Highlights
-
•
The first kinetic characterization of PputG6PDH-1 is presented.
-
•
The relative production of NADH and NADPH by PputG6PDH-1 is quantified.
-
•
The stoichiometric matrix of in silico metabolic models for Pseudomonas putida must be modified.
Abbreviations: G6P, glucose-6-phosphate; G6PDH, glucose-6-phosphate dehydrogenase; PputG6PDH-1, glucose-6-phosphate dehydrogenase encoded by the gene zwf-1 from Pseudomonas putida KT2440; GND, 6-phospho-gluconate dehydrogenase; EDP, Entner–Doudoroff pathway
Keywords: Pseudomonas putida, Glucose-6-phosphate dehydrogenase, NAD(H), NADP(H), Entner–Doudoroff pathway
Abstract
Despite the lack of biochemical information, all available in silico metabolic models of Pseudomonas putida KT2440 consider NADP as the only cofactor accepted by the glucose-6-phosphate dehydrogenases. Because the Entner–Doudoroff pathway is the main glycolytic route in this bacterium, determining how much NADH and NADPH are produced in the reaction catalyzed by these enzymes is very important for the correct interpretation of metabolic flux distributions. To determine the actual cofactor preference of the glucose-6-phosphate dehydrogenase encoded by the zwf-1 gene (PputG6PDH-1), the major isoform during growth on glucose, we purified this protein and studied its kinetic properties. Based on simple kinetic principles, we estimated the in vivo relative production of NADH and NADPH during the oxidation of glucose-6-phosphate (G6P). Contrary to the general assumption, our calculations showed that the reaction catalyzed by PputG6PDH-1 yields around 1/3 mol of NADPH and 2/3 mol of NADH per mol of oxidized G6P. Additionally, we obtained data suggesting that the reaction catalyzed by the 6-phosphogluconate dehydrogenase is active during growth on glucose, and it also produces NADH. These results indicate that the stoichiometric matrix of in silico models of P. putida KT2440 must be corrected and highlight the importance of considering the physiological concentrations of the involved metabolites to estimate the actual proportion of NADH and NADPH produced by a dehydrogenase.
1. Introduction
Pseudomonas putida KT2440 is a promising bacterium for the biotechnological industry due to its metabolic flexibility [1], low maintenance requirements [2] and low pathogenicity. However, further information about its enzymes is required to expand the basic knowledge of this organism and to improve its biotechnological applications.
In the particular case of the glucose-6-phosphate dehydrogenase (G6PDH, E.C. 1.1.1.49), the first step of the Entner–Doudoroff pathway (EDP), there are three alleles (zwf-1, zwf-2 and zwf-3) annotated as encoders for this enzyme in the genome of P. putida KT2440 [3]. While the protein encoded by zwf-1 (PputG6PDH-1) is regarded the major form during the growth on glucose [4], [5], the biological roles of zwf-2 and zwf-3 remain unknown. Interestingly, all the published in silico models consider that NADP and NADPH are the only cofactors participating in the reaction catalyzed by the G6PDHs [6], [7], [8], [9], [10]. However, to the best of our knowledge, no kinetic studies of the G6PDHs from P. putida KT2440 are available so far.
Although in P. putida KT2440 glucose is converted into 6-phospho-gluconate by three different pathways, the combination of 13C-labeling, gene deletions and gene expression experiments have shown that, during the growth on glucose as the sole carbon source, a flux of 3 to 6 mmol gDW−1 h−1 [2], [4] could pass through the reaction catalyzed by PputG6PDH-1. As a matter of comparison, in Escherichia coli K-12 MG1655, the flux through the G6PDH is only around 2 mmol gDW−1 h−1 [11]. Therefore, reliable data are necessary to assess how much NADH and/or NADPH is produced in this reaction, enabling a better interpretation of the metabolic fluxes distributions and the right calculation of the ATP maintenance cost. Moreover, the use of NAD and/or NADP by certain dehydrogenases had been associated with important traits of the metabolic networks [12], [13], helping to understand the evolution and systemic properties of the metabolic pathways.
To obtain a quick insight into the properties of PputG6PDH-1, we measured the NAD and NADP dependent G6PDH activities in crude cellular extracts from P. putida KT2440. Because our results suggested that both NADH and NADPH are produced in vivo during the oxidation of glucose-6-phosphate (G6P), we cloned the gene encoding for PputG6PDH-1, we expressed that gene in E. coli, we purified the recombinant protein and we performed a kinetic characterization of that protein using both NAD and NADP as cofactors. Afterwards, we used the obtained kinetic parameters to quantify the amount of PputG6PDH-1 present in the crude cellular extracts, but our results suggested that at least another G6PDH might be also present in the extracts.
To quantify the relative production of NADH and NADPH due to the activity of PputG6PDH-1 in the physiological conditions, we employed a kinetic model based on simple assumptions. We finally discussed what could be the metabolic impact of the simultaneous generation of NADH and NADPH in the reaction catalyzed by PputG6PDH-1 and how this could help to understand some results obtained in previous studies.
2. Materials and methods
2.1. Strains and plasmids
Cells of P. putida KT2440 (NCBI Taxonomy ID: 160488) were kindly donated by Dr. Henry Valentin, from the Institute of Molecular Microbiology and Biotechnology, Westfälische Wilhelms-Universität, Münster, Germany. Amplification and sequencing of the chromosomal region encoding for the rRNA subunit 16S using the primers 5′-AGAGTTTGATCMTGGCTC-3′ and 5′-CGGTGTGTACAAGACC-3′ confirmed the identity of the bacterium. E. coli MG1655 cells were obtained from the Yale E. coli Genetic Stock Center; while DH10B cells, BL21DE3 cells and plasmid pET28A were purchased from Invitrogen.
2.2. Specific activities in crude cellular extracts
Cells of P. putida KT2440 were aerobically grown in defined mineral medium, formulated as previously reported [14], with glucose (5 g/L) as the sole carbon source, with vigorous agitation (200 rpm), at 30 °C. In parallel, cells of E. coli MG1655 were grown in the same conditions but at 37 °C. Samples of biomass from those bacteria were obtained during the exponential growth phase by centrifugation of the broth (5000×g, 5 min, 4 °C) and the cellular extracts were prepared as previously described [13]. To measure the specific activities was employed the buffer Tris 50 mM pH 8.0, MgCl2 10 mM, NaCl 5 mM, glycerol 10% v/v, 2-mercapto-etanol 10 mM supplemented with the corresponding substrates. Activities were recorded monitoring the initial rate of formation of NAD(P)H following the changes in the absorbance at 340 nm at 30 °C. To control the contribution of possible background reactions to the changes in the absorbance, we monitored the effect of adding crude extracts to reaction mixtures with all the components but G6P, 6-phospho-gluconate, NAD or NADP. The enzymatic activities were normalized using the total protein concentration in the respective cellular supernatants. The concentration of protein in the cellular extracts was estimated using the commercial reagent Bio-Rad Protein assay, with BSA as the protein standard.
2.3. Molecular biology
We purified genomic DNA from cells of P. putida KT2440 using the commercial kit DNeasy Blood & Tissue Kit (Qiagen). After the analysis of the annotated genome, primers were designed to amplify the amino acid encoding sequences of the zwf-1 gene (NCBI reference sequence NP_743183.1). On the other hand, using a high fidelity DNA polymerase we obtained, by PCR, copies of the zwf-1 gene flanked by sites for the restriction enzymes NdeI and BamHI. The sequences of the employed primers were:
5′-aaccatcatatgatggccgcaatcagtgtcgaacc-3′
5′-gtaattggatccttcaattcagatatccccatacc-3′
The PCR products were cleaned, digested with NdeI and BamHI and ligated to molecules of pET28A previously digested with the same restriction enzymes. The ligation reaction was catalyzed by the T4 ligase (Thermo). The ligation products were introduced in E. coli cells strain DH10B by electroporation. The plasmids from three independent positive clones were purified. In those plasmids, the fragment of DNA encoding for the PputG6PDH-1 was sequenced. After confirmation of the integrity of the amino acid encoding sequence, the resulting plasmid (pET28A-Pputzwf-1) was introduced, by electroporation, in E. coli BL21-DE3 cells.
2.4. Protein purification
A purification protocol based on the presence of the poly-histidine tag in the N-term extreme of the molecules of PputG6PDH-1 was implemented following the procedure previously described [15]. Purity was checked by electrophoresis SDS–PAGE and staining with Coomassie blue. To rule out the possibility of co-purification of the G6PDH from the E. coli BL21-DE3 host cells, a sample of the purified enzyme was sent to our local protein identification facility, where mass spectrometry analyses were performed. No traces of other G6PDHs beyond the PputG6PDH-1 were detected.
To determine if the six-histidine tail affected the kinetic properties, we took 0.5 mg of the purified enzyme and we removed the tails by proteolysis with thrombin. After the proteolysis, the thrombin was separated from the PputG6PDH-1 using a procedure previously described [15].
2.5. Enzyme kinetics
The glycerol used to preserve the purified enzyme was removed by dialysis against the buffer Tris 50 mM pH 8.0, MgCl2 10 mM, NaCl 5 mM, glycerol 10% v/v, 2-mercapto-etanol 10 mM. Then, the enzyme was dissolved to the desired concentration. Prior to kinetic assays, the specific activity was checked to detect if significant inactivation happened during storage and/or handling. The kinetic assays were performed in the same buffer triggering the reactions with the addition of G6P. For the cofactor NADP, we explored concentrations from 15 μM to 900 μM, combined with concentrations of G6P from 300 μM to 9 mM. For the cofactor NAD, we explored concentrations from 200 μM to 10 mM, combined with concentrations of G6P from 300 μM to 15.23 mM. The minimal enzyme concentration that we could safely use during the assays was determined as described by Selwyn [16].
The initial rates were determined at 30 °C monitoring the NAD(P)H formation at 340 nm, in an UV–visible spectrophotometer Synergy H1 (Biotek, VE, USA), using non-binding flat-bottom 96-wells plates (model 655901, Greiner, Oberösterreich, Austria). The substrates were neutralized and titrated as described before [15]. NADPH was always freshly prepared and its concentration was directly quantified after neutralization. The molar extinction coefficient employed for quantification of the concentrations of NAD(P)H was 6220 M−1 cm−1, corresponding to λ = 340 nm. Using a working volume of 300 μl, the path length was around 0.7 cm, which means that a change in absorbance of 0.001 represented a change in the concentration of NAD(P)H of 0.23 μM.
In addition to the initial rates measurements, we performed progress curves experiments. The G6P concentration (12 mM) was above 10-fold KMG6P and more than 50-fold greater than the concentration of the cofactor. The chosen concentration of G6P, temperature and pH of the buffer minimized the reverse reaction.
2.6. Statistical analysis
For the different sets of experiments, all the obtained data were globally adjusted to different models and discrimination analyses were automatically performed to choose the model that best explained the observations. These analyses were performed with the software DynaFit [17]. For all the results we reported the standard errors calculated by the software as a measure of dispersion. Data and scripts are available as Supplementary material.
3. Results and discussion
3.1. The specific activities registered in the cellular extracts point to a non-negligible use of NAD by PputG6PDH-1
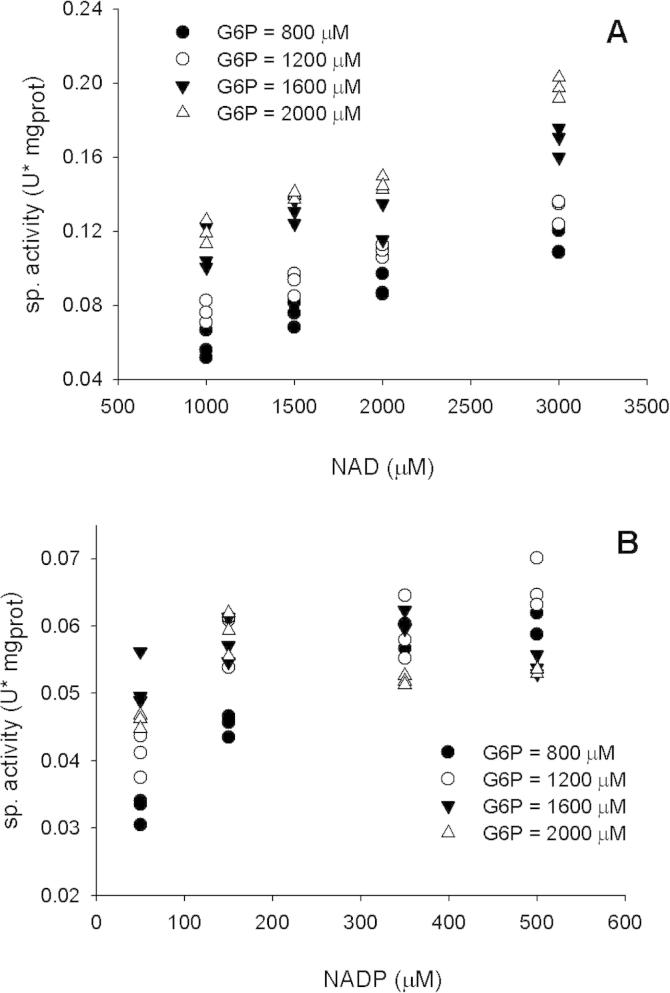
Using cells from P. putida KT2440 grown in mineral medium with glucose as the sole carbon source, we prepared fresh crude cellular extracts. Using these cellular extracts, we measured the enzymatic G6PDH activity using different concentrations of NAD, NADP and G6P. The obtained results indicated that in P. putida KT2440 both NAD and NADP could be used as cofactor for the oxidation of G6P (Fig. 1). These results contrasted with those previously obtained using cellular extracts from E. coli MG1655, where the G6PDH activities registered using NAD as cofactor were negligible [13].
Fig. 1.
Specific G6PDH activities registered in a cellular crude extract from P. putida KT2440 grown in mineral medium with glucose as the sole carbon source, using NAD (A) or NADP (B) as cofactor. The different symbols express values obtained using different concentrations of the co-substrate G6P.
G6PDH activities using NAD as cofactor had been reported in P. putida CSV86 [18]. Moreover, in the other species of the genus Pseudomonas where G6PDHs had been kinetically characterized, these enzymes showed similar preferences for NAD and NADP [19], [20], [21], [22]. Nevertheless, all the so far published in silico metabolic models of P. putida consider the G6PDH reaction as an exclusive NADPH producing step, presumably because the lack of enzyme kinetics information.
As was previously mentioned, in P. putida KT2440 there are at least three genes potentially encoding for proteins with G6PDH activity. Therefore, the registered activities using crude extracts could be the result of the contribution of more than one kind of enzyme, likely with different properties. This could explain why the activities using NADP combined with different concentrations of G6P did not follow the same trend observed when NAD was used as cofactor (Fig. 1).
3.2. PputG6PDH-1 catalyzes the oxidation of G6P using a rapid-equilibrium random mechanism
To get the data necessary to estimate the actual production of NADH and NADPH by PputG6PDH-1, we purified and performed a kinetic study of this enzyme.
Because the his-tagged form of PputG6PDH-1 showed no significant differences respect to the non-tagged form of the same enzyme (data not shown), here we showed the results obtained with the his-tagged form of PputG6PDH-1.
The dependences of kcat and KM on the concentration of G6P were accessed through a global fitting of the data coming from initial rates experiments performed varying the concentration of the cofactor at different but fixed concentrations of G6P (Table 1). The model discrimination indicated that the results could be best explained considering a random-mechanism, under the rapid-equilibrium assumption. The results also showed that PputG6PDH-1 has a specificity constants ratio (kcat/KM)NADP/(kcat/KM)NAD of 3.4, which is close to the value of 9 registered for the G6PDH from Leuconostoc mesenteroides (regarded as an enzyme able to use both NAD and NADP [23]) and it is very different from the value of 410 obtained for the NADP-preferring G6PDH from E. coli [15]. Therefore, PputG6PDH-1 should not be considered a NADP-specific enzyme, as it has been considered so far [6], [7], [8], [9], [10].
Table 1.
Kinetic parameters obtained for PputG6PDH-1. The mechanism that best explained the behavior of the initial rates was, both for NADP and for NAD, the random-ordered under the rapid-equilibrium assumption.
| Parameters | NADP | NAD |
|---|---|---|
| Ki (μM) | 111 ± 12 | 1148 ± 67 |
| KM (μM) | 14 ± 2 | 127 ± 8 |
| KMG6P (μM) | 946 ± 49 | 1137 ± 37 |
| kcat (s−1) | 102 ± 1 | 277 ± 2 |
To study the product inhibition, we set additional initial rates experiments including NADPH at different concentrations in the initial reaction mixtures. In one set of experiments, NADP was the varying substrate while the concentration of G6P was fixed and saturating (10 mM). The mixed-type inhibition was the model that best explained those observations (KMNADP = 16 ± 1.5 μM, kcat = 107 ± 1 s−1, KicNADPH = 35 ± 5 μM, Kiu = 877 ± 120 μM). It is possible that NADPH interacts with some part of the enzyme different from the active site, but this interaction seems to be weak in comparison with the interaction with the active site (Kiu ≈ 25 × KicNADPH). In the other set of experiments, G6P was the varying substrate while the concentration of NADP was fixed and saturating (200 μM), being the competitive inhibition the model that best explained the results (KMG6P = 723 ± 41 μM, kcat = 105 ± 1 s−1, KicNADPH = 163 ± 16 μM). For a full rapid-equilibrium random mechanism, it is expected that NADPH inhibits competitively when NADP is the varying substrate and non-competitively when G6P is the varying substrate [24]. We believe that the disagreement between the expected and the observed patterns of inhibition could be explained considering that some kinetic steps do not follow the rapid-equilibrium mechanism. However, for the sake of simplicity, for further analyses we considered the values obtained considering only competitive inhibition (Table 2), which are in good agreement with the values obtained without adding NADPH (Table 1).
Table 2.
Kinetic parameters obtained in the NADPH inhibition experiments, assuming a competitive model.
| Varying NADP | Varying G6P | |
|---|---|---|
| aKM (μM) | 9.8 ± 1.2 | 723 ± 41 |
| kcat (s−1) | 100 ± 1 | 105 ± 1 |
| Kic (μM) | 18 ± 3 | 163 ± 16 |
KM: correspond to KMNADP when NADP is varying and to KMG6P when G6P is varying.
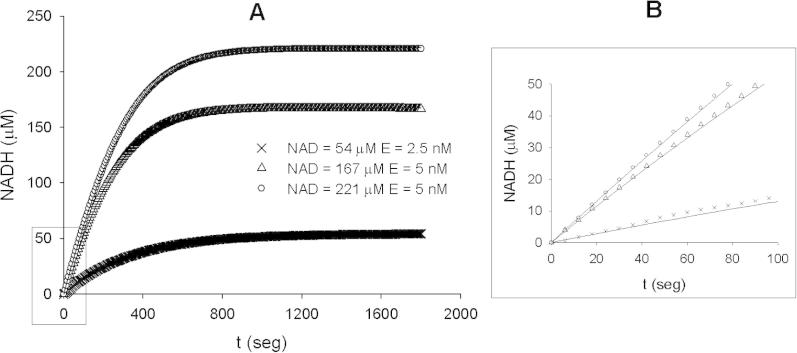
On the other hand, to study the inhibition by NADH, we set up progress-curves experiments (Fig. 2). In that case, NADH was not initially added to the reaction mixture but it was formed during the progress of the reaction. The model discrimination analysis showed that competitive inhibition was the mechanism that best explained the results (kcat = 254 ± 2 s−1, KMNAD = 412 ± 8 μM, KicNADH = 480 ± 7 μM). To study the inhibition by NADH, we chose this approach as a way to avoid an optical artifact known as stray light, previously described as the cause of spurious inhibitory effects of NADH [25].
Fig. 2.
(A) Progress curves representing some reactions catalyzed by PputG6PDH-1, using NAD and the enzyme at different concentrations. The concentration of G6P was saturating during all the progress of the reactions. The triangles, circles and crosses represent the experimental values while the curves represent the projection of the competitive product inhibition model evaluated with the best-fitted parameters. (B) Inset representing the first 100 s of the reaction progress.
We were unable to perform product inhibition experiments using 6-phosphogluconolactone, the other product of the reaction catalyzed by PputG6PDH-1. This compound is highly unstable in the conditions of the assays and it is not commercially available.
It is important to bear in mind that the differences among the kinetic parameters obtained assuming different possible kinetic mechanisms do not affect dramatically the rate of the reaction, as has been clearly demonstrated before [26]. However, the potential effects on the rate of other metabolites beyond the substrates and products need to be studied in the future.
3.3. PputG6PDH-1 is not the only form of G6PDH expressed during the growth on glucose
According with Northern blot analyses previously published, the proteins encoded by the genes zwf-2 and zwf-3 play minor roles in glucose metabolism [5]. Therefore, if PputG6PDH-1 is the predominant form of G6PDH expressed in P. putida KT2440 during the growth on glucose, with the obtained kinetic parameters and the initial velocities measured with crude cellular extracts, it would be possible to estimate the cytoplasmic concentration of the enzyme. To accomplish this, we rearranged the equation describing the initial rates for a bi-substrate random-ordered process under the rapid-equilibrium assumption
where A and B could be G6P or NAD(P), E is the enzyme concentration and v is the initial rate [24].
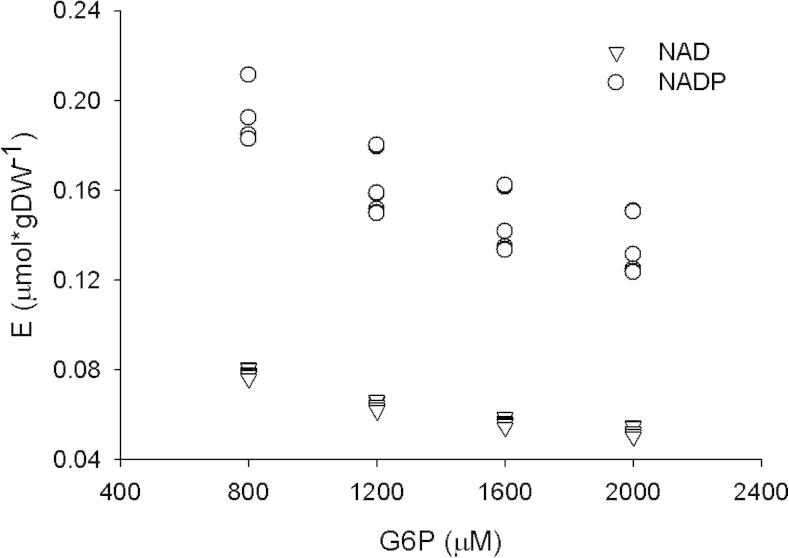
However, the results obtained using the above-described approach gave estimates for E strongly biased in relation with the concentration of G6P and the kind of nucleotide employed in the reaction (Fig. 3), which suggests that at least other G6PDH is contributing to the observed activities. Because the genes gnd and zwf-2 are in the same operon, the activity of 6-phosphogluconate dehydrogenase (GND) could indirectly indicate the expression of the protein encoded by zwf-2, annotated as another G6PDH. 13C-labeling experiments had shown that the oxidative branch of the pentose-phosphate pathway is operative in P. putida KT2440 when glucose is the sole carbon source [2], so we decided to measure the activity of GND in the crude cellular extracts (Table 3). The registered GND activities together with the biased estimations of E (Fig. 3) support that zwf-2 could encode for another G6PDH that is active when P. putida KT2440 is growing on glucose. Interestingly, the GND activity registered using NAD was higher than the activity registered using NADP, contrary to the pattern observed in the crude extract from E. coli MG1655, indicating that the cofactor preferences of the GNDs from these bacteria are different. As a matter of fact, the activity of GND in P. putida KT2440 had been pointed as one of the possible causes of the high NAD(P)H generation capacity of this bacterium [2]. A future characterization will help to clarify the contribution of the flux through the reaction catalyzed by GND to the NADH and NADPH pools.
Fig. 3.
Estimations of the enzyme concentration in the cellular crude extract from P. putida KT2440 using as input the data of the specific activities registered in this extract (Fig. 1). The details of the calculations are explained in the text. Contrary to the expected result if only one kind of G6PDH is present in the cellular extract, the estimations were strongly biased in relation with the kind of cofactor and the concentration of G6P employed during the measuring of the specific activities, suggesting the presence of at least another kind of G6PDH.
Table 3.
GND specific activities, using NAD or NADP as cofactors, registered in crude cellular extracts from E. coli MG1655 and P. putida KT2440 grown in mineral medium with glucose as the sole carbon source.
| Bacteria | aActivity with NAD (nmol mgprot−1 min−1) | aActivity with NADP (nmol mgprot−1 min−1) |
|---|---|---|
| E. coli MG1655 | 8 ± 1.5 | 103 ± 0.4 |
| P. putida KT2440 | 25 ± 0.4 | 6 ± 0.4 |
The reaction mixtures contained 2 mM of 6-phospho-gluconate and NAD (3 mM) or NADP (500 μM). The numbers represent average ± standard deviations from three replicates.
3.4. The dual-cofactor-preference of PputG6PDH-1 enables the production of 1/3 mol of NADPH and 2/3 mol of NADH per mol of oxidized G6P
Having the estimates for the most relevant kinetic parameters of the PputG6PDH-1, we used them to calculate the relative production of NADH and NADPH per mol of G6P oxidized in the reaction catalyzed by this enzyme. To accomplish this task, we adapted the kinetic model describing the initial rates when two or more purely competitive inhibitors are present in the reaction [24], [27]
doing the following considerations:
-
–
S was the cofactor NAD or NADP
-
–kcat and KS were the apparent kcat and KM:
-
–
If NAD was the cofactor, we considered NADP, NADH and NADPH as competitive inhibitors.
-
–
If NADP was the cofactor, we considered NAD, NADH and NADPH as competitive inhibitors.
-
–
We did not considerate the inhibition caused by the 6-phospho-gluconolactone. If the inhibition by this metabolite is eventually important, the effect should be similar for the reactions using NAD or NADP.
Overall, we obtained the following general expression, which can be applied for the reactions using NAD or NADP as substrate:
To evaluate the previous expression, the cytoplasmic concentrations of the involved substrates are needed. However, the published estimates of the concentration of NAD(P)(H) in P. putida KT2440 were obtained using whole-cell extraction methods [28], which are hindered by the fact that they cannot distinguish the free NAD(P)(H) from the non-negligible fraction of these cofactors that is bound to other cellular components, mainly proteins. Nevertheless, it is still possible to obtain meaningful results using reasonable approximations, as is explained below.
It had been documented that the concentration ratio NAD/NADH must be between 50 and 100 to make possible the functioning of the EMP in the glycolytic direction [29], [30], and that the concentration ratio NADP/NADPH must be between 0.8 and 1 [31], [32]. We also assumed that, in vivo, PputG6PDH-1 could be operating under saturating conditions for NAD and NADP, following the idea that many enzymes in vivo do so [33]. Thus we considered that the free cytoplasmic concentrations for these cofactors should be around 10 times the KMNAD and KMNADP respectively. Finally, we considered that the sums of the concentrations NAD + NADH and NADP + NADPH must be conserved, so an increment in one of the forms of the pair implied a diminution in the other form. Certainly, with this consideration we are assuming that the contribution of NAD and NADP from the biosynthetic pathways is exactly compensated with the metabolic degradation of these metabolites. This way, to calculate the values for the concentrations of NAD, NADH, NADP and NADPH satisfying the constraints mentioned above, we set up systems of simple algebraic equations.
To calculate the range of concentrations of NAD and NADH were used:
-
I.
NAD + NADH = (10 × 127 = 1270 ≈ 1300 μM) + (1300/50 = 26 μM)
-
II.
NAD/NADH from 1300/26 = 50 to 1300/13 = 100
And to calculate the range of concentrations of NADP and NADPH:
-
I.
NADP + NADPH = (10 × 13.7 = 137 ≈ 140 μM) + (1.2 × 140 = 168 μM)
-
II.
NADP/NADPH from 140/168 = 0.8 to 140/140 = 1
The results were: NAD (from 1300 μM to 1313 μM), NADH (from 13.13 μM to 26 μM), NADP (from 140 μM to 154 μM) and NADPH (from 154 μM to 168 μM).
Although we had not reliable estimations of E, the ratio between the equations used to calculate the production of NADH and NADPH is independent of E because this term is cancelled during the calculation of the ratio. Therefore, it is possible to estimate the relative production of NADH and NADPH due to the activity of the PputG6PDH-1 without having E.
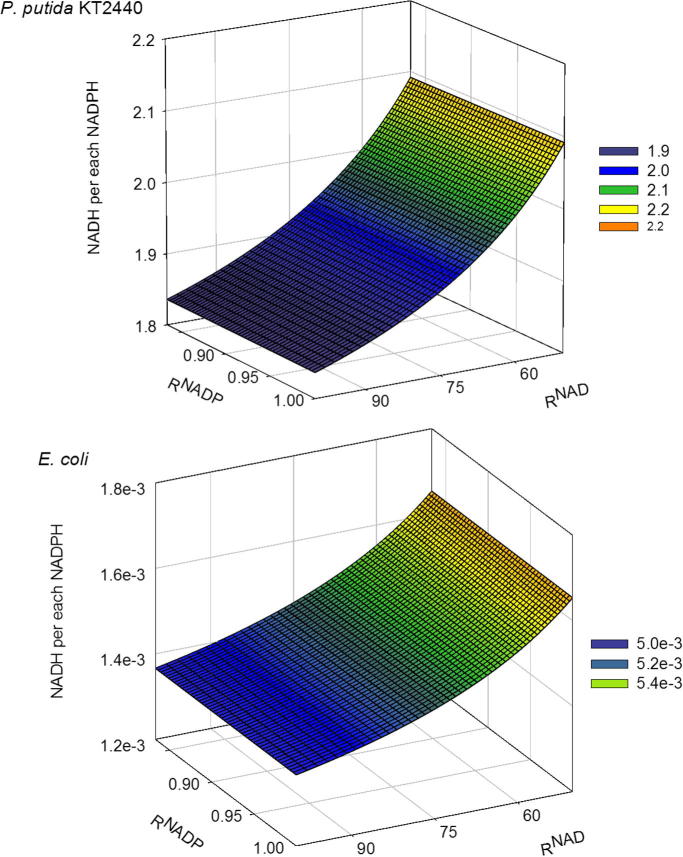
Using a concentration of G6P = 1 mM [34], the relative production of NADH over NADPH was around 2 (Fig.4A). That result showed that the in vivo rate of generation of NADH by PputG6PDH-1 is actually higher than its production of NADPH, which is contrary to the exclusive NADPH production considered in the so far published in silico models [6], [7], [35], [36]. In the unlikely case that the concentration of free NADP in the cells is much greater than that of NAD, then we will be making an overestimate of the NADH production; but if the concentration of free NADP is actually lower than our estimate, then we are making an overestimation of the NADPH production.
Fig. 4.
Relative productions of NADH respect to NADPH during the oxidation of G6P catalyzed by the enzymes PputG6PDH-1 (top) and EcG6PDH (bottom), depending on the concentrations ratios (R) of the oxidized and reduced forms of NAD(H) and NADP(H). The represented values were obtained assuming a concentration of G6P of 1 mM.
To validate the approach proposed here, we applied it to calculate the relative production of NADH and NADPH by the G6PDH from E. coli (EcG6PDH), which is widely accepted as a NADP-preferring dehydrogenase [13], [15], [37]. The results showed that in the case of EcG6PDH, the relative production of NADH over NADPH is 1000 times lower than the case of PputG6PDH-1 (Fig.4B). Therefore, the employed approach seems to be useful for the task of determine the relative production of NADH and NADPH by any dehydrogenase. However, it must be addressed that the results obtained with the method we are proposing should be treated with careful because the actual ‘free’ concentration of the involve metabolites could be hard to determine with accuracy. Although reliable estimations of the NAD/NADH and NADP/NADPH ratios based on thermodynamic considerations are available for eukaryotic microbes [29], [30], [38], this is not the case for the individual nucleotides. The improvements in the methods to quantify the free concentrations of the individual nucleotides will certainly contribute to a more accurate quantification of the relative generation of NADH and NADPH in the reactions catalyzed by dual cofactor preferring dehydrogenases. While these individual estimations are not yet available, sensitivity analyses could help to evaluate the impact of the free nucleotides concentrations on the relative production.
3.5. The metabolic importance of the dual cofactor preference of PputG6PDH-1
It had been recently suggested that the loose cofactor preference of the G6PDHs from some species could be a cofactor balance mechanism to avoid the relative excess of NADPH when the enzyme phosphofructokinase is absent and all the glycolytic flux goes through the G6PDH [13]. As a matter of fact, P. putida KT2440 does not have phosphofructokinase; therefore, the presence of a dual cofactor preferring G6PDH is consistent with that hypothesis.
Our results points to a generation of a non-negligible amount of NADH coupled with the oxidation of G6P. As a “rule of thumb”, and always considering that we used assumed free metabolite concentrations to perform the calculations for the relative production of NADH and NADPH, we suggest the following stoichiometry to represent the oxidation of G6P catalyzed by PputG6PDH-1:
The explicit consideration of the NADH generated in the reaction catalyzed by PputG6PDH-1 could be important to explain the remarkable robustness of the metabolism of P. putida KT2440 to increments in the demand of NADH and ATP [2]. The substitution of the native PputG6PDH-1 by an exclusive NADPH-producer homologous will help to test this hypothesis. On the other hand, our results could be also important for understanding the limitations to survive in anaerobic conditions observed in the mutant strain of P. putida KT2440 upon transformation with the genes encoding for acetate kinase, pyruvate decarboxylase and alcohol dehydrogenase [39]. In that case, the step catalyzed by the alcohol dehydrogenase can re-oxidize the NADH generated in the step catalyzed by the glyceraldehyde-3-phosphate dehydrogenase, but the NADH generated in the step catalyzed by PputG6PDH-1 must be re-oxidized by another enzyme to complete the oxidation of the generated NADH. The introduction of another enzyme(s) to re-oxidize this additional NADH should improve the anaerobic performance of this modified strain. Therefore, the results here presented could help in the design of metabolic engineering strategies based in the use of P. putida KT2440. Finally, the production of NADH in the reaction catalyzed by PputG6PDH-1 must be also considered to recalculate the ATP maintenance requirement according with the glucose consumption rates previously determined [2].
Authors’ contribution
K.O., L.F.S. and J.G.C.G. conceived and supervised the study; K.O., M.M. and H.C.O. designed experiments; K.O., M.M., H.C.O., J.C.R. and F.N.C.V. performed experiments; K.O. analyzed the data; K.O. wrote the manuscript; K.O., H.C.O. and J.G.C.G. made manuscript revisions.
Acknowledgement
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil, grants 2013/50357-2 and 2013/24087-8.
Footnotes
All the raw data used for the determination of the kinetic parameters as well as the scripts to analyze them using the software Dynafit are available as Supplementary material. Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fob.2015.11.002.
Appendix A. Supplementary data
References
- 1.Sudarsan S., Dethlefsen S., Blank L.M., Siemann-Herzberg M., Schmid A. The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2014;80:5292–5303. doi: 10.1128/AEM.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebert B.E., Kurth F., Grund M., Blank L.M., Schmid A. Response of Pseudomonas putida KT2440 to increased NADH and ATP demand. Appl. Environ. Microbiol. 2011;77:6597–6605. doi: 10.1128/AEM.05588-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson K.E. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 4.del Castillo T., Ramos J.L., Rodriguez-Herva J.J., Fuhrer T., Sauer U., Duque E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 2007;189:5142–5152. doi: 10.1128/JB.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J., Jeon C.O., Park W. Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology. 2008;154:3905–3916. doi: 10.1099/mic.0.2008/020362-0. [DOI] [PubMed] [Google Scholar]
- 6.Nogales J., Palsson B., Thiele I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst. Biol. 2008;2:79. doi: 10.1186/1752-0509-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puchalka J., Oberhardt M.A., Godinho M., Bielecka A., Regenhardt D., Timmis K.N., Papin J.A., Martins dos Santos V.A. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput. Biol. 2008;4:e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohn S.B., Kim T.Y., Park J.M., Lee S.Y. In silico genome-scale metabolic analysis of Pseudomonas putida KT2440 for polyhydroxyalkanoate synthesis, degradation of aromatics and anaerobic survival. Biotechnol. J. 2010;5:739–750. doi: 10.1002/biot.201000124. [DOI] [PubMed] [Google Scholar]
- 9.Oberhardt M.A., Puchalka J., Martins dos Santos V.A., Papin J.A. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis. PLoS Comput. Biol. 2011;7:e1001116. doi: 10.1371/journal.pcbi.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poblete-Castro I., Binger D., Rodrigues A., Becker J., Martins Dos Santos V.A., Wittmann C. In-silico-driven metabolic engineering of Pseudomonas putida for enhanced production of poly-hydroxyalkanoates. Metab. Eng. 2013;15:113–123. doi: 10.1016/j.ymben.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu G.P., Golding G.B., Dean A.M. The selective cause of an ancient adaptation. Science. 2005;307:1279–1282. doi: 10.1126/science.1106974. [DOI] [PubMed] [Google Scholar]
- 13.Olavarria K., De Ingeniis J., Zielinski D.C., Fuentealba M., Muñoz R., McCloskey D., Feist A.M., Cabrera R. The metabolic impact of a NADH-producing glucose-6-phosphate dehydrogenase in Escherichia coli. Microbiology. 2014 doi: 10.1099/mic.0.082180-0. [DOI] [PubMed] [Google Scholar]
- 14.Fuhrer T., Sauer U. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 2009;191:2112–2121. doi: 10.1128/JB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olavarria K., Valdes D., Cabrera R. The cofactor preference of glucose-6-phosphate dehydrogenase from Escherichia coli-modeling the physiological production of reduced cofactors. FEBS J. 2012;279:2296–2309. doi: 10.1111/j.1742-4658.2012.08610.x. [DOI] [PubMed] [Google Scholar]
- 16.Selwyn M.J. A simple test for inactivation of an enzyme during assay. Biochim. Biophys. Acta. 1965;105:193–195. doi: 10.1016/s0926-6593(65)80190-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 18.Basu A., Phale P.S. Inducible uptake and metabolism of glucose by the phosphorylative pathway in Pseudomonas putida CSV86. FEMS Microbiol. Lett. 2006;259:311–316. doi: 10.1111/j.1574-6968.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 19.Lessie T., Neidhard F.C. Adenosine triphosphate-linked control of Pseudomonas aeruginosa glucose-6-phosphate dehydrogenase. J. Bacteriol. 1967;93:1337. doi: 10.1128/jb.93.4.1337-1345.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessmann D., Schimz K.L., Kurz G. d-glucose-6-phosphate dehydrogenase (Entner–Doudoroff enzyme) from Pseudomonas fluorescens – purification, properties and regulation. Eur. J. Biochem. 1975;59:545–559. doi: 10.1111/j.1432-1033.1975.tb02481.x. [DOI] [PubMed] [Google Scholar]
- 21.Benbassat A., Goldberg I. Purification and properties of glucose-6-phosphate-dehydrogenase (NADP+–NAD+) and 6-phosphogluconate dehydrogenase (NADP+–NAD+) from methanol-grown Pseudomonas-C. Biochim. Biophys. Acta. 1980;611:1–10. doi: 10.1016/0005-2744(80)90036-4. [DOI] [PubMed] [Google Scholar]
- 22.Anderson B.M., Anderson C.D. Purification and characterization of Azotobacter vinelandii glucose-6-phosphate dehydrogenase: dual coenzyme specificity. Arch. Biochem. Biophys. 1995;321:94–100. doi: 10.1006/abbi.1995.1372. [DOI] [PubMed] [Google Scholar]
- 23.Vought V., Ciccone T., Davino M.H., Fairbairn L., Lin Y., Cosgrove M.S., Adams M.J., Levy H.R. Delineation of the roles of amino acids involved in the catalytic functions of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase. Biochemistry. 2000;39:15012–15021. doi: 10.1021/bi0014610. [DOI] [PubMed] [Google Scholar]
- 24.Segel I.H. John Wiley & Sons, Incorporated; New York: 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme System. [Google Scholar]
- 25.Cavalieri R.L., Sable H.Z. Pitfalls in the study of steady state kinetics of enzymes: spurious inhibition patterns due to stray light errors. Anal. Biochem. 1974;59:122–128. doi: 10.1016/0003-2697(74)90016-5. [DOI] [PubMed] [Google Scholar]
- 26.Rohwer J.M., Hanekom A.J., Crous C., Snoep J.L., Hofmeyr J.H. Evaluation of a simplified generic bi-substrate rate equation for computational systems biology. Syst. Biol. (Stevenage) 2006;153:338–341. doi: 10.1049/ip-syb:20060026. [DOI] [PubMed] [Google Scholar]
- 27.Schäuble S., Stavrum A.K., Puntervoll P., Schuster S., Heiland I. Effect of substrate competition in kinetic models of metabolic networks. FEBS Lett. 2013;587:2818–2824. doi: 10.1016/j.febslet.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 28.Borrero-de Acuña J.M., Bielecka A., Häussler S., Schobert M., Jahn M., Wittmann C., Jahn D., Poblete-Castro I. Production of medium chain length polyhydroxyalkanoate in metabolic flux optimized Pseudomonas putida. Microb. Cell Fact. 2014;13:88. doi: 10.1186/1475-2859-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kummel A., Panke S., Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2006;2(2006):0034. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canelas A.B., van Gulik W.M., Heijnen J.J. Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol. Bioeng. 2008;100:734–743. doi: 10.1002/bit.21813. [DOI] [PubMed] [Google Scholar]
- 31.Andersen K.B., von Meyenburg K. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 1977;252:4151–4156. [PubMed] [Google Scholar]
- 32.Henry C.S., Broadbelt L.J., Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys. J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowry O.H., Carter J., Ward J.B., Glaser L. Effect of carbon and nitrogen sources on level of metabolic intermediates in Escherichia coli. J. Biol. Chem. 1971;246:6511. [PubMed] [Google Scholar]
- 35.Chavarría M., Nikel P.I., Pérez-Pantoja D., de Lorenzo V. The Entner–Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 2013;15:1772–1785. doi: 10.1111/1462-2920.12069. [DOI] [PubMed] [Google Scholar]
- 36.Nikel P.I., Martínez-García E., de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014;12:368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- 37.Sanwal B.D. Regulatory mechanisms involving nicotinamide adenine nucleotides as allosteric effectors. 3. Control of glucose 6-phosphate dehydrogenase. J. Biol. Chem. 1970;245:1626–1631. [PubMed] [Google Scholar]
- 38.Zhang J., ten Pierick A., van Rossum H.M., Seifar R.M., Ras C., Daran J.M., Heijnen J.J., Wahl S.A. Determination of the cytosolic NADPH/NADP ratio in Saccharomyces cerevisiae using shikimate dehydrogenase as sensor reaction. Sci. Rep. 2015;5:12846. doi: 10.1038/srep12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikel P.I., de Lorenzo V. Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3-dichloroprop-1-ene. Metab. Eng. 2013;15:98–112. doi: 10.1016/j.ymben.2012.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.