Abstract
The centrosome plays a pivotal role in a wide range of cellular processes and its dysfunction is causally linked to many human diseases including cancer and developmental and neurological disorders. This organelle contains more than one hundred components, and yet many of them remain uncharacterised. Here we identified a novel centrosome protein Wdr8, based upon the structural conservation of the fission yeast counterpart. We showed that Wdr8 constitutively localises to the centrosome and super resolution microscopy uncovered that this protein is enriched at the proximal end of the mother centriole. Furthermore, we identified hMsd1/SSX2IP, a conserved spindle anchoring protein, as one of Wdr8 interactors by mass spectrometry. Wdr8 formed a complex and partially colocalised with hMsd1/SSX2IP. Intriguingly, knockdown of Wdr8 or hMsd1/SSX2IP displayed very similar mitotic defects, in which spindle microtubules became shortened and misoriented. Indeed, Wdr8 depletion resulted in the reduced recruitment of hMsd1/SSX2IP to the mitotic centrosome, though the converse is not true. Together, we propose that the conserved Wdr8-hMsd1/SSX2IP complex plays a critical role in controlling proper spindle length and orientation.
Keywords: Bipolar spindle, Centriolar satellites, Centriole, Centrosome, Fission yeast, Spindle pole body (SPB), Super resolution microscopy, WD repeats
Highlights
-
•
Human Wdr8 is a centrosomal protein enriched in the proximal end of the centriole.
-
•
Wdr8 and hMsd1/SSX2IP form a complex conserved in fungi.
-
•
Depletion of Wdr8 results in shorter, tilted spindle microtubules.
-
•
Depletion phenotypes of Wdr8 are very similar to those of hMsd1/SSX2IP knockdown.
1. Introduction
The centrosome plays crucial roles in a myriad of biological processes, including cell division and proliferation, differentiation, tissue and body development [1], [2], [3]. In a cell, this organelle comprises the major microtubule organising centre (MTOC). In fungi, the centrosome-equivalent structure is called the spindle pole body (SPB). During mitosis, bipolar spindle microtubules emanate and assemble from the centrosome, which is required for accurate chromosome segregation and cell division. During interphase, the centrosome organises cytoplasmic microtubules, thereby acting as a hub for a wide range of events such as polarised protein and RNA transport, cell motility and polarity [4]. In many cell types, upon serum starvation, the centrosome translocates and is docked to the plasma membrane, where it becomes the basal body. The basal body then assembles the primary cilium, a critical sensory organelle serving as a cellular antenna [4], [5]. Moreover, recent advances in the field have highlighted the everlasting importance of the centrosome as not only MTOCs but also a focus point coordinating multifaceted pathways involved in the cell cycle and signal transduction [2].
While the animal centrosome consists of two orthogonally arranged centrioles and the pericentriolar materials, the fungal SPB does not contain centrioles. Instead it is composed of a membrane-embedded laminar configuration [3]. Despite these structural differences, the central role of the centrosome and SPB as the MTOC is universal. Mitotic bipolar spindle microtubules emanate from the centrosome, in which the minus end of the microtubules are anchored to this organelle; otherwise nucleating spindle microtubules are dissociated from the centrosome and perturb structural integrity. This leads to chromosome segregation defects and aneuploidy, a hallmark of cancer [1], [5]. We recently identified in fission yeast, zebrafish and human beings a conserved protein family (Msd1 in fission yeast, TINA in Aspergillus nidulans and Msd1/SSX2IP in zebrafish and humans) that localises to the centrosome/SPB. Further analysis uncovered that these proteins safeguard the anchorage of the minus end of spindle microtubules to the centrosome/SPB [6], [7], [8]. We also showed that fission yeast Msd1 forms a stable complex with another conserved protein Wdr8/WRAP73 [9], [10] and these two proteins play an essential role in spindle anchoring in concert [11]. In this study, we have addressed whether the analogous complex exists and is operational in higher eukaryotes. We show that the Msd1-Wdr8 complex indeed is conserved in human beings. Furthermore, we have found that this complex localises to the centrosome and plays a critical role in proper spindle assembly.
2. Materials and methods
2.1. Cell cultures
Human cervical cancer HeLa cells, HeLa cells stably expressing GFP-Centrin and osteo-sarcoma U2OS cells were cultured in high-glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS). All cells were cultured in a humidified 5% CO2 incubator at 37 °C.
2.2. RNA interference
Synthetic siRNA oligonucleotides were obtained from Dharmacon-GE Healthcare (Lafayette). The siRNA sequences were 5′- GACAGACAGUUACAAUGUA-3′ (hMsd1 siRNA; Dharmacon), 5′-AAUAUGAGAUCGCCUCUGU-3′ (Wdr8 siRNA No.1) or 5′-CCAAGAUAGUGGUGUAUAA -3′ (Wdr8 siRNA No.2). Control depletion was carried out using siGENOME non-targeting siRNA (Dharmacon). For RNAi experiments, cells were transfected with 40 nM of dsRNA using Lipofectamine RNAi-MAX (Invitrogen), and cells were fixed 48 h after siRNA treatment unless otherwise stated.
2.3. Plasmid construction and DNA transfection
pVenus-Wdr8 was constructed into pVenus-C1 (Clontech). For the construction of RNAi-resistant versions, we introduced 5 or 6 silent substitutions within the Wdr8 siRNA-target region. The Wdr8 siRNA No.1 target region, 5′-AATATGAGATCGCCTCTGT -3′, was changed to 5′-AATACGAAATTGCTACTGT -3′ using site-directed mutagenesis with the primers, 5′-taaggagactgggacggaagcaatttcgtatttactctctgagctcgggagagggc-3′ and 5- gccctctcccgagctcagagagtaaatacgaaattgcttccgtcccagtctcctta -3′. The Wdr8 siRNA No.2 target region, 5′-CCAAGATAGTGGTGTATAA-3′, was changed to 5′- CTAAAATCGTCGTTTACAA -3′ using site-directed mutagenesis with the primers, 5′- gtgggctcttctcggcctccttgtaaacgacgattttaggatcattaatggctgcaggatgc -3′ and 5- gcatcctgcagccattaatgatcctaaaatcgtcgtttacaaggaggccgagaagagcccac -3′.
Cells were treated with siRNAs for 48 h and observed under the microscope. For double transfection experiments, cells were treated with siRNAs for 48 h, followed by the second transfection with various plasmids. Cells were observed under the microscope 24 h later.
2.4. Antibodies
Rabbit polyclonal anti-Wdr8 antibody was produced and affinity-purified (Eurogenetec Co.); a peptide (288-CLSFPPPRAGAGPLPSSES-307) was used as antigen. The following antibodies were also used: chicken anti-GFP (ab13970; Abcam), rabbit anti-Cep135 (ab75005; abcam), mouse anti-C-Nap1 (611374; BD), rabbit anti-SSX2IP (HPA027306; Sigma–Aldrich), rabbit anti-γ-tubulin (T5192; Sigma–Aldrich), mouse anti-γ-tubulin (T6557; Sigma–Aldrich) and mouse anti-α-tubulin (T9026, Sigma–Aldrich). Secondary antibodies were Alexa Fluor 488-coupled anti-rabbit, Alexa Fluor 594-coupled anti-rabbit, Alexa Fluor 594-coupled anti-mouse, Alexa Fluor 488-coupled anti-mouse, Alexa Fluor 488-coupled anti-chicken, Alexa Fluor 647-coupled anti-rabbit, or Cy3-coupled anti-mouse antibodies (all used at 1:1,500, Molecular Probes).
2.5. Mass spectrometry
1.5 mg of total protein extracts were prepared from HeLa cell cultures transfected with empty vector or pVenus-Wdr8, and immunoprecipitation performed using GFP-trap (ChromoTec). Colloidal coomassie-stained bands were cut out from gels and subject to trypsin digestion and Q Exactive LC-MS analysis (Thermo Fisher Scientific). The data was searched against human database using the Andromeda search engine and MaxQuant (Version 1.3.0.5) [12], as well as Mascot Daemon search engines (version 2.4.0, Matrix Science).
2.6. Immunofluorescence microscopy, super resolution microscopy and image analysis
Immunofluorescence microscopy with DeltaVision image acquisition software (softWoRx 3.3.0; Applied Precision Co.) equipped with Coolsnap-HQ digital CCD camera or Cascade EMCCD 512B camera (Roper Scientific) was performed as described previously [6], [7].
Super resolution microscopy was performed using a structured-illumination microscopy system (DeltaVision OMX V3; Applied Precision). A 100 × , 1.4 NA, oil objective (Olympus) was used with 488 nm, 593 nm and 642 nm laser illumination and standard excitation and emission filter sets. 125-nm z-steps were applied to acquire raw images, which were reconstructed in 3D using SoftWoRx software (Applied Precision) and Imaris (Bitplane). Captured images were processed with Adobe Photoshop CS3 (version 10.0).
2.7. Immunoprecipitation
For coimmunoprecipitation, 1 mg cell lysate was incubated with 30 μl GFP-Trap (ChromoTek) in lysis buffer (25 mM Tris–HCl, pH 7.0, 1 mM EDTA, 300 mM NaCl, 10% Glycerol, 1% NP-40, 1 mM DTT, 10 mM NaF, 25 mM DMSF and EDTA-free protease inhibitor tablet (Complete: Roche)) overnight at 4 °C. After washing with lysis buffer, the beads were denatured at 95 °C in NuPAGE buffer (Invitrogen) and run on SDS-PAGE, followed by immunoblotting.
2.8. Spindle length and orientation assay
Procedures previously described were followed [6].
2.9. Quantification and fluorescence signal intensity measurement
For fluorescence signal intensity measurement, fluorescence signals were quantified using maximum intensity, after subtracting background signals in the vicinity of the fluorescent spot. The SoftWoRx software was used for analysis. At least 200 cells were counted in each sample, independently, three times, from which standard deviations and P-values were calculated.
2.10. Statistical data analysis
All data represent the mean of multiple experiments ± SD. Experiment sample numbers and the number of replicates used for statistical testing have been reported in the corresponding figure legends. All p-values are from two-tailed unpaired student t-tests. Unless otherwise stated, we followed this key for asterisk placeholders for p-values in the figures: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
3. Results and discussion
3.1. Human Wdr8 is a constitutive component of the centrosome enriched in the proximal end of the mother centriole
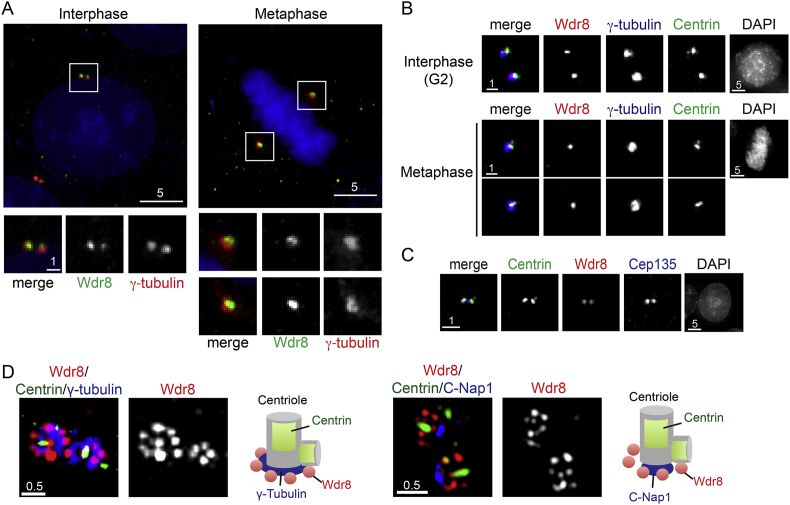
In order to examine the cellular localisation of Wdr8 during the cell cycle, we generated a polyclonal antibody against human Wdr8 (see Materials and methods and Supplementary Fig. S1A). Immunofluorescence microscopy in HeLa cells showed that Wdr8 signals colocalised with centrosome marker γ-tubulin [13], [14] during both interphase and mitotic metaphase (Fig. 1A). Closer inspection of these signals indicated that Wdr8 dots were almost always smaller than those of γ-tubulin. To further define the precise localisation of Wdr8 within the centrosome, we performed a triple immunostaining of Wdr8, γ-tubulin and Centrin (centriole marker) [15]. As shown in Fig. 1B, Wdr8 colocalised with Centrin; in particular it colocalised with the mother centriole, the larger part of the two closely situated Centrin dots. Cep135 is a conserved centriolar protein that localises to the proximal region of centrioles specifically within a cartwheel-shaped structure of the centriole [16]. Immunostaining showed that these two proteins mostly colocalised (Fig. 1C).
Fig. 1.
Wdr8 is a constitutive centrosomal protein enriched in the proximal end of the mother centriole. (A) HeLa cells were immunostained with antibodies against Wdr8 (green) and γ-tubulin (red). DAPI (4′,6-diamidino-2-phenylindole, blue) was used to visualise chromosomes. Enlarged images of the centrosomal region (squares on the top panels) are shown on the bottom. (B) Immunofluorescence images of HeLa cells stably expressing centrin-GFP using antibodies against Wdr8 (red), γ-tubulin (blue) and GFP (Centrin, green) during interphase (top) and mitosis (bottom) are shown. Only the centrosomal regions are dissected. Images of chromosomes (DAPI) are shown in the far-right panels. (C) Immunofluorescence images of HeLa cells stably expressing centrin-GFP using antibodies against Wdr8 (red), GFP (Centrin, green) and Cep135 (blue) are shown. Images of chromosomes (DAPI) are shown in the far-right panels. (D) Super resolution microscopy (OMX) images. HeLa cells stably expressing centrin-GFP were immunostained with antibodies against Wdr8 (left, red), GFP (Centrin, green) and γ-tubulin (blue) or Wdr8 (right, red), GFP (Centrin, green) and C-Nap (blue). Schematic presentation of the deduced localisation of individual proteins around the centrosomal region is shown on the right. Scale bars, 0.5 μm (D), 1 μm (A, bottom row and B and C), 5 μm (A, top row).
We further investigated the location of Wdr8 within the centrosome using super resolution microscopy. A triple staining of Wdr8, γ-tubulin and Centrin showed that Wdr8 is situated in close vicinity of the proximal part of the mother centriole (Fig. 1D, left), consistent with the apparent colocalisation with Cep135. This notion was substantiated by staining with an antibody against C-Nap1 that localises to the proximal end of the centrioles [17], though Wdr8 occupied a broader region than C-Nap1 and appeared to encompass the C-Nap1-localising proximal site (Fig. 1D, right). Together, Wdr8 is a centrosome protein enriched in the proximal end of the mother centriole.
3.2. hMsd1/SSX2IP forms a complex and partially colocalises with Wdr8
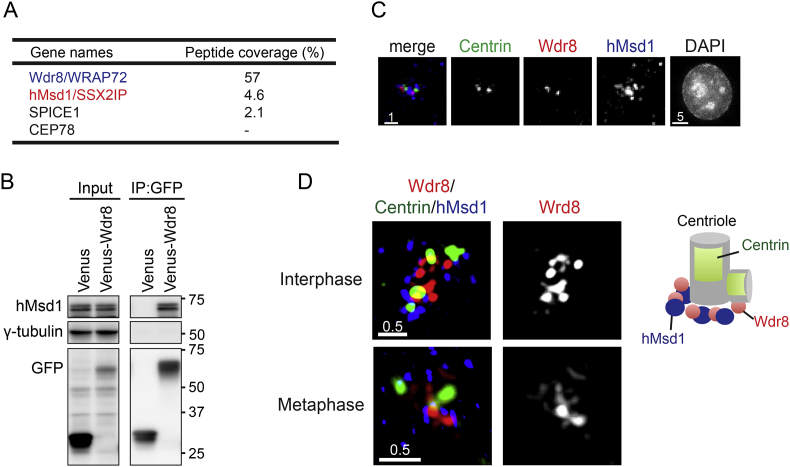
In order to identify Wdr8-interactors, plasmids producing Venus-Wdr8 were introduced into HeLa cells and associated proteins were pulled down. Colloidal coomassie-stained bands were cut out from gels, digested with trypsin and analysed by liquid chromatography–mass spectrometry. Among a number of centrosomal proteins pulled down, we identified hMsd1/SSX2IP [6], [18] (Fig. 2A and Supplementary Fig. S2). Immunoprecipitation showed that ectopically produced Venus-Wdr8 indeed interacted with hMsd1/SSX2IP (Fig. 2B).
Fig. 2.
Wdr8 forms a complex with hMsd1/SSX2IP. (A) Identification of hMsd1/SSX2IP as one of Wdr8 interacting proteins. The list of centrosomal interactors is shown with the percentage of peptide coverage. (B) Wdr8 interacts with hMsd1/SSX2IP in a cell. HeLa cells were transfected with plasmids producing Venus-Wdr8 and total cell extracts immunoprecipitated with an anti-GFP antibody. Immunoblotting were performed with the indicated antibodies. Positions of molecular weight markers are shown on the right. (C) HeLa cells stably expressing centrin-GFP were immunostained with antibodies against GFP (Centrin, green), Wdr8 (red) and hMsd1/SSX2IP (blue). Chromosomes were stained with DAPI. Representative images of the centrosomal region during interphase are show. (D) Super resolution microscopy (OMX) images. HeLa cells stably expressing centrin-GFP were immunostained with antibodies against Wdr8 (red), Centrin (green) and hMsd1/SSX2IP (blue). Representative images of the centrosomal regions during interphase (top) and mitosis (bottom) are shown. Schematic presentation of the deduced localisation of individual proteins around the centrosomal region (interphase) is shown on the right. Scale bars, 0.5 μm (D), 1 μm (C).
Immunostaining using antibodies against hMsd1/SSX2IP and Wdr8 indicated that the localisation of these two proteins appears not the same (Fig. 2C), as hMsd1/SSX2IP localises to both the centrosome and centriolar satellites, numerous peripheral particles locating around the centrosome [6], [18], [19], which hindered the evaluation of colocalisation between Wdr8 and hMsd1/SSX2IP to the centrosome. To precisely evaluate the localisation of these two proteins, we again implemented super resolution microscopy. As shown in Fig. 2D, we observed partial colocalisation of these two proteins around the centrosome during both interphase (top) and mitosis (bottom) at the proximal region of the mother centriole. Hence, the interaction and localisation of Wdr8 and Msd1 homologues are conserved from fungi to human beings.
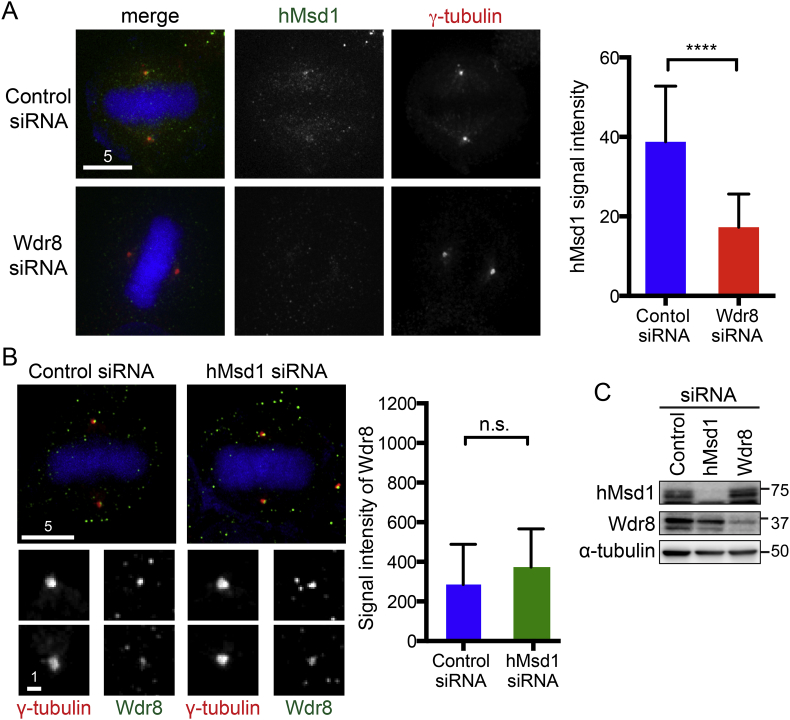
3.3. Proper mitotic localisation of hMsd1/SSX2IP to the centrosome requires Wdr8
In fungi, both Wdr8 and hMsd1/SSX2IP homologues localise to the SPB during mitosis, and furthermore their localisation is interdependent [11], [20]. Given the evolutionary conservation of physical interaction between these two proteins, we next addressed the localisation patterns of hMsd1/SSX2IP upon Wdr8 depletion using siRNA oligonucleotides and vice versa. We first examined hMsd1/SSX2IP localisation in HeLa cells after Wdr8 was knocked down (Supplementary Fig. S1A), and found that intensities of hMsd1/SSX2IP at the centrosomal region was substantially reduced (Fig. 3A). In contrast, depletion of hMsd1/SSX2IP did not lead to the reduction of Wdr8 signals at the mitotic centrosome (Fig. 3B). Under either condition, the total amount of the hMsd1/SSX2IP protein (upon Wdr8 depletion) or that of Wdr8 (upon hMsd1/SSX2IP depletion) was not noticeably reduced (Fig. 3C). The reduction of hMsd1/SSX2IP signals upon Wdr8 depletion was also observed in U2OS cells (Supplementary Figs. S1B and S2A). In contrast to the mitotic localisation, during interphase, depletion of either Wdr8 or hMsd1/SSX2IP did not result in the decreased localisation of the other protein (Supplementary Fig. S2B and S2C).
Fig. 3.
Wdr8 knockdown leads to reduced recruitment of hMsd1/SSX2IP to the mitotic centrosome. (A) HeLa cells were treated with control or Wdr8 siRNA and immunostained with antibodies against hMsd1/SSX2IP (green) and γ-tubulin (red). Chromosomes were stained with DAPI (blue). Quantification of signal intensities of hMsd1/SSX2IP around the mitotic centrosome is shown on the right. At least 150 cells were counted (n = 3). ****p < 0.0001. (B) HeLa cells were treated with control or hMsd1/SSX2IP siRNA and immunostained with antibodies against Wdr8 (green) and γ-tubulin (red). Chromosomes were stained with DAPI (blue). Representative immunofluorescence images of mitotic cells are shown. Quantification of signal intensities of Wdr8 around the mitotic centrosome is shown on the right. At least 150 cells were counted (n = 3). n.s., not significant. (C) Whole cell extracts were prepared from HeLa cells treated with control, hMsd1/SSX2IP or Wdr8 siRNA and immunoblotting were performed with antibodies against hMsd1/SSX2IP, Wdr8 and α-tubulin (loading control).
These results highlighted the spatial regulation of these two proteins between fungi and humans. While the requirement of Wdr8 for hMsd1/SSX2IP localisation to the mitotic centrosome is well conserved, the converse relationship is not. As shown earlier (see Fig. 2D), unlike fungi [11], [20], Wdr8 did not display complete colocalisation with Wdr8. We surmise that the recruitment of Wdr8 to the mitotic centrosome also is regulated by pathway(s) independent of hMsd1/SSX2IP. In line with this hypothesis, phylogenetic analysis indicates that although many organisms contain both homologues (eg. humans, fish, plants and fungi), there are several species, such as Chlamydomonas reinhardtii and Tetrahymena thermophile, that contain only Wdr8 homologues on the genome, but not those of Msd1. Interestingly, no organisms have been found that contain Msd1 homologues but not those of Wdr8 [11].
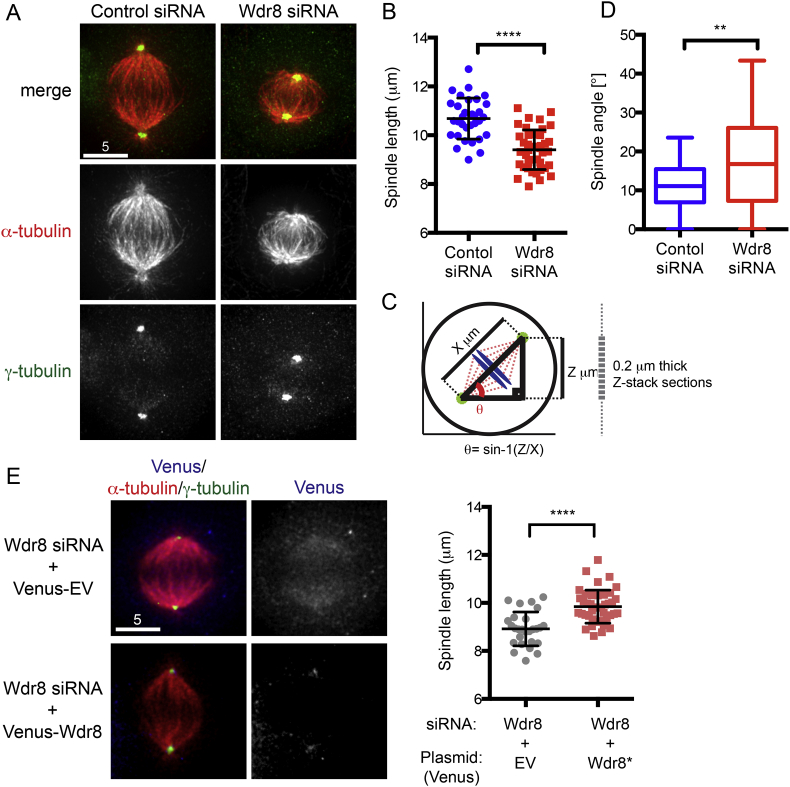
3.4. Depletion of Wdr8 leads to shorter, tilted mitotic spindles, reminiscent of hMsd1/SSX2IP knockdown
During measurement of hMsd1/SSX2IP signal intensities in mitotic cells depleted of Wdr8, we noticed that overall spindle morphologies were not normal; they were shorter and looked somehow distorted (Fig. 4A). Quantification of spindle length and 3-D spindle orientation (angles) in HeLa cells clearly indicated that both parameters were abnormal; spindle length of Wdr8 depleted cells was shorter than that of control cells by ∼20–25% (Fig. 4A and B) and spindles were tilted relative to the planar surface (∼11° in control siRNA vs ∼16° with Wdr8 siRNA, Fig. 4C and D). Additionally, these defects were also observed in U2OS cells (Supplementary Fig. S4A and B). Intriguingly, compared to wild type cells, mitotic spindles in cells depleted of Wdr8 exhibited very scarce, if not absent, astral microtubules (Fig. 4A), which are important to maintain proper spindle orientation [21]. It is worth noting that the defective phenotypes of shorter and tilted spindles were very similar, if not identical, to those induced by the deletion of hMsd1/SSX2IP [6]. Together, the emergence of the defective spindles upon Wdr8 depletion is consistent with the previous result showing that Wdr8 depletion led to the reduced levels of hMsd1/SSX2IP at the mitotic centrosome (see Fig. 3A).
Fig. 4.
Wdr8 depletion results in shorter and tilted mitotic spindles. (A) Representative images of mitotic spindle morphologies of HeLa cells treated with control or Wdr8 siRNA are shown. Immunofluorescence microscopy was performed with antibodies against α-tubulin (red) and γ-tubulin (green). (B) Quantification of spindle length. Data derived from (A) is shown. At least 150 cells were counted (n = 3). ****p < 0.0001. (C, D) Schematic presentation of spindle angle (C, θ represents angle °) and quantification of spindle orientation (D) are shown. Data derived from (A) is shown. At least 150 cells were counted (n = 3). **p < 0.01. (E) Rescue of defective spindle morphologies by introduction of siRNA-resistant Wdr8 (Wdr8*) constructs. HeLa cells depleted of Wdr8 were transfected with plasmids producing siRNA-resistant Wdr8 and immunofluorescence microscopy was performed as in (A). Images of Venus (Venus only on the top and Venus-Wdr8 on the bottom) are shown. Note that Venus-Wdr8 localises to the centrosomal area (bottom). Quantification data is presented in the right-hand side. At least 150 cells were counted (n = 3). ****p < 0.0001. Scale bars, 5 μm.
To confirm that defective spindle morphologies seen in Wdr8 depleted cells were indeed derived from Wdr8 dysfunction, siRNA-resistant Wdr8 constructs were introduced into cells in which endogenous Wdr8 was depleted. As shown in Fig. 4E, shorter spindle defects were effectively rescued by the introduction of siRNA resistant Wdr8. Therefore, Wdr8 plays a critical role in bipolar spindle formation and acts in concert with hMsd1/SSX2IP.
3.5. Evolutionary conservation and diversification of the centrosomal Msd1-Wdr8 complex
This study has demonstrated that the conserved centrosomal protein Wdr8 forms a complex with hMsd1/SSX2IP, which is critical to ensure proper spindle length and orientation. This complex is conserved in fungi, in which it localises to the mitotic SPB and ensures proper spindle assembly; in particular it is required for the anchoring of the minus end of spindle microtubules to the SPB [11], [20]. We previously showed that hMsd1/SSX2IP is also required for spindle anchoring and its depletion led to the emergence of shorter, tilted spindles accompanied with dissociation of astral microtubules from the mitotic centrosome. As shown in this study, Wdr8 depletion led to very similar spindle defects as well as the reduced recruitment of hMsd1/SSX2IP to the centrosome. These results strongly suggest that the Msd1-Wdr8 complex is a universal regulator of spindle anchoring at the mitotic centrosome/SPB.
Our recent work in fission yeast showed that Msd1 and Wdr8 further form a tertiary complex with kinesin-14 Pkl1, thereby antagonising with the opposing force generated by kinesin-5 Cut7 [11]. As kinesin-5 and kinesin-14 are widely conserved throughout eukaryotes [22], it would be of great interest to explore whether the human Msd1/SSX2IP-Wdr8 complex interacts with kinesin-14 (HSET/KIFC1). Furthermore, previous systematic proteomic studies identified Wdr8 as a potential centrosomal component [23], [24], and it associates with Cep135 [25]. As shown here, the localisation of Wdr8 within the centrosome overlaps with that of Cep135 (Fig. 1C). It would be of interest to explore any functional relationship between these two proteins and the possible involvement of hMsd1/SSX2IP in the future.
While fungi Msd1 and Wdr8 are not recruited to the SPB during interphase, hMsd1/SSX2IP is a component of centriolar satellites [6], [18] and Wdr8 localises to the centrosome during interphase. Whether or not Wdr8 is also a component of centriolar satellites remains to be established, though our immunofluorescence data did not clearly show this. Further work will be necessary to explore regulatory pathway(s) that is responsible for Wdr8 localisation to the interphase centrosome and what is a role for Wdr8 during interphase.
4. Conclusion
We have identified the human centrosomal complex, Wdr8-hMsd1/SSX2IP, that is conserved from fungi to human beings. Furthermore, we have shown that the mitotic role of this complex is also conserved; it is required for the assembly of proper bipolar spindles. In particular these two proteins play an essential role in the maintenance of proper spindle length and orientation probably through anchoring the minus end of the mitotic spindles to the centrosome.
Acknowledgements
We thank Michel Bornens for reagents and Anne Vaahtokari for help with the OMX microscope. We are grateful to Akiko Toda for critical reading of the manuscript. T.T. and A.P.S were supported by Cancer Research UK.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.10.169.
Author contribution
The experiments were designed by A.H. and T.T., A.H. and A.M performed the majority of the experiments and data analysis, C.I. participated in the initial stage of the project and prepared anti-Wdr8 antibody, and D.F. performed LC-MS and together with A.P.S. identified Wdr8 interactors. A.H. and T.T. wrote the paper with suggestions from other authors.
Conflict of interest
The authors declare no conflict of interest arising from this work.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 2.Bornens M. The centrosome in cells and organisms. Science. 2012;335:422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 3.Fu J., Hagan I.M., Glover D.M. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 2015;7:a015800. doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg E.A., Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nigg E.A., Raff J.W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Hori A., Ikebe C., Tada M., Toda T. Msd1/SSX2IP-dependent microtubule anchorage ensures spindle orientation and primary cilia formation. EMBO Rep. 2014;15:175–184. doi: 10.1002/embr.201337929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hori A., Peddie C.J., Collinson L.M., Toda T. Centriolar satellite- and hMsd1/SSX2IP-dependent microtubule anchoring is critical for centriole assembly. Mol. Biol. Cell. 2015;26:2005–2019. doi: 10.1091/mbc.E14-11-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toya M., Sato M., Haselmann U., Asakawa K., Brunner D., Antony C., Toda T. γ-Tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat. Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- 9.Koshizuka Y., Ikegawa S., Sano M., Nakamura K., Nakamura Y. Isolation, characterization, and mapping of the mouse and human WDR8 genes, members of a novel WD-repeat gene family. Genomics. 2001;72:252–259. doi: 10.1006/geno.2000.6475. [DOI] [PubMed] [Google Scholar]
- 10.Mahmoudi S., Henriksson S., Corcoran M., Mendez-Vidal C., Wiman K.G., Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Yukawa M., Ikebe C., Toda T. The Msd1-Wdr8-Pkl1 complex anchors microtubule minus ends to fission yeast spindle pole bodies. J. Cell Biol. 2015;290:549–562. doi: 10.1083/jcb.201412111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Y., Jung M.K., Oakley B.R. γ-Tubulin is present in Drosophila melanogaster and Homo sapiens and Is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- 14.Stearns T., Evans L., Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- 15.Salisbury J.L. Centrin, centrosomes, and mitotic spindle poles. Curr. Opin. Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 16.Ohta T., Essner R., Ryu J.H., Palazzo R.E., Uetake Y., Kuriyama R. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 2002;156:87–99. doi: 10.1083/jcb.200108088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry A.M., Mayor T., Meraldi P., Stierhof Y.-D., Tanaka K., Nigg E.A. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barenz F., Inoue D., Yokoyama H., Tegha-Dunghu J., Freiss S., Draeger S., Mayilo D., Cado I., Merker S., Klinger M., Hoeckendorf B., Pilz S., Hupfeld K., Steinbeisser H., Lorenz H., Ruppert T., Wittbrodt J., Gruss O.J. The centriolar satellite protein SSX2IP promotes centrosome maturation. J. Cell Biol. 2013;202:81–95. doi: 10.1083/jcb.201302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen K.F., Osmani S.A. Regulation of mitosis by the NIMA kinase involves TINA and its newly discovered partner An-WDR8 at spindle pole bodies. Mol. Biol. Cell. 2013;24:3842–3856. doi: 10.1091/mbc.E13-07-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyoshima F., Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence C.J., Dawe R.K., Christie K.R., Cleveland D.W., Dawson S.C., Endow S.A., Goldstein L.S., Goodson H.V., Hirokawa N., Howard J., Malmberg R.L., McIntosh J.R., Miki H., Mitchison T.J., Okada Y., Reddy A.S., Saxton W.M., Schliwa M., Scholey J.M., Vale R.D., Walczak C.E., Wordeman L. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 24.Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L.G., Bennetzen M., Westendorf J., Nigg E.A., Uhlen M., Hyman A.A., Andersen J.S. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchins J.R., Toyoda Y., Hegemann B., Poser I., Heriche J.K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M.M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A.A., Peters J.M. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.