ABSTRACT
Engineering industrial microorganisms for ambitious applications, for example, the production of second-generation biofuels such as butanol, is impeded by a lack of knowledge of primary metabolism and its regulation. A quantitative system-scale analysis was applied to the biofuel-producing bacterium Clostridium acetobutylicum, a microorganism used for the industrial production of solvent. An improved genome-scale model, iCac967, was first developed based on thorough biochemical characterizations of 15 key metabolic enzymes and on extensive literature analysis to acquire accurate fluxomic data. In parallel, quantitative transcriptomic and proteomic analyses were performed to assess the number of mRNA molecules per cell for all genes under acidogenic, solventogenic, and alcohologenic steady-state conditions as well as the number of cytosolic protein molecules per cell for approximately 700 genes under at least one of the three steady-state conditions. A complete fluxomic, transcriptomic, and proteomic analysis applied to different metabolic states allowed us to better understand the regulation of primary metabolism. Moreover, this analysis enabled the functional characterization of numerous enzymes involved in primary metabolism, including (i) the enzymes involved in the two different butanol pathways and their cofactor specificities, (ii) the primary hydrogenase and its redox partner, (iii) the major butyryl coenzyme A (butyryl-CoA) dehydrogenase, and (iv) the major glyceraldehyde-3-phosphate dehydrogenase. This study provides important information for further metabolic engineering of C. acetobutylicum to develop a commercial process for the production of n-butanol.
IMPORTANCE
Currently, there is a resurgence of interest in Clostridium acetobutylicum, the biocatalyst of the historical Weizmann process, to produce n-butanol for use both as a bulk chemical and as a renewable alternative transportation fuel. To develop a commercial process for the production of n-butanol via a metabolic engineering approach, it is necessary to better characterize both the primary metabolism of C. acetobutylicum and its regulation. Here, we apply a quantitative system-scale analysis to acidogenic, solventogenic, and alcohologenic steady-state C. acetobutylicum cells and report for the first time quantitative transcriptomic, proteomic, and fluxomic data. This approach allows for a better understanding of the regulation of primary metabolism and for the functional characterization of numerous enzymes involved in primary metabolism.
INTRODUCTION
Clostridium acetobutylicum is a Gram-positive, spore-forming anaerobic bacterium capable of converting various sugars and polysaccharides to organic acids (acetate and butyrate) and solvents (acetone, butanol, and ethanol). Due to its importance in the industrial production of the bulk chemicals acetone and butanol (1–3) and its potential use in the production of n-butanol, a promising biotechnology-based liquid fuel with several advantages over ethanol (4, 5), much research has focused on understanding (i) the regulation of solvent formation (6–13), (ii) the ability to tolerate butanol (14–17), and (iii) the molecular mechanism of strain degeneration in C. acetobutylicum (18, 19). The complete genome sequence of C. acetobutylicum ATCC 824 has been published (20), and numerous transcriptomic and proteomic studies have been performed to date (21–26). Although most of these transcriptomic studies have been performed using two-color microarrays (25, 27–29), RNA deep sequencing (RNA-seq) has recently been used, allowing a more accurate quantification of transcripts as well as the determination of transcription start sites and 5′ untranscribed regions (5′ UTRs) (17, 30). With regard to proteomic studies of C. acetobutylicum, 2-dimensional gel electrophoresis (2-DGE) (22, 24, 31–33) is typically employed. 2-DGE is popular and generates substantially valuable data; however, limitations of this method, such as low reproducibility, narrow dynamic range, and low throughput, remain (34). Recently, more quantitative approaches have been developed using two-dimensional liquid chromatography–tandem mass spectrometry (2D-LC-MS/MS) (35) or iTRAQ tags (36).
In general, transcriptomic and/or proteomic studies of C. acetobutylicum have been focused on understanding (i) the transcriptional program underlying spore formation (21, 23), (ii) the response or adaptation to butanol and butyrate stress (14–17), and (iii) the regulation of primary metabolism (21–23, 25, 35, 37).
Furthermore, to elucidate the molecular mechanisms of endospore formation, microarrays (21, 23) have been used extensively in combination with the downregulation of sigma factors by antisense RNA (23) or inactivation by gene knockout (38, 39). Initially, investigations of the response of C. acetobutylicum to butanol and butyrate stress employed microarrays (14–16) followed by RNA deep sequencing (RNA-seq) to quantify both mRNA and small noncoding RNAs (sRNA) (17), and quantitative transcriptomic and proteomic approaches were later combined (40). Based on one of these studies (16), regulons and DNA-binding motifs of stress-related transcription factors as well as transcriptional regulators controlling stress-responsive amino acid and purine metabolism and their regulons have been identified. Furthermore, integrative proteomic-transcriptomic analysis has revealed the complex expression patterns of a large fraction of the proteome that could be explained only by involving specific molecular mechanisms of posttranscriptional regulation (40).
The regulation of solvent formation in C. acetobutylicum has been extensively studied in batch cultures using transcriptomic (21, 23, 25) and/or proteomic (24, 35) approaches. Despite the valuable insights achieved in those studies, many physiological parameters, such as specific growth rates, specific glucose consumption rates, pH, and cellular differentiation, as well as butyrate and butanol stress, change with time, making it difficult to understand many details of the expression pattern.
In phosphate-limited chemostat cultures, C. acetobutylicum can be maintained in three different stable metabolic states (6, 8–10, 41) without cellular differentiation (37): acidogenic (producing acetate and butyrate) when grown at neutral pH on glucose, solventogenic (producing acetone, butanol, and ethanol) when grown at low pH on glucose, and alcohologenic (forming butanol and ethanol but not acetone) when grown at neutral pH under conditions of high NAD(P)H availability. Indeed, because the cells are maintained under steady-state conditions with constant endogenous and exogenous parameters such as a specific growth rate and specific substrate consumption rate, chemostat culture is the preferred fermentation method by which to achieve standardized conditions with a maximum degree of reproducibility. Transcriptional analysis of the transition from an acidogenic to a solventogenic state (37), as well as transcriptomic and proteomic analyses of acidogenic and solventogenic (22) phosphate-limited chemostat cultures, has already been performed using two-color microarrays for transcriptomic analysis and 2-DGE for proteomic analysis, methods that are semiquantitative. However, a systems biology approach, combining more than two quantitative “omic” analyses of chemostat cultures of C. acetobutylicum, has never been performed.
Therefore, the aim of this study was to apply a quantitative system-scale analysis to acidogenic, solventogenic, and alcohologenic steady-state C. acetobutylicum cells to provide new insight into the metabolism of this bacterium. We first developed an improved genome-scale model (GSM), including a thorough biochemical characterization of 15 key metabolic enzymes, to obtain accurate fluxomic data. We then applied quantitative transcriptomic and proteomic approaches to better characterize the distribution of carbon and electron fluxes under different physiological conditions and the regulation of C. acetobutylicum metabolism.
RESULTS AND DISCUSSION
Improving upon current GSMs for metabolic flux analysis.
The iCac967 model for C. acetobutylicum ATCC 824 spans 967 genes and includes 1,058 metabolites participating in 1,231 reactions (Table 1; also see Data Set S1 in the supplemental material). All reactions are elementally and charge balanced. The iCac967 model is the result of an extensive literature analysis associated with the biochemical characterization of many key metabolic enzymes in an attempt to better understand the distribution of carbon and electron fluxes. The previously uncharacterized butyryl coenzyme A (butyryl-CoA) dehydrogenase (BCD) encoded by bcd-etfB-etfA (CA_C2711, CA_C2710, and CA_C2709, respectively) (42) was biochemically characterized via homologous expression of the encoding operon in C. acetobutylicum and the purification of the enzyme complex (Table 2; see also Fig. S1). We demonstrated that the butyryl-CoA dehydrogenase of C. acetobutylicum is a strictly NADH-dependent enzyme and that ferredoxin is needed for the reaction to proceed. To study the stoichiometry of the reaction, the concentrations of NADH (see Fig. S1A) and crotonyl-CoA (see Fig. S1B) were modulated using constant concentrations of purified ferredoxin (CA_C0303) and hydrogenase (CA_C0028). Based on the initial slope in Fig. S1B in the supplemental material, it was calculated that in the presence of excess crotonyl-CoA, 2.15 mol of NADH was required for the formation of 1 mol of H2; from the initial slope in Fig. S1A in the supplemental material, it was calculated that in the presence of excess NADH, 1.25 mol of crotonyl-CoA was required for the formation of 1 mol of H2. The results indicate that under fully coupled conditions, approximately 1 mol of ferredoxin is reduced by 2 mol of NADH and 1 mol of crotonyl-CoA, similar to the butyryl-CoA dehydrogenase of Clostridium kluyveri (43). Although the possibility that this enzyme might consume 2 mol of NADH and produce 1 mol of reduced ferredoxin in C. acetobutylicum was previously presented as a hypothesis (44), it has not been demonstrated to date, nor has it been integrated in the recently published GSMs (45, 46). This result has strong implications for the distribution of electron fluxes, as discussed below in the metabolic flux analysis section.
TABLE 1 .
Comparison of GSMs of C. acetobutylicuma
| Model statistic | No. of genes, reactions, or metabolites in GSM: |
||||
|---|---|---|---|---|---|
| Senger et al. (56, 57) | Lee et al. (58) | McAnulty et al. (46) | Dash et al. (45) | iCac967 | |
| Genes | 474 | 432 | 490 | 802 | 967 |
| Reactions | 552 | 502 | 794 | 1,462 | 1,231 |
| Metabolites | 422 | 479 | 707 | 1,137 | 1,058 |
The numbers of genes, reactions, and metabolites present in four previous GSMs of C. acetobutylicum and iCac967 are shown.
TABLE 2 .
Activities of purified key metabolic enzymes
| Locus no. | Gene name | Enzyme activity | Activity (U/mg)a |
|---|---|---|---|
| CA_C3299 | bdhA | Butanol dehydrogenase | NADH (0.15 ± 0.05), NADPH (2.57 ± 0.45) |
| CA_C3298 | bdhB | Butanol dehydrogenase | NADH (0.18 ± 0.02), NADPH (2.95 ± 0.36) |
| CA_C3392 | bdhC | Butanol dehydrogenase | NADH (0.24 ± 0.04), NADPH (2.21 ± 0.41) |
| CA_P0162 | adhE1 | Butanol dehydrogenase | NADH (0.04 ± 0.02), NADPH (not detected) |
| CA_P0035 | adhE2 | Butanol dehydrogenase | NADH (4.8 ± 0.42), NADPH (0.12 ± 0.01) |
| CA_P0162 | adhE1 | Butyraldehyde dehydrogenase | NADH (2.27 ± 0.21), NADPH (0.08 ± 0.01) |
| CA_P0035 | adhE2 | Butyraldehyde dehydrogenase | NADH (2.5 ± 0.31), NADPH (0.07 ± 0.01) |
| CA_C2711-CA_C2709 | bcd-etfB-etfA | Butyryl-CoA dehydrogenase | NADH (0.569 ± 0.08), NADPH (not detected) |
| CA_C1673-CA_C1674 | gltA/gltB | Glutamate synthase | NADH (0.61 ± 0.16), NADPH (0.051 ± 0.01) |
| CA_C0737 | gdh | Glutamate dehydrogenase | NADH (41.2 ± 3.4), NADPH (0.12 ± 0.01) |
| CA_C0970 | citA | Re-citrate synthase | 1.9 ± 0.14 |
| CA_C0971 | citB | Aconitase | 6.5 ± 0.52 |
| CA_C0972 | citC | Isocitrate dehydrogenase | NADH (104 ± 6.8), NADPH (7.1 ± 0.43) |
| CA_C1589 | malS1 | Malic enzyme | NADH (156 ± 9.6), NADPH (3.4 ± 0.24) |
| CA_C1596 | malS2 | Malic enzyme | NADH (142 ± 12.7), NADPH (2.9 ± 0.34) |
One unit is the amount of enzyme that consumes 1 µmol of substrate per min.
The second key enzyme that remained uncharacterized was the bifunctional alcohol-aldehyde dehydrogenase (AdhE1 or Aad, encoded by CA_P0162), an enzyme involved in the last two steps of butanol and ethanol formation during solventogenic culturing of C. acetobutylicum (47, 48). First, adhE1 and adhE2 (as a positive control) were individually heterologously expressed in Escherichia coli, after which AdhE1 and AdhE2 were purified as tag-free proteins (Table 2) for biochemical characterization. We demonstrated that in vitro, AdhE1 possesses high NADH-dependent butyraldehyde dehydrogenase activity but surprisingly very low butanol dehydrogenase activity with both NADH and NADPH; in contrast, AdhE2 possesses both high butyraldehyde and butanol dehydrogenase activities with NADH. The three potential alcohol dehydrogenases, BdhA, BdhB, and BdhC (49), encoded by bdhA, bdhB, and bdhC (CA_C3299, CA_C3298, and CA_C3392), respectively, were heterologously expressed in E. coli and then characterized after purification as tag-free proteins (Table 2). The three enzymes were demonstrated to be primarily NADPH-dependent butanol dehydrogenases, results which do not agree with the previous characterizations of BDHI and BDHII (later demonstrated to be encoded by bdhA and bdhB), which were reported to be NADH dependent (49, 50). However, in agreement with our findings, all of the key amino acids of the two GGGS motifs at positions 37 to 40 and 93 to 96 involved in the NADPH binding of YqhD, a strictly NADPH-dependent alcohol dehydrogenase (51), are perfectly conserved in the three C. acetobutylicum alcohol dehydrogenases. Furthermore, these results are also in line with previously published data from two different research groups (9, 52) showing that in a crude extract of solventogenic C. acetobutylicum cultures, the butanol dehydrogenase activity measured in the physiological direction is mainly NADPH dependent. As discussed below, C. acetobutylicum must utilize at least one of these alcohol dehydrogenases to produce butanol and ethanol under solventogenic conditions, which implies that 1 mol of NADPH is needed for each mole of butanol and ethanol produced under solventogenic conditions.
The cofactor specificity of the ammonium assimilation pathway that proceeds via glutamine 2-oxoglutarate aminotransferase (GOGAT) encoded by gltA and gltB (CA_C1673 and CA_C1674, respectively) and glutamate dehydrogenase (GDH) encoded by gdh (CA_C0737) was also characterized. The gltA-gltB and gdh genes were expressed in C. acetobutylicum and E. coli, respectively, and GOGAT and GDH were purified (Table 2). Both enzymes were found to be NADH dependent, in contrast to the corresponding enzymes in E. coli, which are NADPH dependent (53, 54).
The functions of the three genes (CA_C0970, CA_C0971, and CA_C0972) proposed (55) to encode the first three steps of the oxidative branch of the tricarboxylic acid (TCA) cycle were unambiguously characterized. CA_C0970, CA_C0971, and CA_C0972 were individually expressed in E. coli, and their gene products were purified (Table 2); the genes were demonstrated to encode an Re-citrate synthase (CitA), an aconitase (CitB), and an NADH-dependent isocitrate dehydrogenase (CitC), respectively.
Finally, we characterized the cofactor specificity of the two malic enzymes encoded by CA_C1589 and CA_C1596, two almost-identical genes that differ by only two nucleotides. Not surprisingly, the specific activities of the two purified enzymes are almost identical, and both enzymes are NADH dependent (Table 2).
The iCac967 model statistics and those of all other published models for C. acetobutylicum (45, 46, 56–58) are shown in Table 1. iCac967 has 20% more genes than the most recently published model by Dash et al. (45) but fewer metabolites and reactions, as some reactions described by these authors were not validated by our extensive literature analysis or were inappropriate in the context of anaerobic metabolism, for example, R0013 (NADPH + O2 + H+ + 2-octaprenylphenol → H2O + NADP+ + 2-octaprenyl-6-hydroxyphenol) and R0293 (H2O + O2 + sarcosine → H2O2 + glycine + formaldehyde). Furthermore, we applied our GSM to the butyrate kinase knockout mutant (59) and the M5 degenerate strain (60) (which has lost the pSOL1 plasmid) and successfully predicted their phenotypes (see Table S1 in the supplemental material).
Quantitative transcriptomic and proteomic analyses of C. acetobutylicum under stable acidogenic, solventogenic, and alcohologenic conditions. (i) General considerations.
Quantitative transcriptomic and proteomic analyses were performed on phosphate-limited chemostat cultures of C. acetobutylicum maintained in three different stable metabolic states: acidogenic, solventogenic, and alcohologenic (6, 7, 9, 10). The total amount of DNA, RNA, and protein contents (expressed in grams per gram of dry cell weight [DCW]) and the number of cells per gram of DCW were experimentally determined for each steady-state condition under phosphate limitation at a dilution rate of 0.05 h−1. These numbers were not significantly different among the steady-state conditions, in agreement with previous studies (61, 62) on E. coli that have shown that the biomass composition is not dependent on the carbon source but is strictly dependent on the specific growth rate. According to all of the values, the average contents of DNA (1.92 ± 0.03), mRNA ([9.41 ± 0.94] × 103), and protein ([6.26 ± 0.18] × 106) molecules per cell were calculated. Noticeably, the total number of mRNA molecules per cell was only 2.4 times higher than the total number of open reading frames (ORFs) (3,916). In E. coli, the situation was even worse, with a total number of mRNA molecules per cell (1,380) 3 times lower than the total number of ORFs (4,194) (61).
For each gene, we sought to quantify the number of mRNA molecules per cell. For this purpose, we used Agilent’s one-color microarray-based gene expression analysis, as a recent study (63) demonstrated a linear relationship between the amounts of transcript determined by this method and those determined by the RNA-seq method. The minimum number of mRNA molecules per cell detected was around 0.06 while the maximum number was around 80. It was observed that a large number of genes have less than 0.2 mRNA molecule per cell (for 37.1%, 36.8%, and 37.2% of the genes under acidogenic, solventogenic, and alcohologenic conditions, respectively). This result indicates that for these genes, there is either (i) heterogeneity among different cells, such that some cells contain one transcript and others do not, or (ii) a high mRNA degradation rate. Genes that showed a value of mRNA molecules per cell of <0.2 under all three conditions were excluded from further analysis.
The purpose of this study was also to quantify the number of cytoplasmic protein molecules per cell. Different quantitative methods using either 2D-protein gels (26) or peptide analysis by two-dimensional high-performance liquid chromatography (2D HPLC) coupled with tandem mass spectrometry (MS/MS) with peptide labeling (36, 40) have been developed for C. acetobutylicum. In collaboration with the Waters Company, we adapted a recently published method (64) using label-free peptide analysis after shotgun trypsin hydrolysis of cytosolic proteins. For approximately 700 cytosolic proteins, it was possible to quantify the number of protein molecules per cell in at least one of the three steady states. This number is approximately 4 times higher than the number of cytosolic proteins detected in phosphate-limited acidogenic and solventogenic chemostat cultures by Janssen et al. (22) but similar to the number of cytosolic proteins detected by Venkataramanan et al. (40) by iTRAQ. Furthermore, the minimum number of protein molecules per cell detected was around 200 while the maximum number was approximately 300,000. For 96% of the cytosolic proteins that could be quantified, a linear relationship was obtained, with an R2 value of >0.9, when the numbers of protein molecules per cell were plotted against the numbers of mRNA molecules per cell (see Data Set S2 in the supplemental material). This result indicated that for steady-state continuous cultures run at the same specific growth rate and with the same total amount of carbon supplied, the rate of protein turnover is proportional to the mRNA content for 96% of the genes. This result is not necessarily surprising, as it has previously been shown for other microorganisms such as E. coli (65) that the numbers of ribosomes and tRNAs per cell are dependent on the specific growth rate and not on the carbon source. The absolute protein synthesis rates for approximately 700 genes were calculated by assuming that the rate of protein degradation is negligible compared to the rate of protein synthesis (see Data Set S2). These values varied from 0.0007 s−1 for CA_C3723 (ssb encoding a single-stranded DNA-binding protein) to 0.95 s−1 for CA_C0877 (cfa encoding a cyclopropane fatty acid synthase). Interestingly, the rate of protein synthesis appears to correlate inversely with the average number of mRNA molecules per cell (see Data Set S2).
(ii) Comparison of solventogenic versus acidogenic steady-state cells.
Solventogenic cells were first comprehensively compared to acidogenic cells via quantitative transcriptomic and proteomic analyses. The complete transcriptomic and proteomic results are provided in Data Set S2 in the supplemental material. A similar study in phosphate-limited chemostat cultures was previously performed by Janssen et al. (22) using semiquantitative transcriptomic (two-color microarrays) and proteomic (2-DGE) methods. Among the 95 genes shown by Janssen et al. to be upregulated, we qualitatively confirmed upregulation for 68; among the 53 genes shown by Janssen et al. to be downregulated, we qualitatively confirmed downregulation for 27. What might explain the differences between the two studies? First, the culture conditions were slightly different in terms of dilution rate (0.075 h−1 for the work of Janssen et al., 0.05 h−1 in our study), phosphate limitation (0.5 mM for the work of Janssen et al., 0.7 mM in our study), and the pH of the acidogenic culture (5.7 for the work of Janssen et al., 6.3 in our study), leading to a larger amount of glucose consumed and thus a larger amount of products formed in our study. We are confident regarding the validity of our results because we found agreement quantitatively with the transcriptomic data whenever proteins were detected by our method, and thus, quantitative proteomic data were available. Below, we discuss these data in more detail, and striking differences in mRNA molecules per cell are highlighted in Fig. S2A in the supplemental material.
In total, 64 genes matched the significance criteria of ≥4.0-fold-higher expression in solventogenesis than in acidogenesis as well as >0.2 mRNA molecules per cell under at least one of the two conditions (see Table S2 in the supplemental material). In particular, high values (~80- to 150-fold) were documented for the sol operon genes (CA_P0162-CA_P0164) and confirmed by the proteomic analysis, in agreement with (i) the requirement of AdhE1 and CoA-transferase subunits for the production of solvents under solventogenic conditions (12, 47, 48, 66) and (ii) the previous study by Janssen et al. (22). Elevated upregulation (4- to 40-fold) of genes involved in serine biosynthesis (CA_C0014-CA_C0015), seryl-tRNA synthesis (CA_C0017), and arginine biosynthesis (CA_C2388) was detected at the mRNA level and confirmed by the proteomic analysis, in agreement with a previous metabolomic study in batch culture (67), which reported higher intracellular concentrations of serine and arginine in solventogenic cells. Interestingly, all these genes were previously shown to be upregulated in response to butanol stress (16), although these results were not confirmed by proteomic analysis (40). In addition, an ~4- to 8-fold upregulation of genes involved in purine biosynthesis (CA_C1392-CA_C1395, CA_C1655, and CA_C2445) was detected at the mRNA level and confirmed by the proteomic analysis. Similar to the results in the study by Janssen et al. (22), an ~5-fold upregulation of a gluconate dehydrogenase (CA_C2607) was detected; however, as this protein was not detected, this was not confirmed by proteomic analysis.
As reported in previous studies (22, 37), we found elevated upregulations (~4- to 16-fold) of the genes involved in the production of (i) a nonfunctional cellulosome (CA_C0910-CA_C0918 and CA_C0561) (20, 68) and (ii) noncellulosomal pectate lyase-encoding genes (CA_P0056 and CA_C0574) at the mRNA level. However, these results could not be verified by proteomic analysis, as exoproteome analysis was not performed in this study. All these genes, except CA_P0056, were also shown to be upregulated in response to a butanol stress (16).
Importantly, spo0A (CA_C2071), encoding a regulator of sporulation and solvent production (69–71), showed an increase in expression at the level of both mRNA and protein molecules per cell. This increased expression does not agree with previous chemostat culture studies by Grimmler et al. (37) and Janssen et al. (22) but does agree with batch culture studies (21, 25) and also supports the common notion of Spo0A acting as a master regulator of solventogenesis. hsp18 (CA_C3714), encoding a gene product involved in solvent tolerance (72), also exhibited an ~4.5-fold increase in mRNA and protein molecules per cell, in agreement with a previous butanol stress study (40). A striking difference between the study by Janssen et al. and ours was observed with regard to the level of this chaperone, which in contrast to our study showing an ~4.5-fold increase under solventogenesis, was decreased (~5-fold) in the study by Janssen et al. (22). Nonetheless, this difference appears to be due to the limitation of 2-DGE, because 3 different proteins could be detected in the “Hsp18 spot” and transcriptional changes in hsp18 did not correlate with the proteomic data (22); in contrast, our quantitative transcriptomic and proteomic data showed good correlation (R2 > 0.9).
The detailed results of the 45 ORFs that exhibited ≥4.0-fold decreases in numbers of mRNA molecules per cell under solventogenic versus acidogenic conditions and of a number with >0.2 mRNA molecules per cell under at least one of the two conditions are given in Table S2 in the supplemental material. Significantly, in this metabolic state, various genes involved in the assimilation of different carbon sources were downregulated. For example, the highest decrease (~6- to 70-fold) at the mRNA level was observed for genes (CA_C0422-CA_C0426) involved in sucrose transport, metabolism, and the regulation of these genes, which was confirmed by the proteomic analysis. In addition, two genes involved in mannan (CA_C0332) and maltose (CA_C0533) metabolism exhibited 4- and 10-fold decreases, respectively, in their mRNA levels. Because acidogenic culture reached glucose limitation but a small amount of glucose remained in solventogenic culture (similar to our previous publication [9]), this phenomenon can be explained by a release of catabolite repression in acidogenic cultures. The similar high expression observed for CA_C0422-CA_C0426, CA_C0332, and CA_C0533 in alcohologenic and acidogenic cultures that were glucose limited is in agreement with this hypothesis. Two genes located on the megaplasmid pSOL1 (CA_P0036 and CA_P0037), encoding a cytosolic protein of unknown function and a potential transcriptional regulator, respectively, exhibited particularly high scores corresponding to an ~60- to 70-fold decrease, which is in good agreement with the proteomic data and the previous study by Janssen et al. (22). Interestingly, under all conditions, these two proteins are present at a 1:1 molar ratio. Furthermore, three genes involved in cysteine (CA_C2783) and methionine (CA_C1825 and CA_C0390) biosynthesis exhibited ~5-fold decreases in their numbers of mRNA and protein molecules per cell in agreement with a previous metabolomics study by Amador-Noguez et al. (67), showing an ~5-fold decrease in intracellular methionine in solventogenesis.
(iii) Comparison of alcohologenic versus acidogenic steady-state cells.
Alcohologenic cells were comprehensively compared to acidogenic cells by quantitative transcriptomic and proteomic analyses. The complete transcriptomic results are listed in Data Set S2, and striking differences are highlighted in Fig. S2B, both in the supplemental material. In total, 52 genes matched the significance criterion of ≥4.0-fold-higher expression in alcohologenesis than in acidogenesis as well as >0.2 mRNA molecules per cell under at least one of the two conditions (see Table S3). In particular, high values (~55- to 520-fold) were documented for the gene cluster coding for glycerol transport and utilization (CA_C1319-CA_C1323) and confirmed by the proteomic analysis, in agreement with the requirement of GlpK (glycerol kinase) and GlpAB (glycerol-3-phosphate dehydrogenase) for glycerol utilization in alcohologenic metabolism (6, 9). High upregulation (160-fold) of adhE2 (CA_P0035), which is involved in alcohol production under alcohologenic conditions (66), was detected and correlated with a high AdhE2 protein concentration. Interestingly, CA_C3486, which encodes a multimeric flavodoxin, was also highly expressed (~6-fold) and may participate in redistribution of the electron flux in favor of butanol under alcohologenic conditions. Of note, ~20- to 70-fold upregulation of a gene cluster involved in sulfate transport, reduction, and incorporation to produce cysteine (CA_C0102-CA_C0110); ~4-fold upregulation of cysK (CA_C2235), which is also involved in cysteine synthesis; and ~7- to 10-fold upregulation of an operon (CA_C3325-CA_C3327) involved in cysteine transport were detected at the mRNA level and confirmed by the proteomic analysis for the cytosolic proteins detected (CA_C0102-CA_C0104, CA_C0107, CA_C0109-CA_C0110, CA_C2235, and CA_C3327). All of these genes/operons were shown to possess a CymR-binding site in their promoter regions, and some have been shown to be upregulated in response to butanol stress (16).
An ~3- to 5-fold upregulation of an operon involved in histidine synthesis and histidyl-tRNA synthesis (CA_C0935-CA_C0943) and 5-fold upregulation of a gene involved in arginine biosynthesis (CA_C2388) were also detected at the mRNA level and confirmed by the proteomic analysis. These genes were also shown to be upregulated under solventogenic conditions (this study) and in response to butanol stress (16).
The detailed results of the 64 ORFs that exhibited a ≥4.0-fold decrease in transcript levels under alcohologenic versus acidogenic conditions and >0.2 mRNA molecules per cell under at least one of the two conditions are given in Table S3 in the supplemental material. The highest decrease (~70-fold) at the mRNA level was observed for an operon (CA_C0427-CA_C0430) involved in glycerol-3-phosphate transport and coding for a glycerophosphoryl diester phosphodiesterase, which was confirmed by the cytosolic protein analysis. As observed under solventogenic conditions, CA_P0036 and CA_P0037 exhibited ~40- to 50-fold-lower expression levels, which agrees well with the proteomic data. Furthermore, an operon involved in phosphate uptake (CA_C1705-CA_C1709), an operon encoding an indolepyruvate ferredoxin oxidoreductase (CA_C2000-CA_C2001), and a gene encoding a pyruvate decarboxylase (CA_P0025) exhibited ~80- to 350-fold, ~4- to 5-fold, and ~4-fold decreases, respectively, at the mRNA level, confirmed by the proteomic analysis. Additionally, two clusters of genes involved in fatty acid biosynthesis/degradation (CA_C2004-CA_C2017) exhibited ~3.5- to 6-fold decreases at the mRNA level, a result that could not be confirmed by the proteomic analysis as the corresponding proteins were not detected.
Metabolic flux analysis of C. acetobutylicum under stable acidogenic, solventogenic, and alcohologenic conditions.
To perform a metabolic flux analysis of C. acetobutylicum under stable acidogenic, solventogenic, and alcohologenic conditions, iCac967 was combined with our transcriptomic and proteomic data. As a first simple example, we present how the gene responsible for pyruvate ferredoxin oxidoreductase (PFOR) activity was identified. This gene encodes a key enzyme in the glycolytic pathway that decarboxylates pyruvate to produce reduced ferredoxin, CO2, and acetyl-CoA. Two putative PFOR-encoding genes (CA_C2229 and CA_C2499) were identified in our GSM (see Data Set S1 in the supplemental material). Under all conditions, only CA_C2229 was transcribed (average of 56 mRNA molecules per cell) and translated (average of 166,000 protein molecules per cell).
As a second simple example, we present how the main enzyme responsible for crotonyl-CoA reduction to butyryl-CoA was identified. Two different enzymes can potentially catalyze this reaction: the BCD complex encoded by bcd, etfB, and etfA (CA_C2711, CA_C2710, and CA_C2709, respectively) which consumes 2 moles of NADH and produces 1 mole of reduced ferredoxin (see Fig. S1 in the supplemental material) and TER (trans-2-enoyl-CoA reductase) encoded by CA_C0642, which consumes only 1 mole of NADH (73). Under all conditions, bcd was much more highly transcribed than CA_C0642 (67 versus 1.2 mRNA molecules per cell) and in terms of proteins BCD was detected at higher levels (average of 113,000 protein molecules per cell), whereas TER was below the detection limit of the method.
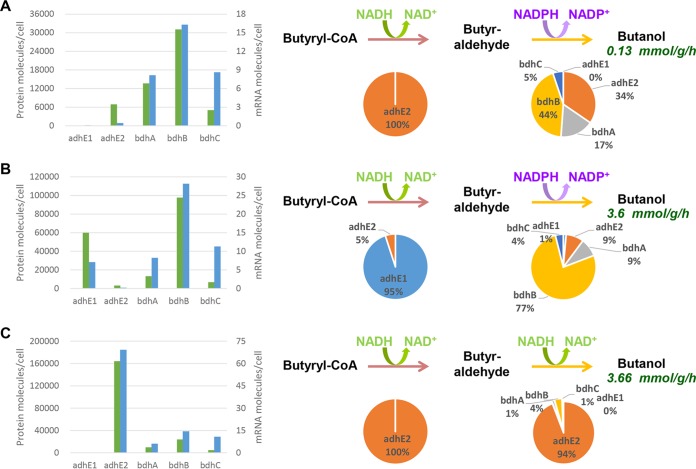
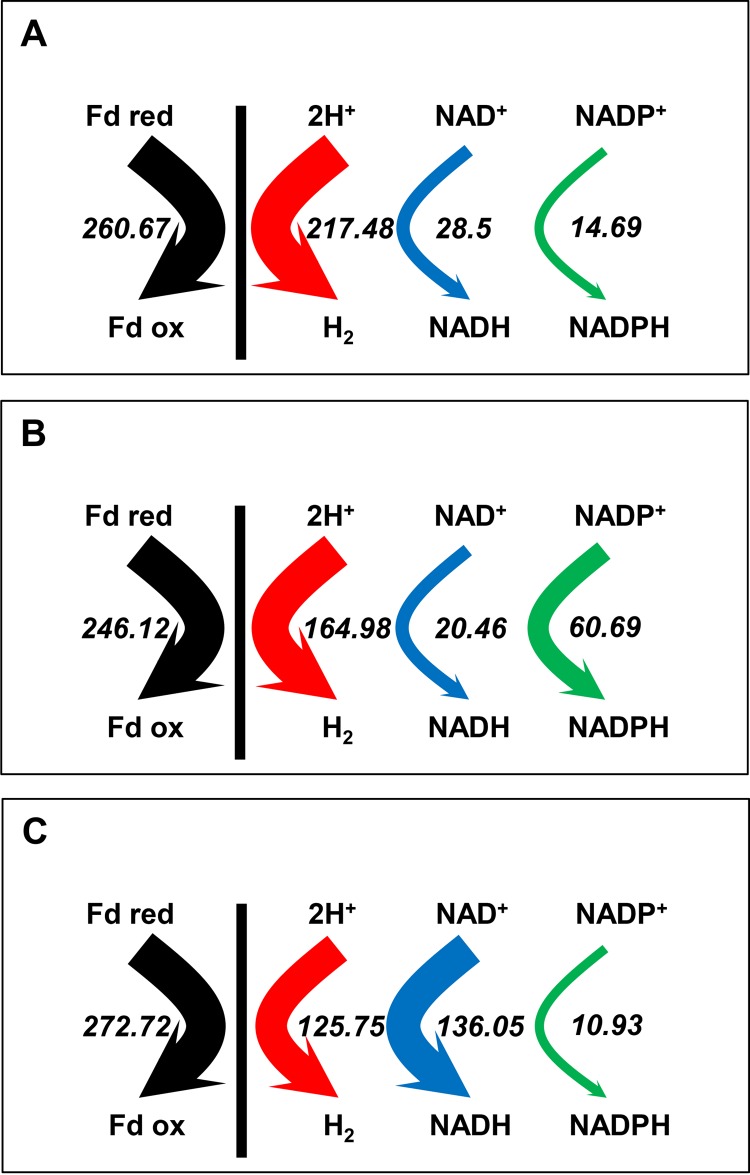
As a complex example, we also present the actors in the different butanol pathways and their cofactor specificities. Five proteins could potentially be involved in the last two steps of butanol formation. AdhE1 retains only NADH-dependent aldehyde dehydrogenase activity, whereas AdhE2 is a bifunctional NADH-dependent aldehyde-alcohol dehydrogenase (66); BdhA, BdhB, and BdhC are NADPH-dependent alcohol dehydrogenases. For each of the three conditions and for each of the abovementioned genes and their corresponding proteins, the number of mRNA molecules per cell and the number of protein molecules per cell were measured. The percentage of the total butanol flux due to each of the five enzymes was calculated by assuming that all five enzymes function at their Vmax and using the amount of each protein per cell. The results are presented in Fig. 1. Under acidogenic conditions, the entire butyraldehyde dehydrogenase flux is due to AdhE2, whereas the butanol dehydrogenase flux is primarily due to BdhB and BdhA. Under solventogenic conditions, the butyraldehyde dehydrogenase flux is largely due to AdhE1, whereas the butanol dehydrogenase flux is primarily due to BdhB, BdhA, and BdhC, in decreasing order of activity. Finally, under alcohologenic conditions, all of the flux of butyraldehyde dehydrogenase activity and most of that of butanol dehydrogenase activity are due to AdhE2. In summary, the last two steps of butanol production consume 1 mole of NADH and 1 mole of NADPH under acidogenic and solventogenic conditions and 2 moles of NADH under alcohologenic conditions (Fig. 1). These results have strong implications for the distribution of electron fluxes and the use of reduced ferredoxin under the respective studied conditions. Under acidogenic conditions, reduced ferredoxin is primarily used to produce hydrogen, and only a small fraction is used to produce the NADH needed for butyrate formation and the NADPH needed for anabolic reactions (Fig. 2A). However, under alcohologenic conditions, reduced ferredoxin is primarily used to produce the NADH needed for alcohol formation (Fig. 2C); under solventogenic conditions, although reduced ferredoxin is predominantly utilized for hydrogen production, a significant amount is used for the NADPH formation needed for the final step of alcohol formation by BdhB, BdhA, and BdhC, as C. acetobutylicum has no oxidative pentose phosphate pathway (zwf, encoding glucose 6-phosphate-dehydrogenase, is absent) to produce NADPH (Fig. 2B and 3). Although the enzymes converting reduced ferredoxin to NADPH or NADH, namely, ferredoxin-NADP+ reductase and ferredoxin-NAD+ reductase, and their corresponding genes are unknown, they likely play key roles in alcohol formation under solventogenic and alcohologenic conditions, respectively.
FIG 1 .
Butanol pathway analysis under acidogenesis (A), solventogenesis (B), and alcohologenesis (C). (Left) Numbers of mRNA (blue) and protein (green) molecules per cell for the five enzymes potentially involved in butanol production. (Right) Activity distributions of the five enzymes are shown for each step under the arrows. The primary cofactors used for each step are shown over the arrows. Butanol flux is indicated under the word “Butanol.”
FIG 2 .
Electron flux map: acidogenesis (A), solventogenesis (B), and alcohologenesis (C). The hydrogenase (red), ferredoxin-NAD+ reductase (blue), and ferredoxin-NADP+ (green) in vivo fluxes are presented. All values are normalized to the flux of the initial carbon source (millimoles per gram [DCW] per hour). Glucose flux is normalized as 100 for acidogenesis and solventogenesis, and the sum of glucose and half of the glycerol is normalized as 100 for alcohologenesis.
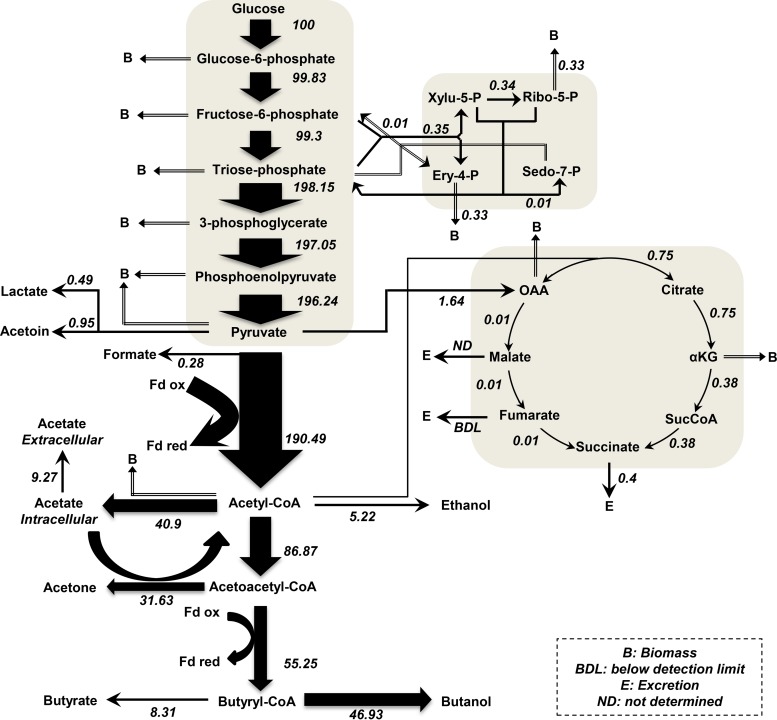
FIG 3 .
Metabolic flux map of C. acetobutylicum in solventogenesis. All values are normalized to the flux of the initial carbon source, glucose (millimoles per gram of DCW per hour). Metabolic flux maps of C. acetobutylicum in acidogenesis and in alcohologenesis are presented in Fig. S3 in the supplemental material.
A fourth example of metabolic flux analysis is the identification of the hydrogen production pathway. Three hydrogenases are potentially involved: two Fe-Fe hydrogenases, HydA (encoded by CA_C0028) and HydB (encoded by CA_C3230), and one Ni-Fe hydrogenase, HupSL (encoded by CA_P0141-CA_P0142). The hydB and the hupSL genes are not expressed under all three conditions, nor were the HydB and HupSL proteins detected by quantitative proteomic analysis. As HydA is the only hydrogenase present, how can the lower observed flux in H2 production under solventogenic and alcohologenic conditions (compared to acidogenic conditions) be explained? Under solventogenic conditions, there is a 3-fold decrease in the expression of hydA; this is associated with a 2-fold decrease in the expression of fdx1 (CA_C0303), which encodes the primary ferredoxin, the key redox partner for the hydrogenase. As these results were confirmed by the proteomic analysis, they may explain the 1.3-fold decrease in H2 production under solventogenic conditions compared to acidogenic conditions (Fig. 2B). Nonetheless, under alcohologenic conditions a 1.7-fold decrease in H2 production (compared to acidogenic conditions) is associated with a 1.8-fold-higher expression of hydA, a 3-fold decrease in the expression of fdx1, and a 6-fold increase in the expression of CA_C3486, which encodes a multimeric flavodoxin, another potential redox partner for the hydrogenase. In fact, the reduced multimeric flavodoxin may be a better substrate for the ferredoxin-NAD+ reductase than for the primary hydrogenase, as was previously shown for reduced neutral red (8). This result would explain the low flux in hydrogen production and the high flux in ferredoxin-NAD+ reductase production under alcohologenic metabolism obtained through growth either in glucose-glycerol mixtures or in glucose in the presence of neutral red (8).
A fifth example of metabolic flux analysis is the glyceraldehyde-3-phosphate oxidation pathway. Two glyceraldehyde-3-phosphate dehydrogenases are potentially involved: GapC (encoded by CA_C0709) (74), which phosphorylates and produces NADH, and GapN (encoded by CA_C3657) (75), which is nonphosphorylating and produces NADPH. For each of the three conditions and each of the genes studied, the numbers of mRNA molecules and protein molecules per cell were measured. The percentage of the total glycolytic flux due to each of the enzymes was calculated by assuming that both enzymes function at their previously published Vmax levels (74, 75) and using the amount of each protein per cell. Here, results are presented for only solventogenic metabolism, though qualitatively, the conclusions were the same for all conditions: gapN is poorly expressed compared to gapC (0.56 versus 66 mRNA molecules per cell; 3,500 versus 190,000 protein molecules per cell) (see Data Set S2 in the supplemental material), and GapN would be responsible for less than 5% of the glycolytic flux.
Two fluxes involved in anaplerotic reactions, namely, those for pyruvate carboxylase (encoded by CA_C2660) and NADH-dependent malic enzymes (encoded by CA_C1589 and CA_C1596), could not be solved using our GSM analysis coupled with transcriptomic and proteomic analyses. All of the genes studied were transcribed and translated under all conditions, and because all fermentations occurred under a high partial pressure of CO2, malic enzymes could function in both malate production from pyruvate and malate decarboxylation to pyruvate, depending on the NADH/NAD+ and pyruvate/malate ratios. Using 13C labeling in a C. acetobutylicum batch culture, Au et al. (76) demonstrated that malic enzymes function in the malate-to-pyruvate direction but that this flux accounted for less than 5% of the pyruvate carboxylase flux. In Fig. 3 and in Fig. S3 in the supplemental material, the anaplerotic fluxes presented are net anaplerotic fluxes, which were attributed to pyruvate carboxylase.
The flux in the oxidative branch of the TCA cycle was much higher than that in the reductive branch (Fig. 3; see also Fig. S3 in the supplemental material). In agreement with the 13C labeling flux data reported by Amador-Noguez et al. (55), who demonstrated the flux from oxaloacetate to malate, but in contrast to the report by Au et al. (76), in which no flux could be measured through this enzyme, under all three conditions, we measured ~1,000 malate dehydrogenase (CA_C0566) protein molecules per cell that could catalyze the first step of the TCA reductive branch (see Data Set S2).
Conclusion.
In this work, an improved GSM containing new and validated biochemical data was developed in conjunction with quantitative transcriptomic and proteomic analyses to obtain accurate fluxomic data. These “omics” data allowed for (i) the determination of the distribution of carbon and electron fluxes, (ii) the elucidation of the different genes/enzymes involved in the primary metabolism of C. acetobutylicum, and (iii) a better understanding of the regulation of C. acetobutylicum primary metabolism under different physiological conditions. The information provided in this study will be important for the further metabolic engineering of C. acetobutylicum to develop a commercial process for the production of n-butanol.
MATERIALS AND METHODS
Chemicals and other reagents.
All chemicals were of reagent grade and were purchased from Sigma-Aldrich Chimie (Saint-Quentin Fallavier, France) or from VWR Prolabo (Fontenay sous Bois, France). All gases used for gas flushing of the medium and for the anaerobic chamber were of the highest purity available and were obtained from Air Liquide (Paris, France). All restriction enzymes and Crimson Taq DNA polymerase used for colony PCR were supplied by New England Biolabs (MA, USA) and were used according to the manufacturer’s instructions. DNA fragments for vector constructions were amplified using Phusion high-fidelity DNA polymerase (New England Biolabs).
Culture conditions. (i) Batch culture.
All liquid cultures of C. acetobutylicum ATCC 824 ΔCA_C1502 Δupp (P. Soucaille, R. Figge, and R. Croux, 2014, U.S. Patent 8,628,967) were performed in 30-ml or 60-ml glass vials under strict anaerobic conditions in clostridium growth medium (CGM) as described previously (77) or in synthetic medium (MS) as described previously (6). C. acetobutylicum was stored in spore form at −20°C after sporulation in MS medium. Heat shock was performed for spore germination by immersing the bottle in a water bath at 80°C for 15 min.
(ii) Continuous culture.
The conditions described previously by Vasconcelos et al. (6) and Girbal et al. (9) were used for the phosphate-limited continuous culture of C. acetobutylicum fed a constant total carbon amount of 995 mM. The cultures were maintained under acidogenesis (pH 6.3, 995 mM carbon from glucose), solventogenesis (pH 4.4, 995 mM carbon from glucose), and alcohologenesis (pH 6.3, 498 mM carbon from glucose and 498 mM carbon from glycerol).
RNA extraction and microarray.
For transcriptomic analysis, 3-ml samples were collected from chemostat cultures and immediately frozen in liquid nitrogen. The frozen cell cultures were ground promptly with 2-mercaptoethanol in a liquid nitrogen-cooled mortar. RNA was extracted using an RNeasy midikit (Qiagen, Courtaboeuf, France) according to the manufacturer’s instructions with the supplementation of DNase treatment using the RNase-free DNase set (Qiagen). RNA quantity and composition were analyzed using an Agilent 2100 bioanalyzer (Agilent Technologies, Massy, France) and a NanoDrop ND-1000 spectrophotometer (Labtech France, Paris, France) at 260 nm and 280 nm. All microarray procedures were performed according to the manufacturer’s protocol (Agilent one-color microarray-based exon analysis). Briefly, the RNAs were labeled with a low-input Quick Amp labeling kit and hybridized following a one-color microarray-based gene expression analysis protocol. The slides were scanned using a Tecan MS200 scanner and analyzed using Feature Extraction V.11.5.1.1.
Protein extraction and analysis.
For proteomic analysis, 20-ml samples were collected from chemostat cultures and treated according to the standard operating procedures developed by Schwarz et al. (78) for the extraction of intracellular proteins, except that phenylmethylsulfonyl fluoride (PMSF) was not added. Samples of 200 µg of each of the lyophilized protein extracts were dissolved at 80°C in 100 µl of 0.1% RapiGest (Waters) in water. Disulfide bonds were reduced with the addition of dithiothreitol (DTT) at 2 mM and incubation at 60°C for 15 min. Cysteine residues were carboxyamidomethylated with the addition of iodoacetamide to a concentration of 10 mM and incubated in the dark at room temperature. Proteolytic digestion was performed with trypsin (10 µg/ml) at 37°C for 12 h. Protein hydrolysates were acidified with 5 µl of concentrated trifluoroacetic acid (TFA), incubated at 37°C for 20 min, and centrifuged at 18,000 × g for 2 min to remove the RapiGest precipitate. The supernatant was collected. Postdigestion samples at a concentration of 2 µg/µl were mixed at a ratio of 1:1 with 40 fmol/µl phosphorylase b internal standard tryptic digest in 200 mM ammonium formate buffer.
Quantitative two-dimensional reversed-phase liquid chromatography-tandem mass spectrometry (LC/LC-MS/MS) was performed at a high-low-pH reversed-phase/reversed-phase configuration using a nano-Acquity ultraperformance liquid chromatography (UPLC)/UPLC system (Waters Corp.) coupled with a Synapt G2 HDMS mass spectrometer (Waters Corp.) and nano-electrospray ionization, as previously described by Foster et al. (64).
Raw MS data were processed either using a Mascot Distiller (version 2.4.3.1) for peptide and protein identification and isobaric quantification or using a Progenesis QI (Nonlinear Dynamics, United Kingdom) for label-free quantification. The MS/MS spectra were searched against the UniProt Clostridium acetobutylicum database using the Mascot search engine (version 2.4.1) with the following search parameters: full tryptic specificity, up to two missed cleavage sites, carbamidomethylation of cysteine residues as a fixed modification, and N-terminal methionine oxidation as a variable modification.
Determination of DNA, mRNA, and protein contents.
DNA and protein contents were measured in cells grown in a chemostat culture after centrifugation (4,000 × g, 10 min, 4°C) and washed twice with Milli-Q water. Protein content was determined via the Biuret method (79). The DNA content was determined after incubation with perchloric acid (0.5 M, 70 to 80°C, 15 to 20 min), as described by Hanson and Phillips (80). The RNA content was determined using the protocol described above for the microarrays.
Measurement of fermentation parameters.
Biomass concentration was determined both by counting the number of cells per milliliter, as previously described (81), and by the DCW method after centrifugation (16,000 × g, 5 min, room temperature), two washes with Milli-Q water, and drying under vacuum at 80°C. The concentrations of glucose, glycerol, acetate, butyrate, lactate, pyruvate, acetoin, acetone, ethanol, and butanol were determined based on high-performance liquid chromatography (HPLC), as described by Dusséaux et al. (82), except that the concentration of H2SO4 was changed to 0.5 mM, as required for mobile phase optimization. The concentrations of formate and fumarate were measured using a formate assay kit (Sigma-Aldrich) and a fumarate assay kit (Sigma-Aldrich), according to the manufacturer’s instructions.
Metabolic enzyme expression and purification.
Information on metabolic enzyme expression and purification is provided as Text S1 in the supplemental material.
Microarray data accession number.
The microarray data can be accessed at GEO through accession number GSE69973.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods and results. Download
Metabolic networks of C. acetobutylicum. Download
Transcriptomic and proteomic data. Download
H2 formation from NADH catalyzed by purified butyryl-CoA dehydrogenase/Etf complex from C. acetobutylicum in the presence of hydrogenase (HydA from C. acetobutylicum), ferredoxin (Fdx from C. acetobutylicum), and crotonyl-CoA. (A) Amount of H2 formed as a function of the amount of NADH added in the presence of excess amounts of crotonyl-CoA. (B) Amount of H2 formed as a function of the amount of crotonyl-CoA added in the presence of excess amounts of NADH. Download
Overview of the transcript levels during solventogenesis versus acidogenesis (A) and alcohologenesis versus acidogenesis (B). Log expression ratios of solventogenesis to acidogenesis (B) and alcohologenesis to acidogenesis are shown. All genes with log values (as logarithms to the basis of 2) higher than 2 (≥4.0-fold-increased expression) are significantly induced under solventogenesis (A) and alcohologenesis (B), and genes with a negative log of less than −2 (≥4.0-fold-decreased expression) were significantly induced in acidogenesis. According to this definition, all genes between the dashed lines were expected to be not significantly influenced. Download
Metabolic flux map of C. acetobutylicum in acidogenesis (A), solventogenesis (B), and alcohologenesis (C). All values are normalized to the flux of the initial carbon source (millimoles per gram of DCW per hour). Glucose flux is normalized as 100 for acidogenesis and solventogenesis, and the sum of glucose and half of the glycerol is normalized as 100 for alcohologenesis. Download
Carbon source consumption and product profiles of C. acetobutylicum. (A) Carbon source consumption. (B) Product profiles. Each histogram indicates different metabolic states: acidogenesis (red), solventogenesis (green), and alcohologenesis (blue). Download
iCac967 validation on previously published data.
Genes with ≥4.0-fold-increased or -decreased expression in solventogenesis versus acidogenesis.
Genes with ≥4.0-fold-increased or -decreased expression in alcohologenesis versus acidogenesis.
ACKNOWLEDGMENTS
We thank Jean-Louis Uribelarrea, Sophie Lamarre, and Lidwine Trouilh for help with the data analysis.
This work was financially supported by the European Community’s Seventh Framework Program “CLOSTNET” (PEOPLE-ITN-2008-237942) to Minyeong Yoo.
Footnotes
Citation Yoo M, Bestel-Corre G, Croux C, Riviere A, Meynial-Salles I, Soucaille P. 2015. A quantitative system-scale characterization of the metabolism of Clostridium acetobutylicum. mBio 6(6):e01808-15. doi:10.1128/mBio.01808-15.
Contributor Information
Eleftherios T. Papoutsakis, University of Delaware.
Sang Yup Lee, Korea Advanced Institute of Science and Technology.
REFERENCES
- 1.Jones DT. 2001. Applied acetone-butanol fermentation, p 125–168. In Bahl H, Dürre P (ed), Clostridia: biotechnology and medical applications. Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- 2.Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol Res 50:484–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones DT, van der Westhuizen A, Long S, Allcock ER, Reid SJ, Woods DR. 1982. Solvent production and morphological changes in Clostridium acetobutylicum. Appl Environ Microbiol 43:1434–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dürre P. 2007. Biobutanol: an attractive biofuel. Biotechnol J 2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 5.Ni Y, Sun Z. 2009. Recent progress on industrial fermentative production of acetone-butanol-ethanol by Clostridium acetobutylicum in China. Appl Microbiol Biotechnol 83:415–423. doi: 10.1007/s00253-009-2003-y. [DOI] [PubMed] [Google Scholar]
- 6.Vasconcelos I, Girbal L, Soucaille P. 1994. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol 176:1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girbal L, Soucaille P. 1994. Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: role of NADH/NAD ratio and ATP pool. J Bacteriol 176:6433–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girbal L, Vasconcelos I, Saint-Amans S, Soucaille P. 1995. How neutral red modified carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH. FEMS Microbiol Rev 16:151–162. doi: 10.1111/j.1574-6976.1995.tb00163.x. [DOI] [Google Scholar]
- 9.Girbal L, Croux C, Vasconcelos I, Soucaille P. 1995. Regulation of metabolic shifts in Clostridium acetobutylicum ATCC 824. FEMS Microbiol Rev 17:287–297. doi: 10.1111/j.1574-6976.1995.tb00212.x. [DOI] [Google Scholar]
- 10.Girbal L, Soucaille P. 1998. Regulation of solvent production in Clostridium acetobutylicum. Trends Biotechnol 16:11–16. doi: 10.1016/S0167-7799(97)01141-4. [DOI] [Google Scholar]
- 11.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl Environ Microbiol 55:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1989. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol 55:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer U, Dürre P. 1995. Differential induction of genes related to solvent formation during the shift from acidogenesis to solventogenesis in continuous culture of Clostridium acetobutylicum. FEMS Microbiol Lett 125:115–120. doi: 10.1111/j.1574-6968.1995.tb07344.x. [DOI] [Google Scholar]
- 14.Janssen H, Grimmler C, Ehrenreich A, Bahl H, Fischer R. 2012. A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—solvent stress caused by a transient n-butanol pulse. J Biotechnol 161:354–365. doi: 10.1016/j.jbiotec.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz KM, Kuit W, Grimmler C, Ehrenreich A, Kengen SWM. 2012. A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—cellular behavior in adaptation to n-butanol. J Biotechnol 161:366–377. doi: 10.1016/j.jbiotec.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Venkataramanan K, Huang H, Papoutsakis ET, Wu CH. 2013. Transcription factors and genetic circuits orchestrating the complex, multilayered response of Clostridium acetobutylicum to butanol and butyrate stress. BMC Syst Biol 7:120. doi: 10.1186/1752-0509-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkataramanan KP, Jones SW, McCormick KP, Kunjeti SG, Ralston MT, Meyers BC, Papoutsakis ET. 2013. The clostridium small RNome that responds to stress: the paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genomics 14:849. doi: 10.1186/1471-2164-14-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornillot E, Soucaille P. 1996. Solvent-forming genes in Clostridia. Nature 380:489. doi: 10.1038/380489a0. [DOI] [Google Scholar]
- 19.Cornillot E, Nair RV, Papoutsakis ET, Soucaille P. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J Bacteriol 179:5442–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol 183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alsaker KV, Papoutsakis ET. 2005. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J Bacteriol 187:7103–7118. doi: 10.1128/JB.187.20.7103-7118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen H, Döring C, Ehrenreich A, Voigt B, Hecker M, Bahl H, Fischer R. 2010. A proteomic and transcriptional view of acidogenic and solventogenic steady-state cells of Clostridium acetobutylicum in a chemostat culture. Appl Microbiol Biotechnol 87:2209–2226. doi: 10.1007/s00253-010-2741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol 9:R114. doi: 10.1186/gb-2008-9-7-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan L, Bennett GN. 2006. Proteome analysis and comparison of Clostridium acetobutylicum ATCC 824 and Spo0A strain variants. J Ind Microbiol Biotechnol 33:298–308. doi: 10.1007/s10295-005-0050-7. [DOI] [PubMed] [Google Scholar]
- 25.Tomas CA, Alsaker KV, Bonarius HPJ, Hendriksen WT, Yang H, Beamish JA, Paredes CJ, Papoutsakis ET. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol 185:4539–4547. doi: 10.1128/JB.185.15.4539-4547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer S, Isci N, Zickner B, Dürre P. 2002. Changes in protein synthesis and identification of proteins specifically induced during solventogenesis in Clostridium acetobutylicum. Electrophoresis 23:110–121. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Paredes CJ, Senger RS, Spath IS, Borden JR, Sillers R, Papoutsakis ET. 2007. A general framework for designing and validating oligomer-based DNA microarrays and its application to Clostridium acetobutylicum. Appl Environ Microbiol 73:4631–4638. doi: 10.1128/AEM.00144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Servinsky MD, Kiel JT, Dupuy NF, Sund CJ. 2010. Transcriptional analysis of differential carbohydrate utilization by Clostridium acetobutylicum. Microbiology 156:3478–3491. doi: 10.1099/mic.0.037085-0. [DOI] [PubMed] [Google Scholar]
- 29.Grimmler C, Held C, Liebl W, Ehrenreich A. 2010. Transcriptional analysis of catabolite repression in Clostridium acetobutylicum growing on mixtures of d-glucose and d-xylose. J Biotechnol 150:315–323. doi: 10.1016/j.jbiotec.2010.09.938. [DOI] [PubMed] [Google Scholar]
- 30.Tan Y, Liu Z, Liu Z, Zheng H, Li F. 2015. Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol Biosyst 11:1434–1442. doi: 10.1039/C4MB00600C. [DOI] [PubMed] [Google Scholar]
- 31.Mao S, Luo Y, Zhang T, Li J, Bao G, Zhu Y, Chen Z, Zhang Y, Li Y, Ma Y. 2010. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res 9:3046–3061. doi: 10.1021/pr9012078. [DOI] [PubMed] [Google Scholar]
- 32.Mao S, Luo Y, Bao G, Zhang Y, Li Y, Ma Y. 2011. Comparative analysis on the membrane proteome of Clostridium acetobutylicum wild type strain and its butanol-tolerant mutant. Mol Biosyst 7:1660–1677. doi: 10.1039/c0mb00330a. [DOI] [PubMed] [Google Scholar]
- 33.Jang Y, Han M, Lee J, Im JA, Lee YH, Papoutsakis ET, Bennett G, Lee SY. 2014. Proteomic analyses of the phase transition from acidogenesis to solventogenesis using solventogenic and non-solventogenic Clostridium acetobutylicum strains. Appl Microbiol Biotechnol 98:5105–5115. doi: 10.1007/s00253-014-5738-z. [DOI] [PubMed] [Google Scholar]
- 34.Magdeldin S, Enany S, Yoshida Y, Xu B, Zhang Y, Zureena Z, Lokamani I, Yaoita E, Yamamoto T. 2014. Basics and recent advances of two dimensional-polyacrylamide gel electrophoresis. Clin Proteomics 11:16. doi: 10.1186/1559-0275-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivagnanam K, Raghavan VG, Shah M, Hettich RL, Verberkmoes NC, Lefsrud MG. 2011. Comparative shotgun proteomic analysis of Clostridium acetobutylicum from butanol fermentation using glucose and xylose. Proteome Sci 9:66. doi: 10.1186/1477-5956-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou S, Jones SW, Choe LH, Papoutsakis ET, Lee KH. 2013. Workflow for quantitative proteomic analysis of Clostridium acetobutylicum ATCC 824 using iTRAQ tags. Methods 61:269–276. doi: 10.1016/j.ymeth.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Grimmler C, Janssen H, Krauβe D, Fischer R, Bahl H, Dürre P, Liebl W, Ehrenreich A. 2011. Genome-wide gene expression analysis of the switch between acidogenesis and solventogenesis in continuous cultures of Clostridium acetobutylicum. J Mol Microbiol Biotechnol 20:1–15. doi: 10.1159/000320973. [DOI] [PubMed] [Google Scholar]
- 38.Tracy BP, Jones SW, Papoutsakis ET. 2011. Inactivation of sigmaE and sigmaG in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J Bacteriol 193:1414–1426. doi: 10.1128/JB.01380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones SW, Tracy BP, Gaida SM, Papoutsakis ET. 2011. Inactivation of sigmaF in Clostridium acetobutylicum ATCC 824 blocks sporulation prior to asymmetric division and abolishes sigmaE and sigmaG protein expression but does not block solvent formation. J Bacteriol 193:2429–2440. doi: 10.1128/JB.00088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkataramanan KP, Min L, Hou S, Jones SW, Ralston MT, Lee KH, Papoutsakis ET. 2015. Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol Biofuels 8:1–29. doi: 10.1186/s13068-015-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahl H, Andersch W, Gottschalk G. 1982. Continuous production of acetone and butanol by Clostridium acetobutylicum in a two-stage phosphate limited chemostat. Eur J Appl Microbiol Biotechnol 15:201–205. doi: 10.1007/BF00499955. [DOI] [Google Scholar]
- 42.Boynton ZL, Bennet GN, Rudolph FB. 1996. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol 178:3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sillers R, Chow A, Tracy B, Papoutsakis ET. 2008. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab Eng 10:321–332. doi: 10.1016/j.ymben.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Dash S, Mueller TJ, Venkataramanan KP, Papoutsakis ET, Maranas CD. 2014. Capturing the response of Clostridium acetobutylicum to chemical stressors using a regulated genome-scale metabolic model. Biotechnol Biofuels 7:144. doi: 10.1186/s13068-014-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McAnulty MJ, Yen JY, Freedman BG, Senger RS. 2012. Genome-scale modeling using flux ratio constraints to enable metabolic engineering of clostridial metabolism in silico. BMC Syst Biol 6:42. doi: 10.1186/1752-0509-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nair RV, Bennett GN, Papoutsakis ET. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol 176:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer RJ, Helms J, Durre P. 1993. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol 175:6959–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter KA, Bennett GN, Papoutsakis ET. 1992. Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J Bacteriol 174:7149–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch RW, Rudolph FB, Papoutsakis ET. 1989. Purification and characterization of the NADH-dependent butanol dehydrogenase from Clostridium acetobutylicum (ATCC 824). Arch Biochem Biophys 273:309–318. doi: 10.1016/0003-9861(89)90489-X. [DOI] [PubMed] [Google Scholar]
- 51.Sulzenbacher G, Alvarez K, Van Den Heuvel RHH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M. 2004. Crystal structure of E.coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342:489–502. doi: 10.1016/j.jmb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 52.Durre P, Kuhn A, Gottwald M, Gottschalk G. 1987. Enzymatic investigations on butanol dehydrogenase and butyraldehyde dehydrogenase in extracts of Clostridium acetobutylicum. Appl Microbiol Biotechnol 26:268–272. doi: 10.1007/BF00286322. [DOI] [Google Scholar]
- 53.Schmidt CNG, Jervis L. 1980. Affinity purification of glutamate synthase from Escherichia coli. Anal Biochem 104:127–129. doi: 10.1016/0003-2697(80)90286-9. [DOI] [PubMed] [Google Scholar]
- 54.Sakamoto N, Kotre AM, Savageau MA. 1975. Glutamate dehydrogenase from Escherichia coli: purification and properties. J Bacteriol 124:775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amador-Noguez D, Feng X-, Fan J, Roquet N, Rabitz H, Rabinowitz JD. 2010. Systems-level metabolic flux profiling elucidates a complete, bifurcated tricarboxylic acid cycle in Clostridium acetobutylicum. J Bacteriol 192:4452–4461. doi: 10.1128/JB.00490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senger RS, Papoutsakis ET. 2008. Genome-scale model for Clostridium acetobutylicum: part I. Metabolic network resolution and analysis. Biotechnol Bioeng 101:1036–1052. doi: 10.1002/bit.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senger RS, Papoutsakis ET. 2008. Genome-scale model for Clostridium acetobutylicum: part II. Development of specific proton flux states and numerically determined sub-systems. Biotechnol Bioeng 101:1053–1071. doi: 10.1002/bit.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J, Yun H, Feist AM, Palsson BØ, Lee SY. 2008. Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network. Appl Microbiol Biotechnol 80:849–862. doi: 10.1007/s00253-008-1654-4. [DOI] [PubMed] [Google Scholar]
- 59.Harris LM, Desai RP, Welker NE, Papoutsakis ET. 2000. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnol Bioeng 67:1–11. doi:. [DOI] [PubMed] [Google Scholar]
- 60.Lee JY, Jang Y, Lee J, Papoutsakis ET, Lee SY. 2009. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnol J 4:1432–1440. doi: 10.1002/biot.200900142. [DOI] [PubMed] [Google Scholar]
- 61.Neidhardt FC, Umbarger HE. 1996. Chemical composition of E. coli, p 13–16. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC. [Google Scholar]
- 62.Pramanik J, Keasling JD. 1997. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng 56:398–421. [DOI] [PubMed] [Google Scholar]
- 63.Miller JA, Menon V, Goldy J, Kaykas A, Lee C, Smith KA, Shen EH, Phillips JW, Lein ES, Hawrylycz MJ. 2014. Improving reliability and absolute quantification of human brain microarray data by filtering and scaling probes using RNA-Seq. BMC Genomics 15:154. doi: 10.1186/1471-2164-15-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foster MW, Morrison LD, Todd JL, Snyder LD, Thompson JW, Soderblom EJ, Plonk K, Weinhold KJ, Townsend R, Minnich A, Moseley MA. 2015. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res 14:1238–1249. doi: 10.1021/pr501149m. [DOI] [PubMed] [Google Scholar]
- 65.Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC. [Google Scholar]
- 66.Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol 184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amador-Noguez D, Brasg IA, Feng X-, Roquet N, Rabinowitz JD. 2011. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl Environ Microbiol 77:7984–7997. doi: 10.1128/AEM.05374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabathé F, Bélaïch A, Soucaille P. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol Lett 217:15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x. [DOI] [PubMed] [Google Scholar]
- 69.Harris LM, Welker NE, Papoutsakis ET. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ravagnani A, Jennert KCB, Steiner E, Grunberg R, Jefferies JR, Wilkinson SR, Young DI, Tidswell EC, Brown DP, Youngman P, Morris JG, Young M. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol Microbiol 37:1172–1185. doi: 10.1046/j.1365-2958.2000.02071.x. [DOI] [PubMed] [Google Scholar]
- 71.Thormann K, Feustel L, Lorenz K, Nakotte S, Durre P. 2002. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J Bacteriol 184:1966–1973. doi: 10.1128/JB.184.7.1966-1973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomas CA, Beamish J, Papoutsakis ET. 2004. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186:2006–2018. doi: 10.1128/JB.186.7.2006-2018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu K, Zhao M, Zhang T, Zha M, Zhong C, Jiang Y, Ding J. 2013. Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism. Biochem J 449:79–89. doi: 10.1042/BJ20120871. [DOI] [PubMed] [Google Scholar]
- 74.Schreiber W, Durre P. 1999. The glyceraldehyde-3-phosphate dehydrogenase of Clostridium acetobutylicum: isolation and purification of the enzyme, and sequencing and localization of the gap gene within a cluster of other glycolytic genes. Microbiology 145:1839–1847. doi: 10.1099/13500872-145-8-1839. [DOI] [PubMed] [Google Scholar]
- 75.Iddar A, Valverde F, Serrano A, Soukri A. 2002. Expression, purification, and characterization of recombinant nonphosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Clostridium acetobutylicum. Protein Expr Purif 25:519–526. doi: 10.1016/S1046-5928(02)00032-3. [DOI] [PubMed] [Google Scholar]
- 76.Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. 2014. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for C metabolic flux analysis. Metab Eng 26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 77.Roos JW, McLaughlin JK, Papoutsakis ET. 1985. The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol Bioeng 27:681–694. doi: 10.1002/bit.260270518. [DOI] [PubMed] [Google Scholar]
- 78.Schwarz K, Fiedler T, Fischer R, Bahl H. 2007. A standard operating procedure (SOP) for the preparation of intra- and extracellular proteins of Clostridium acetobutylicum for proteome analysis. J Microbiol Methods 68:396–402. doi: 10.1016/j.mimet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 79.Peterson GL. 1983. Determination of total protein. Methods Enzymol 91:95–119. [DOI] [PubMed] [Google Scholar]
- 80.Hanson R, Phillips J. 1981. Chemical composition, p 328–364. In Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (ed), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 81.Ferras E, Minier M, Goma G. 1986. Acetonobutylic fermentation: improvement of performances by coupling continuous fermentation and ultrafiltration. Biotechnol Bioeng 28:523–533. doi: 10.1002/bit.260280408. [DOI] [PubMed] [Google Scholar]
- 82.Dusséaux S, Croux C, Soucaille P, Meynial-Salles I. 2013. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab Eng 18:1–8. doi: 10.1016/j.ymben.2013.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods and results. Download
Metabolic networks of C. acetobutylicum. Download
Transcriptomic and proteomic data. Download
H2 formation from NADH catalyzed by purified butyryl-CoA dehydrogenase/Etf complex from C. acetobutylicum in the presence of hydrogenase (HydA from C. acetobutylicum), ferredoxin (Fdx from C. acetobutylicum), and crotonyl-CoA. (A) Amount of H2 formed as a function of the amount of NADH added in the presence of excess amounts of crotonyl-CoA. (B) Amount of H2 formed as a function of the amount of crotonyl-CoA added in the presence of excess amounts of NADH. Download
Overview of the transcript levels during solventogenesis versus acidogenesis (A) and alcohologenesis versus acidogenesis (B). Log expression ratios of solventogenesis to acidogenesis (B) and alcohologenesis to acidogenesis are shown. All genes with log values (as logarithms to the basis of 2) higher than 2 (≥4.0-fold-increased expression) are significantly induced under solventogenesis (A) and alcohologenesis (B), and genes with a negative log of less than −2 (≥4.0-fold-decreased expression) were significantly induced in acidogenesis. According to this definition, all genes between the dashed lines were expected to be not significantly influenced. Download
Metabolic flux map of C. acetobutylicum in acidogenesis (A), solventogenesis (B), and alcohologenesis (C). All values are normalized to the flux of the initial carbon source (millimoles per gram of DCW per hour). Glucose flux is normalized as 100 for acidogenesis and solventogenesis, and the sum of glucose and half of the glycerol is normalized as 100 for alcohologenesis. Download
Carbon source consumption and product profiles of C. acetobutylicum. (A) Carbon source consumption. (B) Product profiles. Each histogram indicates different metabolic states: acidogenesis (red), solventogenesis (green), and alcohologenesis (blue). Download
iCac967 validation on previously published data.
Genes with ≥4.0-fold-increased or -decreased expression in solventogenesis versus acidogenesis.
Genes with ≥4.0-fold-increased or -decreased expression in alcohologenesis versus acidogenesis.