LETTER
The emergence of clades within emm89 Streptococcus pyogenes isolates that rapidly became the dominant lineages expressing this emm type was recently reported in the United Kingdom (1) and in a study that included isolates from the United States, Finland, and Iceland (United States/FI/IC) (2). In the United Kingdom, the emerging clade was associated with the absence of the hasABC locus, responsible for the synthesis of the hyaluronic acid capsule (1). The study from the United States/FI/IC (2) highlighted the strict association of the emerging clade with an nga promoter variant, also found in contemporary emm1 isolates, which results in increased expression of the nga locus. The study from the United Kingdom also examined this region and found that the nga-ifs-slo locus and surrounding sequences of the emerging clade shared 99% DNA identity with that of contemporary emm1 and emm12 strains, but the authors do not offer any information on the nga promoter (1). The acquisition of this region by emm1 isolates is currently considered the major molecular event triggering the success and enhanced virulence of this clone (3).
Given that the two studies characterized emm89 strains recovered in overlapping time periods, it was possible that the two were documenting the dissemination of the same clade in different geographic areas, although the information presented did not allow this conclusion since the papers analyze different aspects of the strains. We set out to test this hypothesis by reanalyzing the publicly available data, and we also characterized emm89 isolates recovered in Portugal to investigate if this clade could be also emerging in southern Europe. While analyzing this data, an additional paper was published focusing on the interplay between expression of the nga-ifs-slo locus and that of capsule in the virulence of the emm89 strains of the United States/FI/IC study (4).
We analyzed the sequencing reads deposited in public databases from the strains included in the two papers in order to determine the presence of the hasABC locus, the sequence types (STs), the variant of the nga promoter, and possible variations in the nga coding sequence. Briefly, the raw sequencing reads were mapped using Bowtie2 to exemplar sequences of each of the loci together with at least 600 bp of upstream and downstream sequence. A total of 907 strains met the quality standards that we required in our analysis in both the has and nga loci. Multilocus sequence typing (MLST) alleles could be confidently determined for 886 of these strains. A total of 79 isolates presented novel STs due to the presence of new alleles or allelic profiles, which were submitted to the S. pyogenes MLST database (http://pubmlst.org/spyogenes/) and assigned ST791 to ST804. In addition, we analyzed 125 emm89 isolates recovered in Portugal between 2000 and 2009, including 26 pharyngitis isolates and 37 invasive isolates which have been previously characterized (5, 6), as well as 62 isolates recovered from skin and soft tissue infections (SSTI) (unpublished data). The isolates were screened for the presence of the has locus using previously described primers (7). The nga gene and its promoter region were amplified and sequenced in a subset of 95 isolates for which MLST data were available.
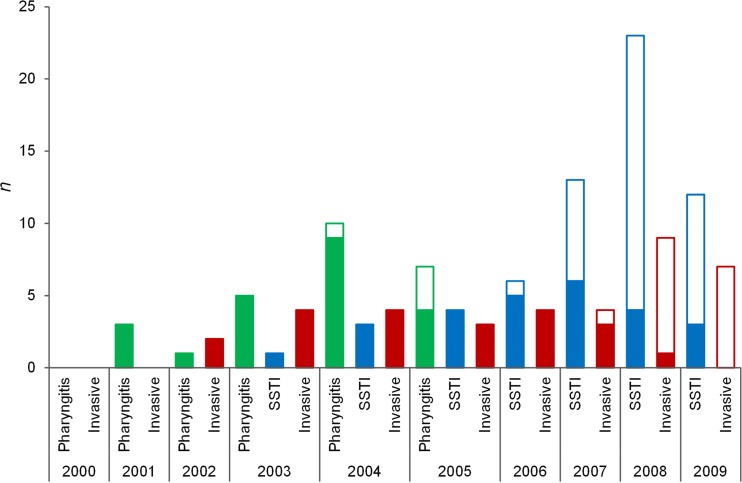
In the three datasets (United Kingdom, United States/FI/IC, and Portugal), all isolates carrying nga promoter variant 3 (2) lacked the hasABC locus (Table 1). Confirming our analysis, the absence of the hasABC locus in strains carrying nga promoter variant 3 in the United States/FI/IC strains was also independently reported (4). In contrast, all isolates harboring the capsular locus presented variant 1 or 2 of the nga promoter. Variant 3 isolates were predominantly of ST101 in all datasets (94%), although 4 United Kingdom isolates and 31 United States/FI/IC isolates presented single-locus variants (SLVs) of ST101. These results indicate that the same clade lacking the hyaluronic acid capsule and carrying an altered nga promoter region (designated emm89-new here) is present in the five countries. In Portugal, emm89-new was first detected among pharyngitis isolates (in 2004) followed by SSTI isolates (2006) and invasive isolates (2007) (Fig. 1) and also rapidly became dominant among emm89 strains, being associated with a significant increase in the incidence of this emm type among isolates from SSTI (data not shown).
TABLE 1 .
Characteristics of 1,002 emm89 strains isolated in the United States/FI/IC (n = 778), United Kingdom (n = 129), and Portugal (n = 125)
| Promoter variant (n) | nga allele (n)a | NADase allele | has locus | ST(s) (n)b |
|---|---|---|---|---|
| 1 (195) | 3 (1) | 2 | + | 407 (1) |
| 4 (2) | 2 | + | 380 (2) | |
| 5 (190) | 2 | + | 407 (135), 803 (44), 795 (2), 799 (1) | |
| 6 (1) | 3 | + | 407 (1) | |
| 11 (1) | 8 | + | 407 (1) | |
| 2 (168) | 7 (160) | 4 | +c | 101 (122), 408 (32), 568 (2), 553 (1), 797 (1) |
| 8 (1) | 5 | + | 101 (1) | |
| 9 (2) | 6 | + | 142 (2) | |
| 10 (2) | 7 | + | 382 (1) | |
| 12 (3) | 4 | + | 101 (3) | |
| 3 (639) | 1 (638) | 1 | − | 101 (593), 801 (22), 580 (3), 791 (2), 792 (1), 793 (1), 794 (1), 796 (1), 798 (1), 800 (1), 802 (1), 804 (1) |
| 2 (1) | 1 | − | 101 (1) |
Nucleotide sequences of the nga alleles can be found at http://dx.doi.org/10.6084/m9.figshare.1573013.
Sequence type information was obtained for a subset of 981 isolates.
The hasB gene was not detected in two isolates (one each from references 1 and 2).
FIG 1 .
Numbers of emm89 isolates with (filled bars) and without (open bars) the has locus recovered in Portugal between 2000 and 2009, classified by infection type. Pharyngeal isolates were studied only in the period of 2000 to 2005; skin and soft tissue infection (SSTI) isolates were studied only in the period of 2003 to 2009.
A total of 12 distinct nucleotide sequences were identified for the nga gene, corresponding to 8 protein variants. Since none included the G330D polymorphism, all variants are predicted to result in NADase proteins with detectable activity (2, 8). Regardless of geographic origin, each promoter variant was largely associated with a specific nga allele, with less than 5% of the isolates of each variant presenting other alleles (Table 1). However, differences were found between the dominant emm89 populations prior to the emergence of emm89-new in the three datasets. Variant 1 of the nga promoter was not found among United Kingdom strains and was identified in a single isolate from Portugal and in a minority of isolates from Finland and Iceland, while it was the dominant variant among the United States strains. All has-positive (has+) emm89 United Kingdom isolates belonged to ST101, while in the United States/FI/IC and Portugal isolates, eight and four other STs were identified, respectively. ST407 and ST408, both SLVs of ST101, were the most frequent STs among the has+ emm89 isolates in the United States/FI/IC study and in Portugal, respectively. The difference between the has+ emm89 populations of the United Kingdom and Portugal is further supported by differences in superantigen profiles, with the majority (49/69, 71%) of the isolates in Portugal carrying the speC gene, in contrast to the situation reported in the United Kingdom (10/48, 21%) (1). Isolates representing emm89-new presented the same dominant profile, including speC, speG, and smeZ in both countries, although other superantigen combinations were also identified, both in the United Kingdom (12/83, 14%) and in Portugal (7/56, 13%).
Taken together, these observations suggest that, in spite of the differences in the structures of the S. pyogenes emm89 populations previously circulating in different countries, the same clade disseminated and outcompeted other emm89 lineages over the first decade of the 21st century in all countries studied and possibly throughout North America and Europe. This successful clade is characterized by the absence of the hasABC locus and the presence of an nga-ifs-slo locus variant associated with increased expression of NADase and SLO, possibly acquired from emm1 or emm12 strains by horizontal gene transfer (1, 2). While virulence studies using animal models indicate an increased virulence of the emm89-new strains (2, 4), other in vitro studies suggest that the novel emm89-new phenotype may be advantageous for environmental persistence and transmission, resulting in an increased number rather than severity of emm89 infections (1). Indeed, epidemiological data from the United Kingdom and Portugal show that an increase in the incidence of disease-associated emm89 isolates has occurred since the emergence of emm89-new. Furthermore, the detection of emm89-new in Portugal occurred sooner among milder infections than in more severe disease, also consistent with an advantage of this lineage in persistence and transmission rather than increased virulence. Additional surveillance will contribute to clarify the relative importance of emm89-new in different types of infection. The emm89-new clade has most probably emerged in an ST101 genetic background, although a minority of isolates presenting SLVs of ST101 were identified. This possibly occurred a limited number of times or even on a single occasion given the extremely limited genetic diversity found among emm89-new isolates. From this one event or from a limited number of events occurring in an unidentified geographic location, emm89-new rapidly disseminated worldwide, displacing a number of other emm89 lineages. Still, the detection of SLVs of ST101, together with the diverse superantigen profiles detected, suggests that emm89-new is already undergoing limited diversification in each of the geographic locations, which may lead to further increases in virulence or transmissibility of this lineage.
Nucleotide sequence accession numbers.
Nucleotide sequences of the nga alleles can be found at http://dx.doi.org/10.6084/m9.figshare.1573013.
Footnotes
Citation Friães A, Machado MP, Pato C, Carriço J, Melo-Cristino J, Ramirez M. 2015. Emergence of the same successful clade among distinct populations of emm89 Streptococcus pyogenes in multiple geographic regions. mBio 6(6):e01780-15. doi:10.1128/mBio.01780-15.
REFERENCES
- 1.Turner CE, Abbott J, Lamagni T, Holden MTG, David S, Jones MD, Game L, Efstratiou A, Sriskandan S. 2015. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. mBio 6:e00622-15. doi: 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L, Olsen RJ, Nasser W, de la Riva Morales I, Musser JM. 2015. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. mBio 6:e01378-15. doi: 10.1128/mBio.01378-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J. 2012. Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. BMC Microbiol 12:280. doi: 10.1186/1471-2180-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friães A, Lopes JP, Melo-Cristino J, Ramirez M, Portuguese Group for the Study of Streptococcal Infections . 2013. Changes in Streptococcus pyogenes causing invasive disease in Portugal: evidence for superantigen gene loss and acquisition. Int J Med Microbiol 303:505–513. doi: 10.1016/j.ijmm.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. 2012. Human disease isolates of serotype M4 and M22 group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio 3:e00413-12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddle DJ, Bessen DE, Caparon MG. 2010. Variation in Streptococcus pyogenes NAD+ glycohydrolase is associated with tissue tropism. J Bacteriol 192:3735–3746. doi: 10.1128/JB.00234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]