ABSTRACT
Peptide-based pheromones are used throughout the fungal kingdom for coordinating sexual responses between mating partners. Here, we address the properties and function of Bar1, an aspartyl protease that acts as a “barrier” and antagonist to pheromone signaling in multiple species. Candida albicans Bar1 was purified and shown to exhibit preferential cleavage of native α pheromone over pheromones from related fungal species. This result establishes that protease substrate specificity coevolved along with changes in its pheromone target. Pheromone cleavage by Bar1 occurred between residues Thr-5 and Asn-6 in the middle of the tridecapeptide sequence. Surprisingly, proteolytic activity was independent of the amino acid residues present at the scissile bond and instead relied on residues at the C terminus of α pheromone. Unlike most aspartyl proteases, Bar1 also exhibited a near-neutral pH optimum and was resistant to the class-wide inhibitor pepstatin A. In addition, genetic analysis was performed on C. albicans BAR1 and demonstrated that the protease not only regulates endogenous pheromone signaling but also can limit interspecies pheromone signaling. We discuss these findings and propose that the unusual substrate specificity of Bar1 is a consequence of its coevolution with the α pheromone receptor Ste2 for their shared peptide target.
IMPORTANCE
Pheromones are important for intraspecies communication across the tree of life. In the fungal kingdom, extracellular proteases play a key role in antagonizing pheromone signaling in multiple species. This study examines the properties and function of Candida albicans Bar1, an aspartyl protease that cleaves and thereby inactivates α pheromone. We demonstrate that Bar1 plays important roles in regulating both intra- and interspecies pheromone signaling. The fungal protease shows preferential activity on the endogenous pheromone, but, surprisingly, cleavage activity is dependent on amino acid residues distal to the scissile bond. We propose that the unusual substrate specificity of Bar1 is a direct result of coevolution with Ste2, the receptor for α pheromone, for recognition of the same peptide target. The novel specificity of Bar1 reveals the complex forces shaping the evolution of mating pathways in fungi and uncovers a protease with potentially important applications in the biotechnology industry.
INTRODUCTION
Pheromone signaling involves the secretion of species-specific chemicals to coordinate cell behavior. Fungi have been intensively studied for their use of sexual pheromones to regulate intercellular signaling and conjugation. In ascomycetes and basidiomycetes, pheromones are peptides or lipopeptides that are secreted into the extracellular milieu and induce morphological and transcriptional responses in target cells. In both of these fungal lineages, mating specificity is determined by the sets of pheromones and pheromone receptors expressed by different cell types (1–6).
In the model ascomycete Saccharomyces cerevisiae, a cells secrete a pheromone and express the α pheromone receptor Ste2, while α cells secrete α pheromone and express the a pheromone receptor Ste3 (7). Pheromone signaling between the 2 cell types activates a conserved mitogen-activated protein kinase (MAPK) signaling cascade, leading to the induction of mating genes and the formation of mating projections or “shmoos” (8–10). The pheromones produced by S. cerevisiae a and α cells show distinct physical properties and are secreted by different mechanisms; α pheromone is an unmodified peptide that is secreted by the traditional secretory pathway, whereas a-factor is farnesylated and carboxymethylated, and transport requires a specific transmembrane translocator (11, 12). The use of both modified and unmodified pheromones is conserved across the ascomycetes, whereas basidiomycetes utilize only lipid-modified pheromones (5, 13).
Candida albicans is a human fungal pathogen related to S. cerevisiae, although these species are as divergent as humans and fish (14). Similar to S. cerevisiae, mating in C. albicans involves pheromone signaling between a and α cells (15–18). However, sexual competency in C. albicans is dependent on cells undergoing a phenotypic switch from the conventional “white” state to the alternative “opaque” state (19, 20). These two states show marked differences in morphology, metabolism, and signaling, including distinct responses to pheromone (21–23). The a and α pheromones are encoded by MFa and MFα genes, respectively, and pheromone signaling between opposite-sex opaque cells leads to the formation of mating projections and conjugation, producing tetraploid a/α cells. Although mating incompetent, C. albicans white cells can respond to pheromones secreted by opaque cells of the opposite mating type. However, rather than forming mating projections, pheromone-treated white cells adhere to inert surfaces and undergo biofilm formation (24–26).
In addition to the secretion of pheromones, yeast cells also produce degradative enzymes that target mating pheromones for destruction. In both S. cerevisiae and C. albicans, a cells secrete an aspartyl protease, Bar1, that acts as a “barrier” to α pheromone signaling by inactivating it (27–30). Bar1 is produced by a cells and sharpens the gradient of α pheromone produced by α cells, thereby facilitating chemotropism and mating between a and α partners (31–33). In addition to increasing mating efficiency, Bar1 promotes higher growth rates in subpopulations of a cells exposed to α pheromone by overcoming pheromone-induced cell cycle arrest (34). Interestingly, loss of Bar1 in C. albicans results in transitioning from a heterothallic to a homothallic (selfing) mode of sexual reproduction. C. albicans a cells secrete low levels of α pheromone (in addition to a pheromone), and loss of Bar1 causes positive feedback of α pheromone on these cell types, triggering same-sex a-a mating (35). The presence of Bar1 therefore prevents autocrine signaling by α pheromone in C. albicans a cells, whereas the high level of α pheromone produced by α cells is sufficient to override Bar1 activity during a-α mating (30, 35). Interestingly, the distantly related archiascomycete Schizosaccharomyces pombe produces a secreted carboxypeptidase, Sxa2, that acts to degrade its own α pheromone-like peptide (36, 37). This indicates that distinct pheromone-degrading proteases have evolved in different ascomycete lineages and supports the assertion that these proteases play key roles in pheromone signaling.
C. albicans Bar1 contains two Asp-(Thr/Ser)-Gly catalytic motifs, as found in other aspartyl proteases, and shares significant homology with well-characterized members of this class (38, 39). Aspartyl proteases have been referred to as acidic proteases, given that they are often optimally active at pH 3 to 5, and most are susceptible to inhibition by the aspartyl protease inhibitor pepstatin A (40). However, S. cerevisiae Bar1 has been shown to have several unusual characteristics for an aspartyl protease, including a pH optimum of 6.5 and resistance to pepstatin A (28, 39). In contrast, the biochemical properties of purified C. albicans Bar1 have not been examined, nor has its specificity for endogenous α pheromone.
In this work, we investigate the biochemical properties of C. albicans Bar1 and reveal that the protease is highly selective in cleaving the endogenous α pheromone. Interestingly, substrate cleavage occurs in the middle of the pheromone, but specificity is largely determined by amino acids present at the C terminus of the sequence. As such, this substrate specificity is highly unusual for an endoprotease and may have arisen due to constraints on Bar1 activity imposed by the pheromone receptor Ste2, which must also recognize α pheromone. We also examine the role of BAR1 in intra- and interspecies pheromone signaling in C. albicans. These experiments reveal that in addition to modulating endogenous α pheromone activity, C. albicans Bar1 also restricts signaling in response to pheromones secreted by related Candida species. Together, our studies reveal that C. albicans Bar1 is an aspartyl protease with novel substrate specificity that acts to regulate both intra- and interspecies signaling events and is a potentially invaluable tool with applications in biotechnology.
RESULTS
Purification and characterization of C. albicans Bar1 protease.
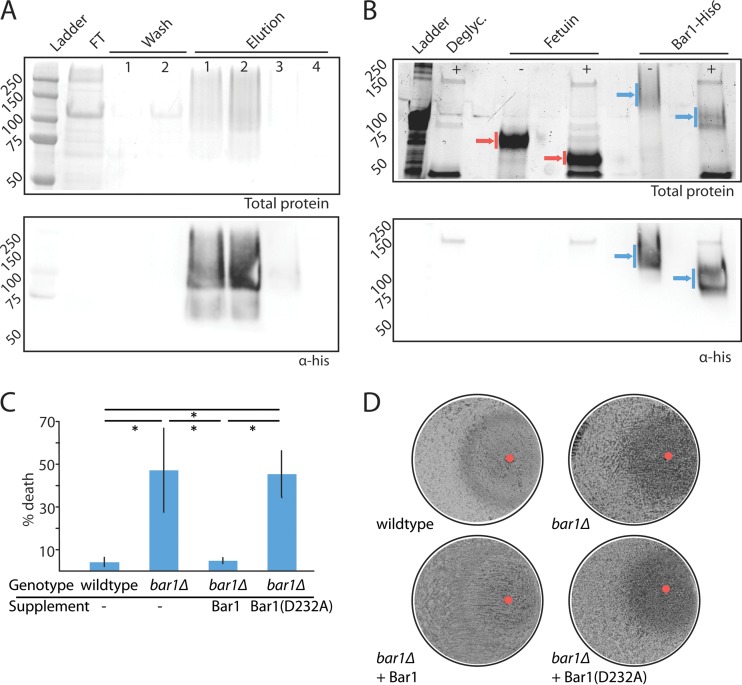
To define the biochemical activity of C. albicans Bar1, a recombinant form of the protein was overexpressed and purified from Pichia pastoris. The C. albicans BAR1 gene was engineered with a C-terminal hexahistidine tag and overexpressed using the PichiaPink expression system (see Materials and Methods). BAR1 was expressed under the control of the P. pastoris AOX1 promoter and contained the S. cerevisiae α pheromone signal peptide sequence to ensure efficient secretion into the supernatant. Secreted Bar1 protein was collected and purified by nickel affinity chromatography (Fig. 1A) to a final yield of 6 mg/liter. The protein migrated as a broad range of high-molecular-weight bands in Western blots, suggestive of protein glycosylation. To test this, C. albicans Bar1 was treated with a commercial mixture of glycosylases that increased protein migration by SDS-PAGE (Fig. 1B), indicating that the recombinant protein was highly glycosylated. Aspartyl proteases often contain two catalytic aspartic acid residues that are present in conserved Asp-(Thr/Ser)-Gly motifs (39). C. albicans Bar1 contains two such DSG motifs at residues 56 to 58 and 232 to 234, and a mutant Bar1 protein was purified that contained an amino acid substitution (D232A) at one of the predicted catalytic aspartyl residues (see Fig. S1 in the supplemental material).
FIG 1 .
Purification and analysis of C. albicans Bar1 protease. (A) Purification of His-tagged C. albicans Bar1 from Pichia pastoris using nickel affinity chromatography. Nickel beads were washed twice (20 mM imidazole), and protein was eluted with 500 mM imidazole. FT, flowthrough. Total protein was assayed by Coomassie staining (top panel), and Bar1 was detected by Western blotting with an anti-His antibody (lower panel). (B) Protein glycosylation was assessed by treatment of Bar1 or fetuin (positive control) with a cocktail of deglycosylation (Deglyc.) enzymes. Blue arrows, Bar1; red arrows, fetuin. The presence or absence of deglycosylase enzymes is noted with ±. Total protein is shown (top panel) using BioRad Stainfree indicator, and Bar1 was detected by Western blotting with an anti-His antibody (lower panel). (C) To evaluate the activity of recombinant Bar1, wild-type and bar1Δ/bar1Δ opaque a strains were treated with or without 300 nM α pheromone for 5 h, together with recombinant Bar1 or Bar1(D232A) at 127 nM. The percentage of death was analyzed by staining with propidium iodide and flow cytometry. Values are means ± standard deviations (SD). n = 3. *, P < 0.05 by t test. (D) Cell cycle arrest was analyzed by plating a lawn of opaque a cells on solid medium and spotting 20 µg α pheromone (red circle) together with Bar1 or Bar1(D232A), followed by imaging of plates after 3 days of culture at 22°C.
To assess the functionality of recombinant Bar1, we tested its ability to complement a C. albicans mutant lacking the BAR1 gene. A quantitative readout of the C. albicans response to pheromone is the pheromone-induced cell death (PID) assay; a significant proportion of opaque a cells responding to α pheromone undergo cell death that can be measured by staining with the vitality dye propidium iodide (41). The pheromone response is dependent on Bar1 activity, as a cells lacking BAR1 undergo PID at lower pheromone concentrations than wild-type Bar1+ cells (41). Thus, wild-type opaque a cells exposed to 10 µM α pheromone showed low levels (<10%) of cell death, whereas bar1Δ/bar1Δ mutants showed ~70% cell death (Fig. 1C). Addition of recombinant Bar1 (12.7 nM) to bar1Δ/bar1Δ mutants reduced cell death to <10% of the population, whereas addition of the Bar1(D232A) mutant failed to restore wild-type levels of PID (Fig. 1C).
A halo assay was also used to verify the functionality of recombinant Bar1. In this assay, α pheromone is spotted onto a lawn of a cells, which are then allowed to grow to confluence. Growth is inhibited in C. albicans cells that respond to the exogenous pheromone, generating “halos” that indicate the strength of the pheromone response (30). Unlike the weak halos produced by wild-type opaque a cells, bar1Δ/bar1Δ mutants produced a more distinct halo in response to α pheromone. Addition of recombinant Bar1 reduced the clarity of the halo formed by bar1Δ/bar1Δ cells (Fig. 1D), whereas the Bar1(D232A) mutant did not show complementation. Together, these results establish that recombinant Bar1 inhibits its physiological target and that mutation of one of the catalytic aspartyl residues abolishes activity consistent with its classification as an aspartyl protease.
Biochemical characterization of C. albicans Bar1.
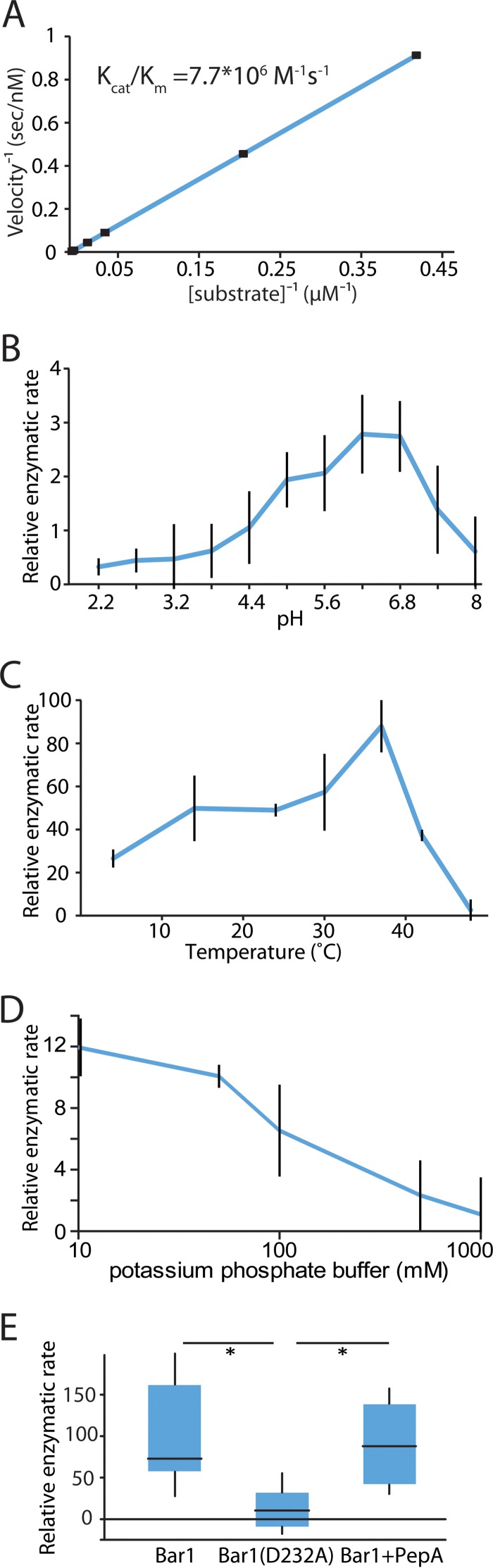
To determine the kinetics of C. albicans Bar1 cleavage, an internally quenched (IQ) version of C. albicans α pheromone (GFRLTNFGYEPG) was utilized. Quenching of the C-terminal EDANS fluorophore occurs by resonance energy transfer to the N-terminal dabcyl quencher, and upon cleavage of the peptide, the quencher is released so that fluorescence is detected. Under defined conditions, Bar1 processed α pheromone with a kcat/Km of ~7.7 × 106 M−1 s−1 (Fig. 2A), whereas the Bar1(D232A) mutant showed no detectable cleavage activity on the IQ substrate. This rate is comparable to that of other secreted Candida aspartyl proteases for their respective substrates (42). Bar1 exhibited maximal activity at a pH of 6.5 and at 37°C, with the greatest activity in low-molarity solutions (Fig. 2B to D). Interestingly, the potent aspartyl protease antagonist pepstatin A did not inhibit Bar1 activity, even at 100 µM (Fig. 2E). S. cerevisiae Bar1 is likewise insensitive to this compound (43). Pepstatin A acts as a transition state analog of aspartyl protease substrates (44) and may be a poor mimic of C. albicans α pheromone, which contains polar, rather than hydrophobic, residues at its cleavage site (see below and reference 40).
FIG 2 .
Characterization of C. albicans Bar1 protease activity. Bar1 activity was determined using an internally quenched peptide corresponding to the native α pheromone sequence of C. albicans, GFRLTNFGYFEPG. (A) Activity was plotted using a Lineweaver-Burk plot to determine enzyme kinetics. (B) pH dependence, (C) temperature dependence, and (D) osmolarity dependence of Bar1 activity. Relative enzymatic rate is the amount of fluorescence per unit of time. The default conditions were 37°C at pH 6.5. Data show means ± SD. (E) Enzymatic activities of Bar1, Bar1(D232A), or Bar1 together with 100 µM pepstatin A were determined by coincubation with the internally quenched C. albicans α pheromone peptide substrate for 90 min at 37°C at pH 6.5. A Tukey box plot is shown. n ≥ 5. *, P < 0.05 by Mann-Whitney U test.
Cleavage of α pheromone peptides by C. albicans Bar1.
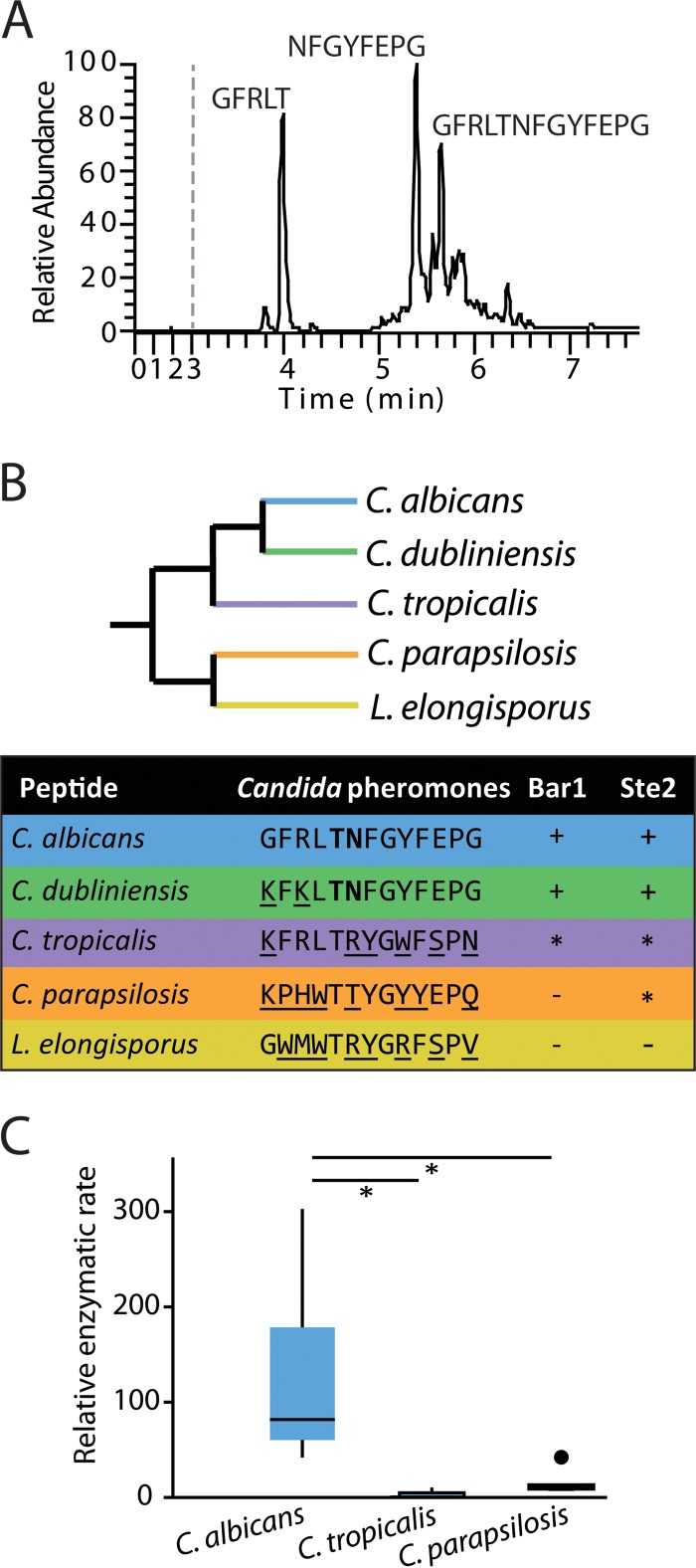
To determine the position at which Bar1 cleaves C. albicans α pheromone, we coincubated recombinant Bar1 with synthetic α pheromone and analyzed the digestion products by liquid chromatography-mass-spectroscopy (LC-MS [see Materials and Methods]). A single cleavage event was detected at the bond between threonine 5 and asparagine 6 (Fig. 3A). In contrast, no cleavage products were generated by the catalytically inactive Bar1(D232A) protein (data not shown), confirming that we were monitoring Bar1 activity and not that of a contaminating protease.
FIG 3 .
Analysis of C. albicans Bar1 protease activity on α pheromones from multiple Candida species. (A) Cleavage of C. albicans α pheromone by C. albicans Bar1 protease. Bar1 and α pheromone were coincubated for 1 h, and the products were analyzed by LC-MS. Mass spectrometry identified two major products (GFRLT and NFGYFEPG), as well as uncleaved pheromone (GFRLTNFGYFEPG). (B) An unrooted, phylogenetic tree of multiple Candida clade species is shown, as well as the activity of Bar1 on α pheromone peptides from each species. The detection of specific degradation products after 1 h of coincubation is indicated by “+” in the Bar1 column. “−” indicates no products were detected, while “*” indicates products were formed only after an extended (24-h) incubation. Boldface residues indicate amino acids adjacent to the cleavage site. The Ste2 column indicates whether the pheromone induced a robust response (+), a weak response (*), or no response (−) in C. albicans MTLa cells. Ste2 data are presented as adapted from Alby and Bennett (49) (see Fig. S2 in the supplemental material). (C) Bar1 activities were compared on C. albicans, C. tropicalis, and C. parapsilosis α pheromones using internally quenched (IQ) peptide substrates. Purified Bar1 was incubated with α pheromones for 90 min at 37°C. Relative enzymatic rate is the amount of fluorescence per unit of time due to cleavage of the fluorophore-conjugated peptide. Shown is a Tukey box plot with outliers noted by a large dot. n ≥ 5. *, P < 0.05 by Mann-Whitney U test.
C. albicans cells were previously shown to respond to several pheromones from related Candida species (41), leading us to test Bar1 activity on each of these peptides using LC-MS (Fig. 3B and C). Bar1 cleaved both Candida dubliniensis and Candida tropicalis α pheromones, and the former, which differs by only two residues from C. albicans α pheromone, was cleaved with similar efficiency to the native substrate. As with C. albicans α pheromone, both C. dubliniensis and C. tropicalis α pheromones were cleaved between residues 5 and 6 (Thr-Asn and Thr-Arg, respectively).
To provide a more sensitive and quantitative analysis of Bar1 activity, IQ peptides were compared representing C. albicans, C. tropicalis, and C. parapsilosis α pheromones. Bar1 cleaved each of the three IQ peptides, although the cleavage rates of C. tropicalis and C. parapsilosis α pheromones were significantly lower (by ~10-fold) than that of the C. albicans α pheromone (Fig. 3C). Together, these results indicate that Bar1 can cleave pheromones besides C. albicans α pheromone, although the endogenous pheromone is, by far, the preferred substrate.
Analysis of the cleavage specificity of C. albicans Bar1.
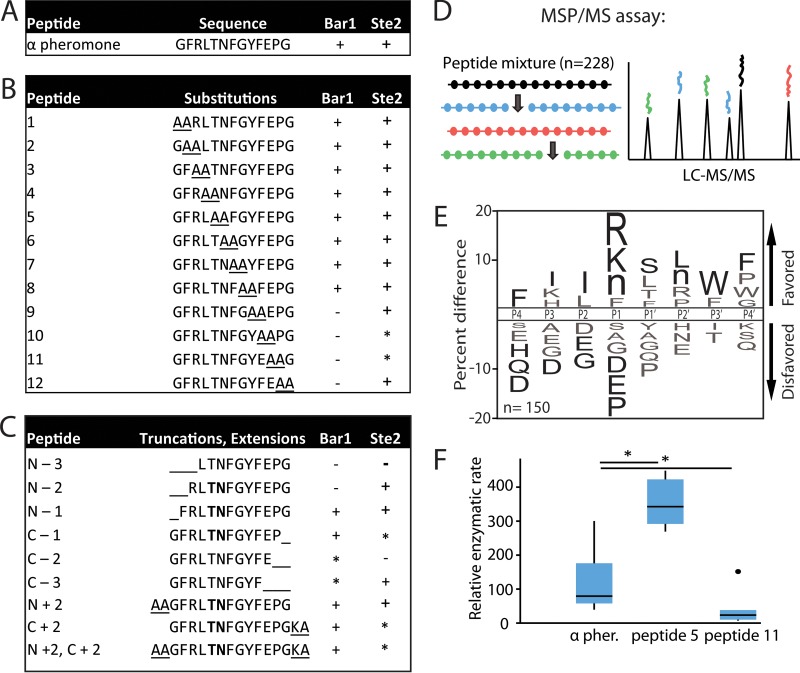
To further determine the substrate specificity of C. albicans Bar1, a series of pheromone-like peptides were incubated with recombinant Bar1, and the products were analyzed by LC-MS. The peptides included scanning di-alanine substitutions across the α pheromone sequence (Fig. 4A and B), as well as N- and C-terminal truncations or extensions of the native sequence (Fig. 4C). Surprisingly, Bar1 cleaved most alanine-substituted peptides, even those with substitutions immediately flanking the scissile bond. However, substitution of several C-terminal residues (residues 10 to 13) effectively blocked cleavage (Fig. 4B). In each case, proteolysis occurred at the same position as in the native peptide (i.e., between the residues at positions 5 and 6). We found that Bar1 did not efficiently degrade peptides in which either the two most N-terminal or C-terminal residues were absent (see peptides N-2 and C-2 in Fig. 4C). In comparison, S. cerevisiae Bar1 shows similar length requirements, as this protease requires 4 amino acids or more on either side of the scissile bond for activity (39).
FIG 4 .
C. albicans Bar1 degradation of α pheromone analogs. Recombinant Bar1 was incubated for 1 h at 37°C with the indicated peptides, and the products were analyzed by mass spectrometry. (A) C. albicans α pheromone; (B) di-alanine substitutions within the C. albicans α pheromone sequence; (C) peptides corresponding to truncated or extended α pheromone. The detection of specific degradation products after 1 h of incubation is indicated by “+” in the Bar1 column. “−” indicates no products were detected, while “*” indicates products were formed only after extended (24-h) incubation. Boldface residues indicate amino acids flanking the cleavage site. The Ste2 column indicates whether the pheromone induced a robust response (+), a weak response (*), or no response (−) in C. albicans MTLa cells using published data in conjunction with data in Fig. S2 in the supplemental material (49). (D) Overview of the multiplex substrate profiling by mass spectrometry (MSP-MS) assay. C. albicans Bar1 was coincubated with 228 unique dodecapaptides, and cleavage products identified using mass spectrometry. (E) MSP-MS data were used to generate an iceLogo identifying amino acids that were enriched or selected against in Bar1 cleavage sites. The percentage of difference is the difference in amino acid frequency surrounding the cleavage sites relative to the frequency of amino acids surrounding all peptide bonds in the library (n = 2,964). Residues above the midline are favored, while those below the midline are disfavored. Residues colored black significantly influence Bar1 activity (P < 0.05), whereas residues colored gray did not reach significance. Methionines were replaced with norleucines in the peptide library and are represented as “n.” (F) Enzymatic activity of Bar1 on substituted forms of α pheromone. Internally quenched peptides included α pheromone with alanine substitutions at positions P1 and P1′ or positions P6′ and P7′ corresponding to peptides 5 and 11 in panel B. The relative enzymatic rate is the amount of fluorescence per unit of time due to cleavage of the fluorophore-conjugated peptide. Rates were normalized to the Bar1(D232A) mutant control. Shown is a Tukey box plot with outliers noted by a large dot. n ≥ 5. *, P < 0.05 by Mann-Whitney U test.
To generate an unbiased profile of protease substrate specificity, Bar1 was tested using a multiplex substrate profiling by mass spectrometry (MSP-MS) approach (45). For this experiment, a physiochemically diverse library of 228 tetradecapeptides was incubated with Bar1 and the products were analyzed at multiple time points by LC-tandem MS (MS/MS) (Fig. 4D and E). Two amino acids positioned close to the scissile bond (usually within positions P4-P4′) are often sufficient for recognition and cleavage by most proteases, so the library of peptides was designed to contain all combinations of neighbor and near-neighbor amino acid pairs (45). Based on the set of cleavage sites identified in the peptide library, a consensus cleavage motif for Bar1 was generated using iceLogo (46), highlighting substrate specificity on either side of the scissile bond. Notably, the iceLogo motif for Bar1 (Fig. 4E) showed little similarity to the sequence of C. albicans α pheromone. The exception was at the P4 position, where a phenylalanine is present in α pheromone, and this amino acid was significantly favored in the MSP-MS assay (Fig. 4E). Taken together, these results suggest that Bar1 recognition of α pheromone is largely independent of the amino acids present near the scissile bond. This contrasts with most other endoproteases, where the majority of substrate selectivity originates from residues at, or very close to, the scissile bond (40, 45, 47, 48).
We further explored the specificity of C. albicans Bar1 using IQ versions of peptides 5 and 11 (Fig. 4B). These peptides have substituted the amino acid residues immediately flanking the scissile bond (P1 and P1′ [peptide IQ-5]) or those within the C-terminal region (P6′ and P7′ [peptide IQ-11]) with alanine residues. Cleavage of peptide IQ-5 was found to be more efficient than that of native α pheromone, whereas cleavage of peptide IQ-11 was significantly lower than that of the natural substrate (Fig. 4F). These experiments further establish that amino acid residues at the C terminus of α pheromone are important for efficient cleavage, whereas those on either side of the scissile bond can both be substituted without negatively affecting Bar1 activity.
Pheromone signaling in C. albicans is mediated by α pheromone binding to the Ste2 receptor on the surface of a cells. We therefore compared the specificity of C. albicans Bar1 with that of Ste2 by examining the ability of different pheromones to activate a cellular response in C. albicans a cells (Fig. 4B) (49). Interestingly, Bar1 substrate specificity mirrored that of Ste2: both C. albicans activities were negatively affected by substitution of amino acid residues at the C terminus of α pheromone but not by residues in the middle of the peptide (see Fig. 4B and C). Furthermore, a significant correlation was found between the pheromone peptides that activate Ste2 and those that are efficiently cleaved by Bar1 (P = 0.00061, Pearson’s rho = 0.51, n = 27), suggesting that Bar1 and Ste2 are dependent on the same residues of α pheromone for activity.
Contribution of Bar1 to pheromone signaling in C. albicans.
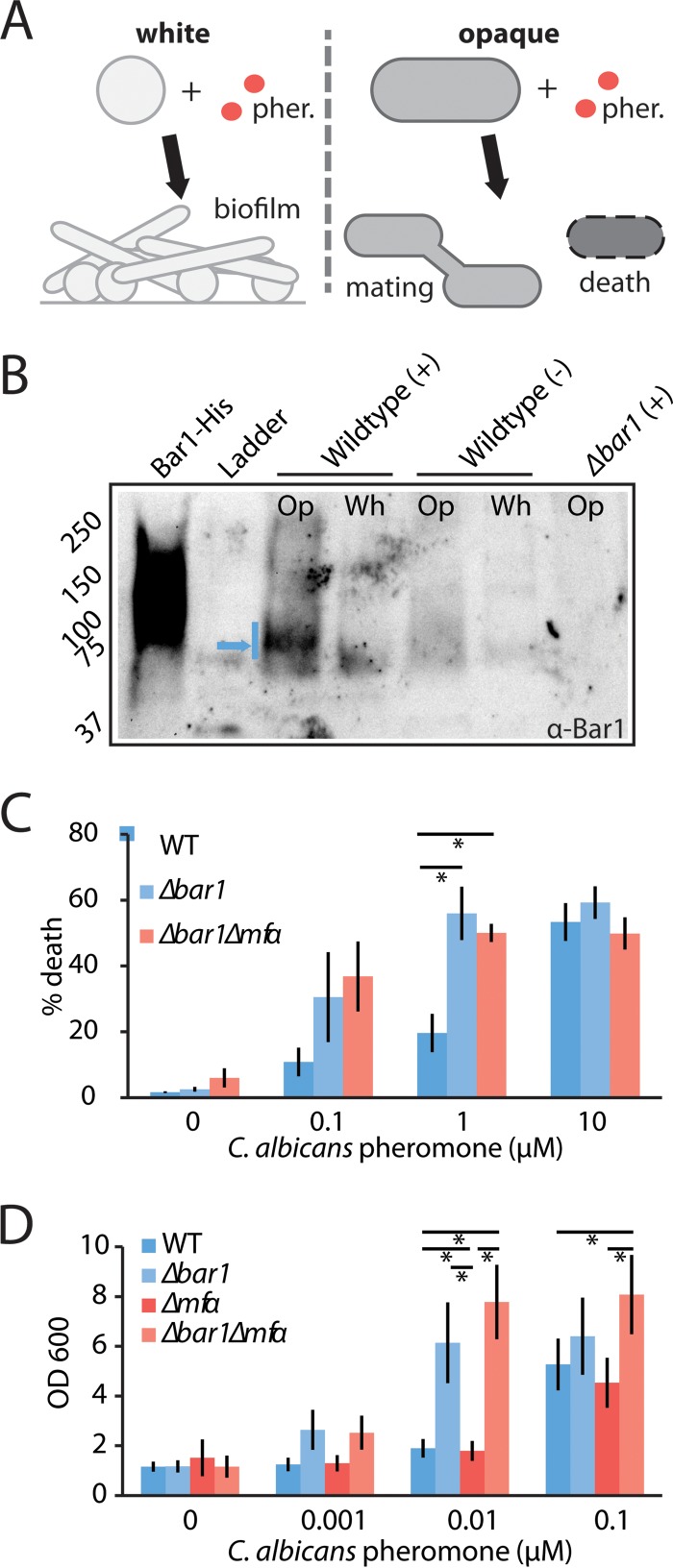
C. albicans a and α cells can exist in two alternative phenotypic states: white and opaque. Mating involves pheromone signaling between cells in the opaque (mating-competent) state, but white cells also respond to pheromone by becoming adhesive and forming biofilms (Fig. 5A) (24–26). In C. albicans a cells, signaling often occurs in response to α pheromone (encoded by the MFα gene) produced by opaque α cells, but it can also be activated by an autocrine loop resulting from α pheromone secreted by opaque a cells (35).
FIG 5 .
Analysis of the role of C. albicans BAR1 on the pheromone response in white and opaque MTLa cells. C. albicans wild-type, bar1Δ/bar1Δ, and bar1Δ/bar1Δ mfαΔ/mfαΔ MTLa strains were compared for their responses to α pheromone. (A) Schematic indicating pheromone (pher.) responses of C. albicans white and opaque a cells. White cells become adherent and form biofilms, whereas a fraction of opaque cells experience cell death. (B) Secretion of Bar1 by white (Wh) or opaque (Op) MTLa cells. Cells were exposed to a vehicle control (−) or 0.3 µM α pheromone (+) for 5 h, and Bar1 protein was detected from the supernatant via Western blotting using an anti-Bar1 antibody. Recombinant Bar1-His protein is used as a positive control, and bar1Δ/bar1Δ opaque cells are used as a negative control. Blue arrow, dominant Bar1 band. (C) Evaluation of gene function in opaque MTLa cells. A pheromone-induced death (PID) assay was used to assess the response of opaque a cells to α pheromone. Cells were treated with the indicated amount of α pheromone for 5 h and stained with propidium iodide, and the percentage of death was determined by flow cytometry. Error bars indicate standard errors (SE). (D) Evaluation of gene function in white MTLa cells. A biofilm assay was used to assess the response of white cells to α pheromone. Cells were incubated with α pheromone for 2 days, and adherent cells were quantified using absorbance at 600 nm. Error bars indicate SD. *, P < 0.05 by t test.
To determine the function of C. albicans Bar1 in both white and opaque cells, we first examined Bar1 levels secreted by cells in the two phenotypic states. A custom antibody was raised against C. albicans Bar1, and Western blots were performed on conditioned media from cultures of white and opaque a cells. Unstimulated cells of both types did not produce Bar1, whereas opaque cells secreted detectable levels of Bar1 when stimulated with α pheromone for 5 h (Fig. 5B). These data are consistent with RNA profiling experiments, as BAR1 is highly induced (>200-fold) upon pheromone challenge in opaque cells (50).
Next, to test the contributions of Bar1 and autocrine pheromone signaling to cellular phenotypes, we constructed bar1Δ/bar1Δ and bar1Δ/bar1Δ mfαΔ/mfαΔ mutants in C. albicans a cells. Pheromone signaling in opaque cells was detected using the PID assay described previously, whereas white-cell responses were quantified by measuring pheromone-induced cell adherence to a plastic substrate (24, 25). Deletion of the BAR1 gene resulted in heightened sensitivity of opaque cells to exogenous α pheromone, in agreement with previous studies (30). Thus, a large proportion of bar1Δ/bar1Δ cells underwent cell death at pheromone concentrations as low as 0.1 µM, while wild-type cells required >1 µM of pheromone to elicit a similar level of cell death (Fig. 5C). Deletion of the MFα gene (encoding α pheromone) prevents autocrine signaling, and yet bar1Δ/bar1Δ mfαΔ/mfαΔ cells showed similar levels of PID to bar1Δ/bar1Δ mutants (Fig. 5C). This result indicates that endogenous α pheromone production and autocrine signaling have a negligible effect on the pheromone response under these conditions. Similar results were obtained with white cells; loss of BAR1 increased the sensitivity of white cells to exogenous α pheromone, and deletion of the MFα gene, either singly or in combination with BAR1, did not show a significant effect on pheromone-induced adherence (Fig. 5D).
Bar1 regulation of interspecies signaling in C. albicans.
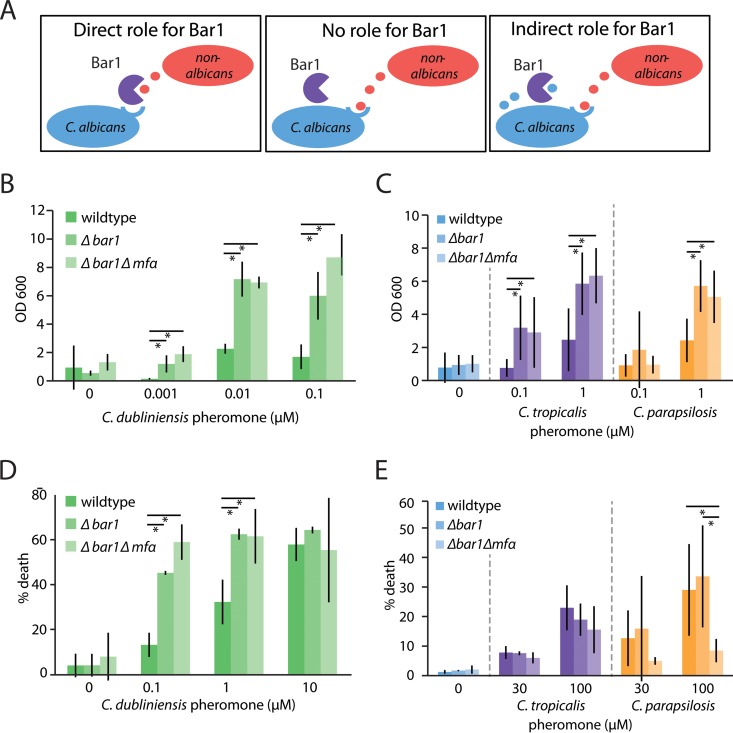
C. albicans a cells have been shown to respond to α pheromones from related Candida species, with opaque cells induced to undergo same-sex mating, whereas white cells exhibit increased biofilm formation (49). These results led us to ask two questions. First, can Bar1 regulate interspecies signaling by degradation of α pheromones secreted by other species? Second, does secretion of endogenous α pheromone from C. albicans a cells enhance interspecies signaling by activating autocrine signaling? There are three scenarios for how pheromone signaling could be influenced by BAR1 and MFα in C. albicans a cells. (i) Signaling between species is unaffected by Bar1 or endogenous α pheromone produced by a cells. (ii) Bar1 degrades non-albicans pheromones, thereby directly restricting interspecies pheromone signaling. (iii) Bar1 does not influence interspecies signaling directly but restricts it by degrading endogenous α pheromone produced by a cells, thereby reducing autocrine signaling (Fig. 6A).
FIG 6 .
Role of C. albicans BAR1 in interspecies pheromone signaling. (A) Three alternative models for how C. albicans Bar1 may affect interspecies signaling between Candida species. (i) Bar1 may affect signaling through direct recognition and degradation of nonnative pheromones, (ii) it may have no effect on interspecies signaling events, or (iii) it may play an indirect role in signaling due to degradation of C. albicans α pheromone (and blocking of autocrine signaling) following receptor activation with a nonnative pheromone. C. albicans wild-type, bar1Δ/bar1IΔ, and bar1Δ/bar1Δ mfαΔ/mfαΔ MTLa white cells were incubated with C. dubliniensis (B), C. tropicalis (C), or C. parapsilosis (D) α pheromones in a biofilm assay. After 2 days, adherent cells were quantified by spectrometry at 600 nm. Mean ± SD. *, P < 0.05 by t test. (E) C. albicans wild-type, bar1Δ/bar1Δ, and bar1Δ/bar1Δ mfαΔ/mfαΔ MTLa opaque cells were treated with C. tropicalis or C. parapsilosis α pheromones. The pheromone response was quantified by determining the percentage of cell death after 5 h of pheromone exposure by flow cytometry. Mean ± SD. *, P < 0.05.
Deletion of the BAR1 gene in C. albicans white a cells led to a heightened response to C. dubliniensis, C. tropicalis, and C. parapsilosis α pheromones (Fig. 6B and C). This is consistent with the biochemical activity of Bar1, which was capable of degrading each of these three pheromones in vitro (Fig. 3B and C). Sensitivity to C. dubliniensis pheromone was considerably higher than that to C. tropicalis and C. parapsilosis pheromones, consistent with the close sequence homology of C. albicans and C. dubliniensis pheromones (Fig. 3B). In contrast to BAR1, deletion of the MFα gene did not affect the response to the three non-albicans pheromones (Fig. 6B and C). This indicates that Bar1 directly affects the response to these pheromones, similar to the result with C. albicans α pheromone (compare Fig. 5C with 6B and C).
Analogous experiments were performed using C. albicans opaque a cells to determine the contributions of BAR1 and MFα to interspecies signaling in this cell type (Fig. 6D and E). C. dubliniensis α pheromone produced a similar response to the native C. albicans α pheromone in opaque cells, and deletion of MFα did not affect the outcome (Fig. 6D). In contrast, deletion of both MFα and BAR1 genes depressed the response of C. albicans opaque a cells to C. parapsilosis α pheromone, indicating that autocrine signaling significantly affects the response to this pheromone (Fig. 6E). Taken together, these results recapitulate our biochemical findings that Bar1 acts most effectively on C. albicans α pheromone but also has the capacity to degrade pheromones from other Candida species, thereby limiting interspecies signaling.
DISCUSSION
Mating in fungi is choreographed by pheromone signaling between cell partners. In this work, we addressed the biochemical properties and function of C. albicans Bar1, an aspartyl protease secreted by a cells that degrades α pheromone. We show that Bar1 cleaves endogenous α pheromone between threonine 5 and asparagine 6, thereby inactivating the 13-amino-acid peptide. This protease exhibits several features common to secreted aspartyl proteases, including extensive glycosylation and two conserved Asp-(Thr/Ser)-Gly motifs (39). Mutation of one of the putative active site aspartyl residues, Asp-232, was sufficient to block Bar1 activity. However, Bar1 also displays unusual properties for an aspartyl protease, including a near-neutral pH optimum and resistance to pepstatin A, a potent aspartyl protease antagonist. In addition to Bar1, C. albicans contains a set of 10 SAP (secreted aspartyl protease) genes, several of which are implicated in pathogenesis (38, 51–53). Most C. albicans Sap proteases exhibit an acidic pH optimum characteristic of other aspartyl proteases (54, 55), although Sap9 and Sap10 exhibit optimal activity between pH 6 and 7 (56). In addition, the majority of C. albicans Saps are inhibited by pepstatin A, with Sap7 being the lone exception (54, 56–58). Notably, S. cerevisiae Bar1 is also an unusual aspartyl protease in that this enzyme displays a near-neutral pH optimum and is not inhibited by pepstatin A (28, 29, 38, 39). It therefore appears that S. cerevisiae Bar1 (ScBar1) and C. albicans Bar1 (CaBar1) (as well as certain Sap proteins) share unusual traits for aspartyl proteases, and this is despite the extensive evolutionary divergence between S. cerevisiae and C. albicans (14).
A comparative analysis of C. albicans Bar1 cleavage of α pheromones from multiple Candida species revealed that Bar1 preferentially cleaved C. albicans α pheromone, whereas α pheromones from closely related species (including C. tropicalis and C. parapsilosis) were also cleaved, albeit at a much reduced rate. Bar1 did not cleave pheromones from more evolutionarily distant fungal species, and unlike most C. albicans Sap proteases (58), showed no detectable activity on bovine serum albumin even when tested at high protein concentrations (data not shown). Thus, C. albicans Bar1 has evolved high substrate specificity toward the endogenous α pheromone. Studies on ScBar1 also suggest a species-specific function, as S. cerevisiae cells were unable to cleave C. albicans α pheromone (59). Together, these experiments establish that Bar1 has evolved selectivity toward its native pheromone in diverse fungal lineages.
An unexpected feature of C. albicans Bar1 cleavage activity was its dependence on amino acid residues located distal to the scissile bond. The specificity of most endoproteases is guided by amino acids located at, or very close to, the scissile bond (45, 48). In contrast, Bar1 cleaved α pheromone by recognition of amino acid residues at the C terminus of the peptide. Thus, alanine substitutions at positions 10 to 13 of α pheromone (P4′ to P7′) inhibited cleavage, whereas substitutions at the scissile bond did not (Fig. 4B and F). The unusual specificity of Bar1 was further supported by an unbiased analysis of activity using MSP-MS, which identifies amino acids close to the scissile bond that are enriched or de-enriched in substrates (45). The amino acids favored on either side of the scissile bond by MSP-MS analysis had little similarity to those present in C. albicans α pheromone. Together, these studies demonstrate that the cleavage specificity of CaBar1 for α pheromone is mediated by residues distal to the scissile bond rather than amino acids close to the site of cleavage.
Why might C. albicans Bar1 have evolved specificity for residues away from the scissile bond? We show that the specificity of CaBar1 exhibits a striking similarity to that of the C. albicans Ste2 receptor for α pheromone. A previous study revealed that substitution of residues at the C terminus of α pheromone (particularly residues 11 and 12) resulted in the largest reduction in pheromone signaling by C. albicans a cells (see the “Ste2” column in Fig. 4B and C) (49). Thus, both C. albicans Bar1 and Ste2 are critically dependent on the same C-terminal residues of α pheromone for activity. We therefore propose that there has been coevolution of these factors due to their dependence for the same peptide target. In particular, as receptor-pheromone interactions diverged between species, Bar1 specificity for α pheromone would have had to evolve in parallel to retain activity. By having Bar1 and Ste2 recognize the same residues in α pheromone, this would have facilitated coevolution of these factors together with their peptide target during speciation (Fig. 7). Studies have also addressed the interaction between S. cerevisiae Ste2 and α pheromone and have shown that ScSte2 makes multiple contacts with α pheromone, although residues near the C terminus of the pheromone (residues 10 to 13) play the most important role in receptor binding (60–63). Thus, in both C. albicans and S. cerevisiae, the carboxyl-terminal residues of α pheromone are critical for receptor function. It will now be of interest to examine pheromone-receptor and pheromone-Bar1 interactions across multiple species to further address how these factors coevolved and if the C terminus of α pheromone has a conserved role in mediating interactions with both of its protein targets across diverse species.
FIG 7 .
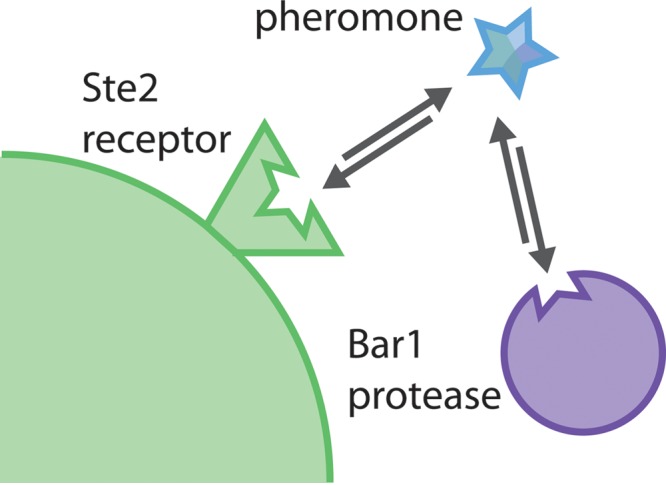
Model of interactions between Bar1, Ste2, and α pheromone. Both Bar1 and Ste2 recognize the same or overlapping regions of α pheromone, thereby facilitating coevolution of their substrate specificities together with pheromone divergence during speciation.
To complement the biochemical analysis of C. albicans Bar1, we performed genetic experiments to define the role of this protease in regulating intra- and interspecies pheromone signaling. Loss of BAR1 sensitized C. albicans white and opaque a cells to native α pheromone, indicating functional secretion of the Bar1 enzyme by both cell types. Deletion of the BAR1 gene also sensitized C. albicans a cells to α pheromones from C. dubliniensis, C. tropicalis, and C. parapsilosis, indicating that Bar1 effectively inhibits signaling to pheromones produced by closely related species. This could be important in nature, as C. albicans is often found cocolonizing the oral cavity with multiple other Candida clade species (64).
We also addressed whether autocrine pheromone production contributes to interspecies signaling, as C. albicans a cells have been shown to secrete α pheromone in addition to the conventional a pheromone (35). To test this, the MFα gene (encoding α pheromone) was deleted in the presence or absence of BAR1, and a cells were challenged with α pheromones. In most cases, loss of MFα did not affect the response of C. albicans a cells to other species’ pheromones. However, the response to C. parapsilosis α pheromone was reduced upon deletion of both BAR1 and MFα genes, indicating that autocrine signaling enhanced interspecies signaling to this pheromone (Fig. 6E). These results are the first to demonstrate that Bar1 acts to restrict interspecies pheromone signaling and also show a role for endogenously produced pheromone in enhancing the response to a pheromone from a related species.
Finally, we note that the unusual specificity of C. albicans Bar1 makes it an attractive candidate for bioengineering a protease with specialized properties. There are a number of potential technical applications for a protease whose specificity is dependent on residues distal to the scissile bond, including the ability to completely remove affinity tags from purified proteins without leaving behind the cleavage site residues. In principle, this is analogous to restriction enzymes (e.g., type III restriction enzymes) that cleave the DNA at a short distance from their recognition sequence. In fact, enteropeptidase is a serine protease whose specificity determinants are located on the C-terminal side of the scissile bond, so that N-terminal tags can be removed, leaving the product with a native N terminus (48). C. albicans Bar1 could be engineered to perform a similar function for the complete removal of C-terminal tags from recombinant proteins. Bar1 can be highly overexpressed, shows strong substrate selectivity, and is active at neutral pH, all of which are properties that would favor its use in many applications. Further analysis of Bar1 homologs from Saccharomycotina species could also help identify the residues in Bar1 that are responsible for determining its substrate specificity, as Bar1 orthologs appear to be under strong selection to evolve in parallel with their pheromone substrates. Such experiments could shed light on the precise mechanism by which α pheromone is recognized and also allow further refinement of the proteolytic activity for important commercial applications.
MATERIALS AND METHODS
Media.
All media were prepared using previously described methods (65, 66). Spider medium contained 1% nutrient broth, 0.4% potassium phosphate, and 2% mannitol (pH 7.2). Solid media were made with 1.35% agar.
Strain and plasmid construction.
The strains used here are listed in Table S1 in the supplemental material. To overexpress C. albicans Bar1 protein, PCR was used to generate a DNA product containing the entire open reading frame (ORF) of BAR1 from C. albicans genomic DNA. The primers used added a 6× His (His6) tag to the end of the ORF and restriction endonuclease cut sites. The PCR product was then cloned into the pPinkα-HC plasmid under the AOX1 promoter (Life Technologies) to yield the plasmid pBar1-His. Site-directed mutagenesis via the QuikChange II kit (Agilent Technologies) was used to mutate position +695 from adenine to cytosine to yield plasmid pBar1(D232A)-His, which contains amino acid substitution D232A in one of the two predicted active sites of the Bar1 protein. Both plasmids were transformed into PichiaPink strains to generate strains CAY794 and CAY800.
To construct the C. albicans MFα and BAR1 deletion strains, plasmids pRB13 and pRB35, respectively, were used with the previously described SAT1-flipper method (67). Briefly, regions flanking the ORF of the gene of interest were PCR amplified together with oligonucleotides containing the ApaI/XhoI (5′ flank) and SacII/SacI (3′ flank) restriction sites. The products were cloned into plasmid pSFS2a. The resulting plasmids were digested with ApaI and SacI and transformed into C. albicans. Correct integration was confirmed by PCR, and the SAT1 marker was excised by growth on maltose medium to induce the FLP recombinase (67). Transformations were then repeated to replace the second copy of the gene of interest. To fix cells in the opaque state, strains were transformed with plasmid pRB99 (pACT1-WOR1) to constitutively express WOR1 (41).
C. albicans Bar1 protein expression and purification.
P. pastoris strains CAY794 and CAY800 were grown in BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base [YNB], 0.00004% biotin, 1% glycerol) for 2 days at 28°C, and then cells were concentrated and switched to BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 0.00004% biotin, 0.5% methanol) to induce expression of Bar1 for 2 days at 28°C. The methanol concentration was maintained by further addition of methanol after 24 h. Cells were centrifuged, and the supernatant was collected, followed by 40-fold concentration using an Amicon cell (EMD Millipore, Darmstadt, Germany). The cell was also used to perform buffer exchange into Ni-nitrilotriacetic acid (NTA) binding buffer (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, pH 8.0). The resulting solution was batch bound to HisPur Ni-NTA resin (Thermo, Fisher Scientific, Waltham, MA) and then washed twice (300 mM NaCl, 50 mM NaH2PO4, 20 mM imidazole, pH 8.0), and the product was eluted (300 mM NaCl, 50 mM NaH2PO4, 500 mM imidazole, pH 8.0). The product was dialyzed into storage buffer (phosphate-buffered saline [PBS], 20% glycerol). The protein concentration was determined using a Bradford protein assay (Thermo Scientific).
Analysis of peptide cleavage by liquid chromatography-mass spectrometry.
Lyophilized peptides (Lifetein LLC) were dissolved in 10% dimethyl sulfoxide (DMSO), mixed with 127 nM C. albicans Bar1 or Bar1(D232A) protein in 50 µM potassium phosphate solution (pH 6.5) for 1 h at 37°C, and then passed through a centrifugal filtration unit to separate peptide fragments from the peptidase. Samples were analyzed using a Shimadzu high-performance liquid chromatograph and Thermo LCQ Deca XP Max ion trap mass spectrometer system.
MSP-MS.
MSP-MS assays were carried out as described previously (45). Briefly, ~60 nM Bar1, Bar1(D232A), or matched no-enzyme control were assayed against a diverse library of 228 tetradecapeptides pooled at 500 nM in 10 mM KH2PO4 (pH 6.5) and incubated at 37°C. After 60, 240, and 1,200 min, 30 µl of assay mixture was removed, quenched with 7.5 µl 2% formic acid, and frozen. Prior to mass spectrometry acquisition, peptide samples were desalted using Millipore C18 ZipTips and rehydrated in 0.2% formic acid. LC-MS/MS data were acquired using a Thermo Scientific LTQ-Orbitrap mass spectrometer, which was equipped with a Thermo Scientific EASY-Spray ion source, an EASY-Spray PepMap C18 column (3 µM, 100 Å), and a Waters nanoACQUITY ultraperformance liquid chromatography (UPLC) system. Mass spectrometry peak lists were generated using PAVA, and data were searched against the 228-member peptide library using Protein Prospector software v.5.12.4 (UCSF). Protein Prospector score thresholds were selected with a minimum protein score of 15 and minimum peptide score of 10. Maximum expectation values of 0.01 and 0.05 were used for protein and peptide matches, respectively. Peptides corresponding to cleavage products were imported into iceLogo to generate substrate specificity profiles as described previously (45). Octapeptides corresponding to P4-P4′ were used as the positive data set, and octapeptides corresponding to all possible cleavages in the MSP-MS library (n = 2,964) were used as the negative data set.
Protein characterization.
To determine Bar1 levels in culture, C. albicans strains were grown at 22°C overnight in synthetic complete dextrose (SCD) medium, and cells were washed. Next, 2 × 108 cells were grown for 5 h in 10 ml SCD medium with or without 0.3 µM synthetic α pheromone. The supernatant and cells were separated by centrifugation, and 40 µl of each was prepared for SDS-PAGE in Laemmli sample buffer. Samples were analyzed by SDS-PAGE and Western blotting. For anti-His blots, a 1:5,000 dilution of Sigma’s horseradish peroxidase (HRP)-conjugated monoclonal anti-hexahistidine (anti-His6) antibody (A7058) was used according to manufacturer’s directions. For anti-Bar1 blots, a 1:1,000 dilution of sera containing a polyclonal anti-Bar1 antibody designed by New England Peptides (project 2095, raised against epitopes Ac-YFINETIRSNDWKC-amide and Ac-CSDDKITSVTSNPQ-amide) was used, followed by a 1:10,000 dilution of goat anti-rabbit Ig-HRP (Jackson Immunoresearch). Proteins were visualized using a chemiluminescent substrate (Pierce) and a Chemidoc XRS+ system (Bio-Rad).
For assessment of glycosylation of C. albicans Bar1, a protein deglycosylation assay was performed on the purified Bar1 protein according to the manufacturer’s directions (New England Biolabs P6039S), and followed by SDS-PAGE and Western blotting as described above. Assessment of the signal peptide sequence of Bar1 was completed using SignalP-4.1 prediction (http://www.cbs.dtu.dk/services/SignalP/).
IQ peptide assays.
IQ peptides were produced by LifeTein, LLC (Hillsborough, NJ) with the dabcyl at the N terminus and EDANS at the C terminus. Unless otherwise noted, all experiments were done at 37°C in 50 µM potassium phosphate buffer, pH 6.5, with a substrate concentration of 50 µM and 60 nM recombinant Bar1 protease or Bar1(D232A) mutant protease. The protease and substrate were combined in a 96-well plate immediately before beginning the experiment, and then fluorescence emission was measured over 90 min using a BioTek Synergy HT plate reader (Winooski, VT). For experiments containing inhibitors, the inhibitor was added to the protease before being combined with the substrate. Each experimental replicate was an average of three technical replicates. Values were normalized to the Bar1(D232A) mutant control when noted. Rates were determined over the period where substrate was in excess of the enzyme.
Pheromone-induced cell death.
C. albicans opaque a cells derived from strain AM2003 (41) were grown in SCD medium overnight at 22°C in a rotating drum. Cultures were washed with water and then resuspended in Spider medium at 2 × 107 cells/ml, and α pheromone was added at the indicated concentration. After 5 h of incubation at 22°C in a rotating drum, 500 µl of culture was removed and washed with water. Then the cells were resuspended in SCD medium containing 1 µg/ml propidium iodide for 15 min. Cells were washed again, resuspended in SCD medium, and analyzed by flow cytometry on a FACSCalibur cell sorter (BD Biosciences). At least 10,000 events were analyzed for fluorescence in channel FL3. For halo assays, 105 opaque cells were plated onto Spider medium. Twenty micrograms of α pheromone was spotted onto the center of the plate, and the plates were incubated at 22°C for 3 days, after which images were acquired.
Pheromone-induced adherence assay.
C. albicans white a cells derived from strain P37005 (68) were grown overnight in Spider medium at 22°C on a rotating drum. Cells were washed with water and then administered to each well of a 12-well plate containing 1 ml of Lee’s plus glucose medium at a concentration of 4 × 107 cells/ml. Pheromones were added at the indicated concentration, and then plates were incubated at 22°C for 2 days with no shaking. Each well was washed gently 3 times with water to remove nonadherent cells. Adherent cells were scraped and resuspended in 1 ml of water, and the optical density of each sample was determined at 600 nm on a Nanodrop 2000c (Thermo Scientific).
Statistical analysis.
Statistics were analyzed using the PAST software package (http://folk.uio.no/ohammer/past/). Using the Anderson-Darling method, data sets were tested for normality, and parametric tests were used when appropriate and possible. Tests between samples were performed using the Student’s t test, Tukey’s test, or Mann-Whitney test, as indicated in the figure legends. Asterisks indicate a P value of 0.05 or less, unless otherwise noted. In order to correlate Bar1 cleavage of the tested peptides and activation of Ste2 signaling by the same peptides, the efficiency of Bar1 cleavage was first binned into three groups (products detected by HPLC-MS after 1 h of incubation with Bar1 or after 24 h of incubation, or products were not detected even after 24 h of coincubation). Next, activation of Ste2 signaling was compared using data from published pheromone-induced biofilm assays (49) and binned into three groups (similar or better response than with the wild-type α pheromone, a lower but detectable response, or no response). The binned data were then correlated using Spearman’s rho.
SUPPLEMENTAL MATERIAL
Purification of mutant Bar1 protease. C. albicans Bar1(D232A) was overexpressed in Pichia pastoris, and the protein was concentrated and purified by nickel affinity chromatography. Flow, 10 mM imidazole. Washes, 20 mM imidazole. Elutions, 500 mM imidazole. (A) Total protein via Coomassie stain. (B) Western blotting using an anti-His6 antibody. Download
C. albicans BAR1 affects C. albicans white cells’ response to truncated α pheromone peptides. A biofilm assay was used to assess the response of white C. albicans cells to truncated versions of C. albicans α pheromone. After 48 h of incubation with the indicated amount of peptide, adherent cells were resuspended in medium and quantified by spectrometry at 600 nm. Mean ± SD. *, P < 0.05 by t test. Download
Strain table.
ACKNOWLEDGMENTS
The National Bio-Organic, Biomedical Mass Spectrometry Resource Center (UCSF) is supported by a grant from the National Institutes of Health (P41GM103481). Work was also supported by National Institutes of Health grants AI081704 and AI112362 (to R.J.B.), by a predoctoral fellowship F31DE022701 (to S.K.J.), and by a PATH award from the Burroughs Wellcome Fund (to R.J.B.).
We thank members of the Bennett and Craik labs for their support and thoughtful input. MSP-MS data were acquired in the National Bio-Organic, Biomedical Mass Spectrometry Resource Center (UCSF). LC-MS data were acquired with the help of Tun-Li Shen at the Mass Spectroscopy Facility in the Department of Chemistry (Brown University).
Footnotes
Citation Jones SK, Jr, Clarke SC, Craik CS, Bennett RJ. 2015. Evolutionary selection on barrier activity: Bar1 is an aspartyl protease with novel substrate specificity. mBio 6(6):e01604-15. doi:10.1128/mBio.01604-15.
REFERENCES
- 1.Bender A, Sprague GF Jr. 1986. Yeast peptide pheromones, a-factor and alpha-factor, activate a common response mechanism in their target cells. Cell 47:929–937. doi: 10.1016/0092-8674(86)90808-1. [DOI] [PubMed] [Google Scholar]
- 2.Stanton BC, Giles SS, Staudt MW, Kruzel EK, Hull CM. 2010. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet 6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender A, Sprague GF Jr. 1989. Pheromones and pheromone receptors are the primary determinants of mating specificity in the yeast Saccharomyces cerevisiae. Genetics 121:463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bölker M, Urban M, Kahmann R. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441–450. doi: 10.1016/0092-8674(92)90182-C. [DOI] [PubMed] [Google Scholar]
- 5.Gonçalves-Sá J, Murray A. 2011. Asymmetry in sexual pheromones is not required for ascomycete mating. Curr Biol 21:1337–1346. doi: 10.1016/j.cub.2011.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seike T, Nakamura T, Shimoda C. 2015. Molecular coevolution of a sex pheromone and its receptor triggers reproductive isolation in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 112:4405–4410. doi: 10.1073/pnas.1501661112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AD. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev 5:552–558. doi: 10.1016/0959-437X(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 8.Jones SK Jr, Bennett RJ. 2011. Fungal mating pheromones: choreographing the dating game. Fungal Genet Biol 48:668–676. doi: 10.1016/j.fgb.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merlini L, Dudin O, Martin SG. 2013. Mate and fuse: how yeast cells do it. Open Biol 3:130008. doi: 10.1098/rsob.130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Sapperstein SK, Choi JD, Michaelis S. 1997. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol 136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchler K, Sterne RE, Thorner J. 1989. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J 8:3973–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casselton LA, Olesnicky NS. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev 62:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich J, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. 2004. Genome evolution in yeasts. Nature 430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 15.Bennett RJ, Miller MG, Chua PR, Maxon ME, Johnson AD. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol Microbiol 55:1046–1059. doi: 10.1111/j.1365-2958.2005.04466.x. [DOI] [PubMed] [Google Scholar]
- 16.Bennett RJ, Uhl MA, Miller MG, Johnson AD. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol 23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockhart SR, Zhao R, Daniels KJ, Soll DR. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell 2:847–855. doi: 10.1128/EC.2.5.847-855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT. 2003. MFalpha1, the gene encoding the alpha mating pheromone of Candida albicans. Eukaryot Cell 2:1350–1360. doi: 10.1128/EC.2.6.1350-1360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MG, Johnson AD. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293–302. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 20.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiger J, Wessels D, Lockhart SR, Soll DR. 2004. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun 72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si H, Hernday AD, Hirakawa MP, Johnson AD, Bennett RJ. 2013. Candida albicans white and opaque cells undergo distinct programs of filamentous growth. PLoS Pathog 9:e1003210. doi: 10.1371/journal.ppat.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol 9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 24.Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J 25:2240–2252. doi: 10.1038/sj.emboj.7601099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. 2013. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog 9:e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell 19:957–970. doi: 10.1091/mbc.E07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprague GF Jr, Herskowitz I. 1981. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J Mol Biol 153:305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- 28.Manney TR. 1983. Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J Bacteriol 155:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKay VL, Welch SK, Insley MY, Manney TR, Holly J, Saari GC, Parker ML. 1988. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci U S A 85:55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer D, Cote P, Whiteway M, Bennett RJ. 2007. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot Cell 6:907–918. doi: 10.1128/EC.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews SS, Addy NJ, Brent R, Arkin AP. 2010. Detailed simulations of cell biology with Smoldyn2.1. PLoS Comput Biol 6:e1000705. doi: 10.1371/journal.pcbi.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barkai N, Rose MD, Wingreen NS. 1998. Protease helps yeast find mating partners. Nature 396:422–423. doi: 10.1038/24760. [DOI] [PubMed] [Google Scholar]
- 33.Jin M, Errede B, Behar M, Mather W, Nayak S, Hasty J, Dohlman HG, Elston TC. 2011. Yeast dynamically modify their environment to achieve better mating efficiency. Sci Signal 4:ra54. doi: 10.1126/scisignal.2001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diener C, Schreiber G, Giese W, del Rio G, Schröder A, Klipp E. 2014. Yeast mating and image-based quantification of spatial pattern formation. PLoS Comput Biol 10:e1003690. doi: 10.1371/journal.pcbi.1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alby K, Schaefer D, Bennett RJ. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladds G, Davey J. 1996. Characterisation of Sxa2, a carboxypeptidase involved in pheromone recovery in fission yeast. Biochem Soc Trans 24:210S. doi: 10.1042/bst024210s. [DOI] [PubMed] [Google Scholar]
- 37.Ladds G, Rasmussen EM, Young T, Nielsen O, Davey J. 1996. The sxa2-dependent inactivation of the P-factor mating pheromone in the fission yeast Schizosaccharomyces pombe. Mol Microbiol 20:35–42. doi: 10.1111/j.1365-2958.1996.tb02486.x. [DOI] [PubMed] [Google Scholar]
- 38.Naglik JR, Challacombe SJ, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKay VL, Armstrong J, Yip C, Welch S, Walker K, Osborn S, Sheppard P, Forstrom J. 1991. Characterization of the bar proteinase, an extracellular enzyme from the yeast Saccharomyces cerevisiae, p 161–172. In Dunn BM. (ed), Structure and function of the aspartic proteinases. Plenum Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 40.Dunn BM. 2001. Overview of pepsin-like aspartic peptidases. Curr Protoc Protein Sci Chapter 21:Unit 21.23. [DOI] [PubMed] [Google Scholar]
- 41.Alby K, Schaefer D, Sherwood RK, Jones SK Jr, Bennett RJ. 2010. Identification of a cell death pathway in Candida albicans during the response to pheromone. Eukaryot Cell 9:1690–1701. doi: 10.1128/EC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fusek M, Smith EA, Monod M, Dunn BM, Foundling SI. 1994. Extracellular aspartic proteinases from Candida albicans, Candida tropicalis, and Candida parapsilosis yeasts differ substantially in their specificities. Biochemistry 33:9791–9799. doi: 10.1021/bi00198a051. [DOI] [PubMed] [Google Scholar]
- 43.Nath R. 1993. Properties of barrier, a novel Saccharomyces cerevisiae acid protease. Biochimie 75:467–472. doi: 10.1016/0300-9084(93)90112-6. [DOI] [PubMed] [Google Scholar]
- 44.Marciniszyn J Jr, Hartsuck JA, Tang J. 1976. Mode of inhibition of acid proteases by pepstatin. J Biol Chem 251:7088–7094. [PubMed] [Google Scholar]
- 45.O’Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, Anderson MO, Derisi JL, McKerrow JH, Burlingame AL, Craik CS. 2012. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods 9:1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. 2009. Improved visualization of protein consensus sequences by iceLogo. Nat Methods 6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 47.Dunn BM, Hung S. 2000. The two sides of enzyme-substrate specificity: lessons from the aspartic proteinases. Biochim Biophys Acta 1477:231–240. doi: 10.1016/S0167-4838(99)00275-7. [DOI] [PubMed] [Google Scholar]
- 48.Waugh DS. 2011. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr Purif 80:283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alby K, Bennett RJ. 2011. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci U S A 108:2510–2515. doi: 10.1073/pnas.1017234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett RJ, Johnson AD. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol Microbiol 62:100–119. doi: 10.1111/j.1365-2958.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- 51.Schaller M, Korting HC, Schafer W, Bastert J, Chen W, Hube B. 1999. Secreted aspartic proteinase (sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol 34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 52.Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schafer W, Hube B. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun 70:3689–3700. doi: 10.1128/IAI.70.7.3689-3700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, Maccallum D, Odds FC, Schafer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem 281:688–694. doi: 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- 54.Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol 28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- 55.Koelsch G, Tang J, Loy JA, Monod M, Jackson K, Foundling SI, Lin X. 2000. Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim Biophys Acta 1480:117–131. doi: 10.1016/S0167-4838(00)00068-6. [DOI] [PubMed] [Google Scholar]
- 56.Schild L, Heyken A, de Groot PWJ, Hiller E, Mock M, de Koster C, Horn U, Rupp S, Hube B. 2011. Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell 10:98–109. doi: 10.1128/EC.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoki W, Kitahara N, Miura N, Morisaka H, Yamamoto Y, Kuroda K, Ueda M. 2012. Candida albicans possesses Sap7 as a pepstatin A-insensitive secreted aspartic protease. PLoS One 7:e32513. doi: 10.1371/journal.pone.0032513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staib P, Lermann U, Blass-Warmuth J, Degel B, Wurzner R, Monod M, Schirmeister T, Morschhauser J. 2008. Tetracycline-inducible expression of individual secreted aspartic proteases in Candida albicans allows isoenzyme-specific inhibitor screening. Antimicrob Agents Chemother 52:146–156. doi: 10.1128/AAC.01072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janiak AM, Sargsyan H, Russo J, Naider F, Hauser M, Becker JM. 2005. Functional expression of the Candida albicans alpha-factor receptor in Saccharomyces cerevisiae. Fungal Genet Biol 42:328–338. doi: 10.1016/j.fgb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Abel MG, Zhang YL, Lu HF, Naider F, Becker JM. 1998. Structure-function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine-scanned analogs. J Pept Res 52:95–106. doi: 10.1111/j.1399-3011.1998.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 61.Lee BK, Khare S, Naider F, Becker JM. 2001. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to alpha-factor pheromone binding. J Biol Chem 276:37950–37961. [DOI] [PubMed] [Google Scholar]
- 62.Naider F, Becker JM. 2004. The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides 25:1441–1463. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 63.Son CD, Sargsyan H, Naider F, Becker JM. 2004. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry 43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 64.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guthrie C, Fink GR. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA. [Google Scholar]
- 66.Liu H, Kohler J, Fink G. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 67.Reuss O, Vik Å, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 68.Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of mutant Bar1 protease. C. albicans Bar1(D232A) was overexpressed in Pichia pastoris, and the protein was concentrated and purified by nickel affinity chromatography. Flow, 10 mM imidazole. Washes, 20 mM imidazole. Elutions, 500 mM imidazole. (A) Total protein via Coomassie stain. (B) Western blotting using an anti-His6 antibody. Download
C. albicans BAR1 affects C. albicans white cells’ response to truncated α pheromone peptides. A biofilm assay was used to assess the response of white C. albicans cells to truncated versions of C. albicans α pheromone. After 48 h of incubation with the indicated amount of peptide, adherent cells were resuspended in medium and quantified by spectrometry at 600 nm. Mean ± SD. *, P < 0.05 by t test. Download
Strain table.