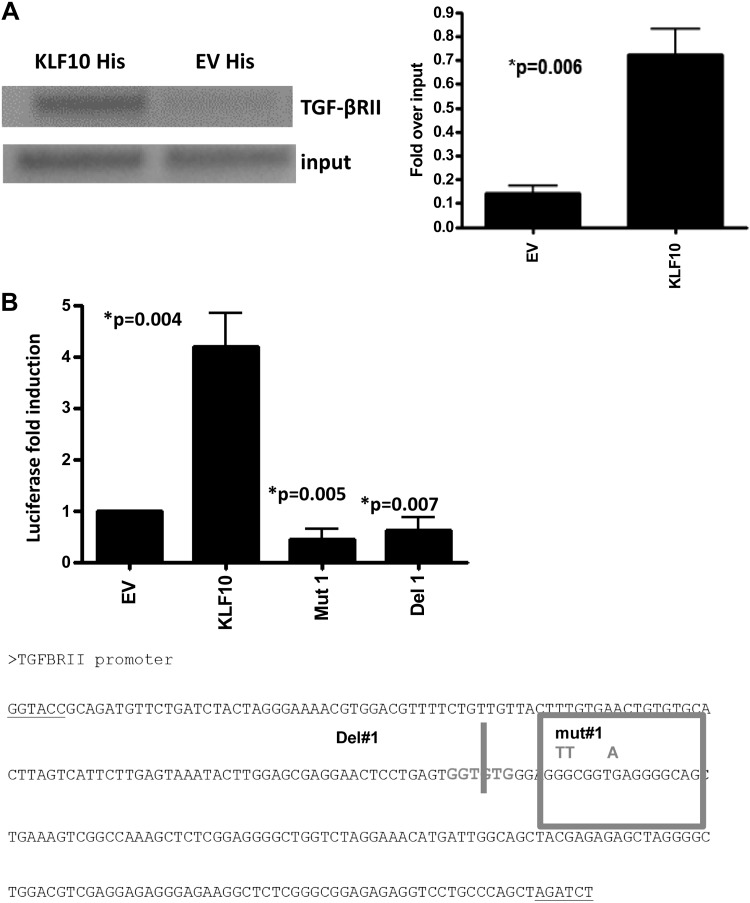
Given the critical role of KLF10 in TGF-β signaling and the importance of TGF-β/TGF-βR pathway in mucosal immune homeostasis (5, 23), we reasoned that KLF10 may regulate TGF-β1 signaling in innate immune cells, including macrophages. To this end we have analyzed colonic macrophages and showed that TGF-βRII expression is lower in KLF10−/− CD11b+ colonic macrophages compared with WT mice (Fig. 4A). Moreover, TGF-β1 signaling in KLF10−/− macrophages led to an early attenuation of Smad-2 phosphorylation compared with WT mice (Fig. 4B), indicating that, in the absence of KLF10, TGF-β signaling in macrophages is impaired. Thus, to further explore the molecular pathways by which KLF10 regulates TGFβRII expression in macrophages, we addressed the notion that KLF10 binds to and transcriptionally activates the TGF-βRII gene promoter in macrophages. To this end we used ChiP assays to confirm binding of the KLF10 transcription factor to the core TGFβRII promoter in macrophages. BMDM from KLF10−/− and WT mice were analyzed for KLF10 binding to the TGF-βRII promoter as described in materials and methods. As shown in Fig. 5A, KLF10 binds to the TGF-βRII core promoter region between −500 to +500 relative to the transcriptional start site.
Fig. 4.
Colonic macrophages in KLF10−/− mice express lower levels of TGFβRII and have altered TGF-β signaling. A: colonic macrophages were isolated and stained for CD45, CD11b, CD64, and CD103. Cells were gated on CD11b+CD64+ cells and analyzed for the expression of TGFβRII. The depicted representative histogram indicates the MFI of TGFβRII expression in WT vs. KLF10−/− colonic macrophages from 3 independent experiments with similar results. B: attenuated early Smad-2 phosphorylation in KLF10−/− macrophages in response to TGF-β1 stimulation. For the analysis of pSmad2, cells were serum starved for 24 h and activated for the indicated time points with TGF-β1 (5 ng/ml) lysed and analyzed for p-Smad2 and total Smad2 by Western blotting. Representative of 4 experiments with similar results is shown. The average ratio of p-Smad2/total Smad2 between WT and KLF10−/− macrophages at the indicated time points from 4 independent experiments is shown at bottom (P < 0.001 by 1-way ANOVA analysis).
Fig. 5.
KLF10 binds to and transactivates the promoter activity of TGFβRII in macrophages. A: chromatin immunoprecipitation (ChiP) assay demonstrating binding of KLF10 to the TGFβRII core promoter locus in BMDM. Semiquantitative PCR analysis of the expression of TGFβRII postimmunoprecipitation for His-tagged KLF10 in BMDM transfected with KLF10-His expression vector demonstrate significant binding of KLF10 to the core promoter. Results presented are controlled to TGF-βRII expression of preimmunoprecipitated sample (input). Left: representative DNA gel for PCR reaction analysis of the expression of TGFβRII in BMDM postimmunoprecipitation for His. Right: data are representative of 3 independent experiments combined as fold over input (P = 0.006). B: THP-1 cells were transfected with empty vector (EV) or 1 μg of KLF10 plus TGF-βRII promoter reporter as described in materials and methods and analyzed by a dual-luciferase assay system. Cotransfection of a TGF-βRII promoter reporter that carries a mutation (Mut1) or a deletion (Del1) of the KLF10 binding site completely abrogates the promoter activity. The results represent the average of 3 transfection experiments performed in duplicate. Bottom: TGF-βRII promoter sequences with the respective deletion or mutation construct used for the transfection experiments.
We next transfected the macrophage cell line THP-1 with KLF10 and TGF-βRII reporter constructs and analyzed luciferase activity. As shown in Fig. 5B, cotransfection of KLF10 with TGFβRII led to a dose-dependent induction of luciferase activity compared with the transfection with empty vector. Transfection with 1 μg of KLF10 and 1 μg of TGFβRII led to a fourfold enhancement of promoter activity relative to the empty vector (n = 4; P = 0.004) (Fig. 5B). Cotransfection of KLF10 with different TGFβRII reporter constructs that carry either a deletion or mutation of the KLF binding sites significantly abrogates luciferase activity (0.35 ± 0.32 for the deletion and 0.057 ± 0.006 for the mutant TGF-βRII promoter, P = 0.005 and P = 0.007, respectively) (Fig. 5B).
Collectively, our data show that KLF10 directly binds to the core TGF-βRII promoter region in BMDM and transcriptionally activates the TGFβRII promoter in macrophages.
KLF10 functions by recruiting the histone acetyl transferase PCAF to the TGF-βRII promoter in macrophages.
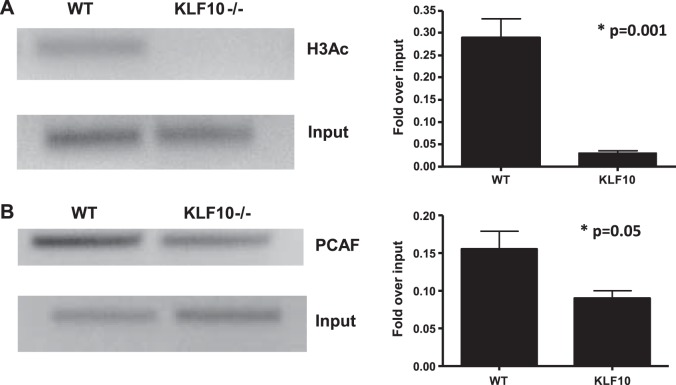
Chromatin remodeling has emerged as one of the most important aspects of gene regulation (13, 15, 16). To further address the chromatin-modifying events by which KLF10 may activate the TGF-βRII promoter in macrophages, we examined histone modifications by ChiP assays using pan-AcH3 as an indicator of open chromatin. We observed that, in the absence of KLF10, there is significantly lower histone H3 acetylation (Fig. 6A). These data suggest that KLF10 is required for the recruitment of an H3-specific acetyltransferase to modify chromatin required for gene activation. Our group have previously discovered a critical interaction between KLF10 and p300/CBP-associated factor (PCAF) in immune cells, in particular the regulation of FOXP3 promoter activity in T cells (37). Therefore, we explored the possibility that KLF10 interacts with PCAF to regulate TGF-βRII promoter activity in macrophages. We indeed observed a significant decrease of PCAF binding to the TGF-βRII core promoter in the absence of KLF10 (Fig. 6B). Taken together, our data provide a novel pathway by which KLF10 specifically recruits PCAF to induce histone H3 acetylation to activate the TGFβRII gene transcription in macrophages.
Fig. 6.
Histone H3 acetylation is a KLF10-dependent event through recruitment of P300/CBP-associated factor (PCAF). A: ChiP assay of BMDM from KLF10−/− or WT mice for acetylated histone H3 (H3A3) was performed as described in materials and methods. We observed lower histone H3 acetylation in the absence of KLF10 in BMDM. Right: data are representative of 4 independent experiments combined as fold over input (P = 0.001). B: PCAF is recruited to the TGFβRII promoter. BMDM were isolated from WT or KLF10−/− mice and ChiP assays were performed by using anti-PCAF antibody. Primers flanking the core TGFβRII promoter were used to amplify the cross-linked DNA by semiquantitative PCR. Right: data are representative of 3 independent experiments combined as fold over input (P = 0.05).
DISCUSSION
In the present article we report several novel observations. We first demonstrate that transfer of KLF10−/− BMDM to WT mice worsens experimental colitis. Secondly, we show that, at the cellular mechanistic level, in the absence of KLF10 intestinal macrophages display an altered phenotype and a proinflammatory cytokine profile in response to LPS stimulation. Moreover, colonic macrophages display lower expression of TGFβRII and attenuated Smad-2 phosphorylation in response to TGF-β stimulation. Third, at the molecular level, exploring the chromatin-modifying events we demonstrate that KLF10 binds to the core TGFβRII promoter and epigenetically regulates TGFβRII gene transcription in macrophages via histone H3 acetylation through recruitment of the histone acetyl transferase PCAF. Thus, in light of these novel observations, we propose that KLF10 is an important transcriptional regulator of intestinal macrophage differentiation and anti-inflammatory function in vivo.
Macrophages are the most abundant leukocytes in the healthy intestinal LP and contribute significantly to gut homeostasis (22, 29) through several mechanisms including phagocytosis, degradation of microorganisms and dead tissue cells, and epithelial cell restitution (22). Importantly, they produce large amounts of IL-10, which blocks Toll-like receptor (TLR)-induced inflammatory responses and enhances the survival and function of local FOXP3+ Tregs (8, 22, 41). Moreover, the response of macrophages to mucosally produced IL-10 is critical in maintaining homeostasis since disruption of IL-10-receptor signaling leads to spontaneous colitis in mice (27, 41).
Small intestinal and colonic macrophages are continuously replenished by chemokine receptor CCR2-dependent influx of Ly6Chi monocytes that differentiate locally into mature, anti-inflammatory macrophages (1, 8, 41). Recent evidence also suggests that TGF-β-signaling contributes to the conditioning of recruited blood macrophages to an anti-inflammatory phenotype (9, 11, 25). For example, mice with a macrophage-specific transgene of TGFβDNRII(CD68TGFβDNRII) have an impaired ability to resolve colitic inflammation and produce less IL-10 but increased levels of IL-33+ macrophage compared with control littermates, suggesting that TGF-β may promote the suppression of intestinal inflammation, at least in part, through direct effects on macrophage function (25). A recent report also demonstrated that TGF-β, primarily the TGFβ2 isoform, suppresses macrophage inflammatory responses in the developing intestine and protects against inflammatory mucosal injury (18). In addition, TGF-β signaling plays a critical role in promoting alternative macrophage activation, thus limiting systemic inflammatory responses (9). Moreover, TGF-β produced by intestinal epithelial cells is required for the tolerogenic condition of intestinal dendritic cells and possibly macrophage (11). Therefore, characterizing the molecular cues by which IL-10 or TGF-β contribute to the regulatory phenotype of intestinal mononuclear phagocyte system, including macrophages, is of critical importance in understanding perturbation of these pathways that may contribute to the development of mucosal inflammation. In this study, we provide novel insight into the chromatin-modifying events that control the regulation of TGF-βRII gene expression in macrophages, namely the requirement of TGF-β-induced KLF10 in recruiting PCAF to the core TGF-βRII promoter and regulating gene transcription. PCAF is one of the histone-modifying enzymes that associates with KLF10 in the TGF-βRII promoter and is consistent with our previous studies showing that KLF10 can recruit PCAF in the FOXP3 promoter to induce Treg differentiation (36, 37). The critical role of KLF10 is highlighted by the significant decrease of histone H3 acetylation in KLF10−/− BMDM and the inability to transactivate the TGF-βRII promoter in THP-1 cells when KLF10 binding sites of the promoter were deleted or mutated. Additional protein partners may be recruited by KLF10 to the core TGF-βRII promoter in macrophages in addition to PCAF that will enable the full gene activation; these possibilities are currently under investigation.
KLF10 has been implicated in cell differentiation, as a target gene for a variety of signaling pathways, and in serving as a potential marker for human diseases such as breast cancer, cardiac hypertrophy, and osteoporosis (32). Certain genetic variants of KFL10 have been described for cardiomyopathy (3) and minor contributions of Smad 7 and KLF10 in Type 2 diabetes (10). Recent report also suggested KLF10 as a susceptibility gene for IgA nephropathy in Han Chinese (17). Higher levels of KLF10 have also been described in human chronic obstructive pulmonary disease and liver cirrhosis (14). There is no known effect of low KLF10 that may affect innate immune response in human disease. However, the relevance of these regulatory pathways in human inflammatory bowel disease, particularly Crohn's disease, is highlighted by the fact that KLF10 is a critical regulator of Smad 7 (12), the latter being an important negative regulator of TGF-β signaling of T cells and other cell types. Indeed, the lower Smad 2 phosphorylation we observed in TGF-β-stimulated KLF10−/− macrophages (Fig. 4) could be partly related to increased Smad 7 expression as the result of absent suppressive role of KLF10 on Smad 7 expression. The role of KLF10 in regulating Smad 7 expression in macrophages is currently under investigation. Recently, Smad 7 has been targeted (Mongersen, Celgene) for the treatment of Crohn's disease with encouraging results (21). Although in theory Mongersen may target Smad 7 in intestinal T cells, its effect on intestinal macrophages may contribute to its therapeutic effect. In this context it would be interesting to see, if indeed Smad 7 is dysregulated in the absence of KLF10, whether targeting of Smad 7 in KLF10−/− mice may ameliorate the severity of colitis.
In summary, we have identified important molecular cues by which TGF-β-induced KLF10 may further enhance TGF-β signaling through regulating the expression of its own receptor in colonic macrophages. These molecular mechanisms include epigenetic modifications of the TGFβRII promoter locus through recruitment of chromatin modifying enzymes, such as PCAF. Dissecting further the molecular pathways by which KLF10 may regulate the acquisition of a regulatory phenotype in colonic macrophages will shed light on unique pathways that may be attractive therapeutic targets to treat mucosal inflammation.
GRANTS
Mayo Clinic Foundation (K. A. Papadakis), NIH RO1 DK52913 (R. A. Urrutia), Career Developmental Award, Mayo Clinic SPORE in Pancreatic Cancer and Career Developmental Award, Mayo Clinic Center for Cell Signaling in Gastroenterology, NIH RO1 CA178627 (G. A. Lomberk), and NIAID RO1 AI089714 (W. A. Faubion).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
K.A.P. and W.A.F. conception and design of research; K.A.P., J.K., P.S., Y.X., and O.F.S. performed experiments; K.A.P., J.K., P.S., Y.X., O.F.S., and G.A.L. analyzed data; K.A.P., G.A.L., R.A.U., and W.A.F. interpreted results of experiments; K.A.P. prepared figures; K.A.P. drafted manuscript; K.A.P. and W.A.F. edited and revised manuscript; K.A.P., J.K., P.S., O.F.S., G.A.L., R.A.U., and W.A.F. approved final version of manuscript.
REFERENCES
- 1.Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 15: 929–937, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, Guilliams M, Malissen B, Agace WW, Mowat AM. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 6: 498–510, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos JM, Subramaniam M, Hawse JR, Christiaans I, Rajamannan NM, Maleszewski JJ, Edwards WD, Wilde AA, Spelsberg TC, Ackerman MJ. TGFbeta-inducible early gene-1 (TIEG1) mutations in hypertrophic cardiomyopathy. J Cell Biochem 113: 1896–1903, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood 116: 4404–4414, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Z, Wara AK, Icli B, Sun X, Packard RR, Esen F, Stapleton CJ, Subramaniam M, Kretschmer K, Apostolou I, von Boehmer H, Hansson GK, Spelsberg TC, Libby P, Feinberg MW. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(-) T cells and T regulatory cells. J Biol Chem 284: 24914–24924, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442: 299–302, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol 104: 15.25:15.25.1-15.25.14, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol 8: 1086–1094, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Gong D, Shi W, Yi SJ, Chen H, Groffen J, Heisterkamp N. TGFbeta signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol 13: 31, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez-Aguilar R, Benmezroua Y, Balkau B, Marre M, Helbecque N, Charpentier G, Polychronakos C, Sladek R, Froguel P, Neve B. Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab 33: 372–378, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Iliev ID, Mileti E, Matteoli G, Chieppa M, Rescigno M. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol 2: 340–350, 2009. [DOI] [PubMed] [Google Scholar]
- 12.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 21: 5783–5790, 2002. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Matsuoka K, Sheikh SZ, Russo SM, Mishima Y, Collins C, deZoeten EF, Karp CL, Ting JP, Sartor RB, Plevy SE. IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J Immunol 189: 1792–1799, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koczulla AR, Jonigk D, Wolf T, Herr C, Noeske S, Klepetko W, Vogelmeier C, von Neuhoff N, Rische J, Wrenger S, Golpon H, Voswinckel R, Luisetti M, Ferrarotti I, Welte T, Janciauskiene S. Kruppel-like zinc finger proteins in end-stage COPD lungs with and without severe alpha1-antitrypsin deficiency. Orphanet J Rare Dis 7: 29, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzarides T. Chromatin modifications and their function. Cell 128: 693–705, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell 131: 822, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Foo JN, Wang JQ, Low HQ, Tang XQ, Toh KY, Yin PR, Khor CC, Goh YF, Irwan ID, Xu RC, Andiappan AK, Bei JX, Rotzschke O, Chen MH, Cheng CY, Sun LD, Jiang GR, Wong TY, Lin HL, Aung T, Liao YH, Saw SM, Ye K, Ebstein RP, Chen QK, Shi W, Chew SH, Chen J, Zhang FR, Li SP, Xu G, Tai ES, Wang L, Chen N, Zhang XJ, Zeng YX, Zhang H, Liu ZH, Yu XQ, Liu JJ. Identification of new susceptibility loci for IgA nephropathy in Han Chinese. Nat Commun 6: 7270, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maheshwari A, Kelly DR, Nicola T, Ambalavanan N, Jain SK, Murphy-Ullrich J, Athar M, Shimamura M, Bhandari V, Aprahamian C, Dimmitt RA, Serra R, Ohls RK. TGF-beta2 suppresses macrophage cytokine production and mucosal inflammatory responses in the developing intestine. Gastroenterology 140: 242–253, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathison A, Grzenda A, Lomberk G, Velez G, Buttar N, Tietz P, Hendrickson H, Liebl A, Xiong YY, Gores G, Fernandez-Zapico M, Larusso NF, Faubion W, Shah VH, Urrutia R. Role for Kruppel-like transcription factor 11 in mesenchymal cell function and fibrosis. PLoS One 8: e75311, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest 121: 4787–4795, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteleone G, Neurath MF, Ardizzone S, Di Sabatino A, Fantini MC, Castiglione F, Scribano ML, Armuzzi A, Caprioli F, Sturniolo GC, Rogai F, Vecchi M, Atreya R, Bossa F, Onali S, Fichera M, Corazza GR, Biancone L, Savarino V, Pica R, Orlando A, Pallone F. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N Engl J Med 372: 1104–1113, 2015. [DOI] [PubMed] [Google Scholar]
- 22.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol 14: 667–685, 2014. [DOI] [PubMed] [Google Scholar]
- 23.Papadakis KA, Krempski J, Reiter J, Svingen P, Xiong Y, Sarmento OF, Huseby A, Johnson AJ, Lomberk GA, Urrutia RA, Faubion WA Jr. Kruppel-like factor KLF10 regulates transforming growth factor receptor II expression and TGF-β signaling in CD8+ T lymphocytes. Am J Physiol Cell Physiol 308: (5) C362–C371, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramalingam R, Larmonier CB, Thurston RD, Midura-Kiela MT, Zheng SG, Ghishan FK, Kiela PR. Dendritic cell-specific disruption of TGF-beta receptor II leads to altered regulatory T cell phenotype and spontaneous multiorgan autoimmunity. J Immunol 189: 3878–3893, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rani R, Smulian AG, Greaves DR, Hogan SP, Herbert DR. TGF-beta limits IL-33 production and promotes the resolution of colitis through regulation of macrophage function. Eur J Immunol 41: 2000–2009, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med 209: 139–155, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, Mascanfroni ID, Al Adham Z, Lavoie S, Ibourk M, Nguyen DD, Samsom JN, Escher JC, Somech R, Weiss B, Beier R, Conklin LS, Ebens CL, Santos FG, Ferreira AR, Sherlock M, Bhan AK, Muller W, Mora JR, Quintana FJ, Klein C, Muise AM, Horwitz BH, Snapper SB. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40: 706–719, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, Bouchard P, George MD, Hu WK, Dandekar S, Smith PD. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem 285: 19593–19604, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbach EC, Plevy SE. The role of macrophages and dendritic cells in the initiation of inflammation in IBD. Inflamm Bowel Dis 20: 166–175, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol 25: 1191–1199, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res 23: 4907–4912, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. Functional role of KLF10 in multiple disease processes. Biofactors 36: 8–18, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D, Bain CC, Mowat AM, Reis e Sousa C, Poulin LF, Malissen B, Guilliams M. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol 42: 3150–3166, 2012. [DOI] [PubMed] [Google Scholar]
- 34.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, Su J, Liu YC. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol 9: 245–253, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan YY, Leon RP, Marks R, Cham CM, Schaack J, Gajewski TF, DeGregori J. Transgenic expression of the coxsackie/adenovirus receptor enables adenoviral-mediated gene delivery in naive T cells. Proc Natl Acad Sci USA 97: 13784–13789, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong Y, Khanna S, Grzenda AL, Sarmento OF, Svingen PA, Lomberk GA, Urrutia RA, Faubion WA Jr. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J Biol Chem 287: 34372–34385, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Svingen PA, Sarmento OO, Smyrk TC, Dave M, Khanna S, Lomberk GA, Urrutia RA, Faubion WA Jr. Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene. Am J Physiol Regul Integr Comp Physiol 307: R608–R620, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Nakatsukasa H, Tu E, Kasagi S, Cui K, Ishikawa M, Konkel JE, Maruyama T, Wei G, Abbatiello B, Wang ZQ, Zhao K, Chen W. PARP-1 regulates expression of TGF-beta receptors in T cells. Blood 122: 2224–2232, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Wang X, Xia X, Liu X, Suo S, Guo J, Li M, Cao W, Cai Z, Hui Z, Subramaniam M, Spelsberg TC, Wang J, Wang L. Klf10 inhibits IL-12p40 production in macrophage colony-stimulating factor-induced mouse bone marrow-derived macrophages. Eur J Immunol 43: 258–269, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou M, McPherson L, Feng D, Song A, Dong C, Lyu SC, Zhou L, Shi X, Ahn YT, Wang D, Clayberger C, Krensky AM. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J Immunol 178: 5496–5504, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, Brenner O, Krauthgamer R, Varol C, Muller W, Jung S. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40: 720–733, 2014. [DOI] [PubMed] [Google Scholar]
- 42.Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol 34: 162–168, 2013. [DOI] [PubMed] [Google Scholar]
- 43.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, Shakhar G, Halpern Z, Jung S. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37: 1076–1090, 2012. [DOI] [PubMed] [Google Scholar]