Abstract
Halophilic organisms are having adaptations to extreme salinity, the majority of them being Archaean, which have the ability to grow at extremely high salt concentrations, (from 3 % to 35 %). Level of salinity causes natural fluctuations in the halophilic populations that inhabit this particular habitat, raising problems in maintaining homeostasis of the osmotic pressure. Samples such as salt and water taken from Turda Salt Mine were analyzed in order to identify the eco-physiological bacterial groups. Considering the number of bacteria of each eco-physiological group, the bacterial indicators of salt quality (BISQ) were calculated and studied for each sample. The phosphatase, catalase and dehydrogenases enzymatic activities were quantitatively determined and the enzymatic indicators of salt quality (EISQ) were calculated. Bacterial isolates were analyzed using 16S rRNA gene sequence analysis. Universal bacterial primers, targeting the consensus region of the bacterial 16S rRNA gene were used. Analysis of a large fragment, of 1499 bp was performed to improve discrimination at the species level.
Keywords: Halophilic bacteria, Salt mine microbiota, Turda salt mine
Introduction
If life exists outside the Earth, it is most likely in forms similar to extremophilic bacteria and Archaea, based on analogy with extreme terrestrial environments. Studying such environments may give us clues about how life might succeed in the outer space.
Halophiles can survive in hypersaline habitats because of their ability to maintain osmotic balance (Kamekura 1998). There are many halophilous species belonging to Archaea but also to Bacteria. Archaea accumulate salts such as sodium chloride or potassium chloride (NaCl or KCl) up to concentrations that are isotonic with the environment (Wang et al. 2009). Accordingly, proteins from halophiles must cope with high salt concentrations. Lately, the potential biotechnological targets of these metabolites and enzymes have attracted much attention (Margesin and Schinner 2001). For instance, Birbir et al. (2004) isolated from salt mines some halophilic bacteria that were able to produce cellulase, but without a biochemical and molecular characterization. Also, Vreeland et al. (1998) isolated some cellulolytic strains from salt mines.
Some halophilic microorganisms are producing halostable exoenzymes, including amylases, proteases and lipases, with potential commercial value in various fields. They are used extensively in the food industry as additives, in biomedical science and in the chemical industry (Sánchez-Porro et al. 2003). It is known that halophilic and halo-tolerant enzymes are of general scientific interest because of their unique adaptation to environments of low water potential (Margesin and Schinner 2001).
In this paper, data on halophilic microorganisms adapted to environments of extreme salinity, collected from Turda Salt Mine are presented. The aim of our work was to determine the cultivable species found in Turda Salt Mine, in order to find halo-tolerant bacteria useful for biotechnological approaches.
Materials and Methods
Site Description and Sample Collection
In the Transylvanian Basin (Romania), salt (NaCl) rich regions are relatively common. Samples of salt and water were collected from Turda Salt Mine, (46º 35′ 0.3″ North; 23º 46′ 47″ East). Salt samples were collected at different depths as follows: from borehole of (A) Iosif Mine (–84 m), from (B) Big Hall of Rudolf Mine (–42 m). Water samples were collected from the lake within (C) Maria Theresia Mine (−112 m) (see http://salinaturda.eu for pictures of the sites (A), (B) and (C)). The samples were collected on sterile conditions, to avoid contamination. All salt samples were grab samples placed into 50 ml falcon tubes, subsequently sealed with parafilm and packed in an ice-chilled cooler for transportation.
Temperature, specific conductance and pH of water and salt samples were determined in the field, with a Multi 340i portable multiparameter. Salt concentration was estimated from specific conductance (SC) using the equation (Williams 1998):
where - CNaCl is the concentration of NaCl in g/l−1 (range 21–311 g/l−1); - SC is the specific conductance of the water sample in mS/cm−1 (range 30–225 mS/cm−1).
Bacterial Community and Enzymatic Activities
Microbiological analyses to identify the eco-physiological bacterial groups were made. In order to perform the bacteriological analysis we prepared a successive serial dilution of 1 g sampled salt (1 ml water for C site) at 9 ml sterile solution of NaCl 20 %. The number of the heterotrophs was determined by counting on agar medium plate (Atlas 2004). The analysis of the ammonifiers was performed on peptone medium and for denitrifiers was used the De Barjac culture medium (Atlas 2004). Except the heterotrophs (where the plate count method was used), the most probable number method was applied to estimate the number of microorganisms, using dilution series and multiple tubes per dilution. The most probable number of bacteria was calculated after incubation, according to the statistical table of Alexander (1965).
The general bacterial potential of the salt was appreciated based on the bacterial indicator of salt quality values (BISQ), considering the number of bacteria belonging to each group (Muntean 1995–1996).
The enzymological analyses consisted in determination of the following enzymatic activities: catalase activity expressed in mg split H2O2/1.5 g salt; phosphatase activity expressed in mg phenol/2.5 g salt; actual/potential dehydrogenase activity expressed in mg formazan/ g salt (Alef and Nannipieri 1995; Carpa et al. 2014).
Considering the absolute values of each enzymatic activity, the enzymatic indicator of salt quality (EISQ) was calculated (Muntean et al. 1996).
Isolation and Identification of Halophilic Bacteria
In order to isolate halophilic bacteria, LMG agar medium 220 (Atlas 2010) was used. 16S rRNA gene was amplified from bacterial isolates by direct PCR technique. The colony PCR protocol was modified, using bacterial suspensions as templates. Pure cultures obtained from a single bacterial colony after 24–48 h incubation were suspended in sterile water at a concentration of approximately 106 CFU/ml (Crăciunaş et al. 2010; Lupan et al. 2014). Universal bacterial primers, targeting the consensus region of the bacterial 16S rRNA gene, were used. Analysis of a large fragment, of 1499 bp was performed to improve discrimination at the species level (Li et al. 2013).
| Primer | Target | Sequence |
| 16S–8F | 16S rRNA gene | 5′-AGAGTTTGATCCTGGCTCAG -3′ |
| 16S–1493R | 5′-ACGGCTACCTTGTTACGACTT -3′ |
PCR mixture (25-μl) contained 2.5 μl 10xPCR reaction buffer (Fermentas), 25 pmol of each primer, 200 μM concentrations of each dNTP, 2 μl MgCl2 25 mM (2 mM final concentration) 0.75 U of Taq polymerase, and 2 μl bacterial suspension. PCR was performed in a Thermocycler, (Gradient Palm-Cycler™, Corbett Life Science).
The PCR program was as follows: denaturing step at 94oC for 5 min, followed by 30 cycles of denaturing for 45 s at 94oC, annealing for 45 s at 56oC and extension for 90 s at 72oC, followed by a final extension at 72oC for 5 min. Amplicons were separated on 1.5 % agarose gel, stained with ethidium bromide.
The presence of 16S rRNA gene was confirmed by amplicon sequence analysis. PCR products were purified using NucleoSpin gel and PCR clean-up kit (Macherey-Nagel), according to the manufacturer instructions. Purified PCR products were sequenced by Sanger method (3730XL, Applied Biosystems) at Macrogen Inc., Netherlands (Kumar et al. 2004). Both primers were used due to the length of fragments. Nucleotide sequences were edited by BioEdit and compared with the known sequences in GenBank, using BLAST of the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/).
Results and Discussion
In the samples originating from the 3 sites (borehole of Iosif Mine, Big Hall of Rudolf Mine, lake of Maria Theresia Mine) the specific conductance is reverse proportional with pH.
The specific conductance (SC) of salt samples was in the range 114–143 mS/cm-1. The pH of the samples was low to medium alkaline (Table 1). The air temperature in the Turda Salt Mine is between 10-12 °C and the water temperature in Maria Theresia Mine is around 8.5 °C.
Table 1.
Chemical analyses of samples from Turda Salt Mine
| Location in Turda Salt Mine | Mine depth (m) | T (°C) | pH | SC (mS/cm-1) | Estimated salt conc. (g/1-1) |
|---|---|---|---|---|---|
| Iosif Mine | −84 m | 10–12 | 7.8 | 143 | 129.93 |
| Big Hall of Rudolf Mine | −42 m | 10–12 | 8.3 | 123 | 102.24 |
| Lake of Maria Theresia Mine | −112 m | 8.5 | 8.7 | 114 | 90.98 |
The salt concentrations (NaCl) of the samples ranged between 90.98 and 129.9 g · l−1 (Table 1). According to these results, the environment can be considered hypersaline, the salt concentration of the water exceeding the concentration of the sea water (35 g · l−1). The specific feature of this habitat is that the salt deposit is the result of layers of dried solutes (evaporites) from ancient seas (Ștefănescu et al. 2000).
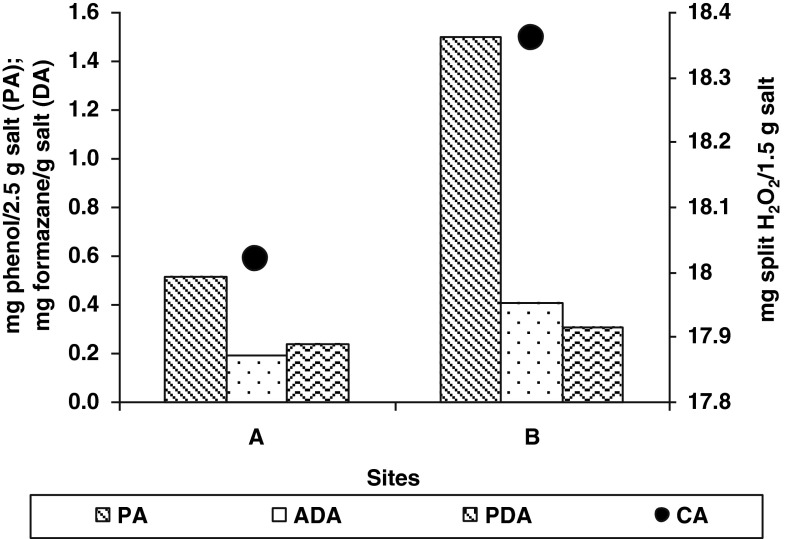
Overall, the assessed microbiological and enzymological activities were low. In samples A from borehole of Iosif Mine, enzymatic activity was lower than samples B from Big Hall of Rudolf Mine (Fig. 1). Phosphatase (PA) (1.4982 mg phenol/2.5 g salt in B site) and also catalase (CA) (18.36 mg split H2O2/1.5 g salt) activities had higher values because these enzymes are accumulating in time (Carpa and Butiuc-Keul 2009).
Fig. 1.
Enzymological analyses of salt samples from A and B sites (Turda Salt Mine). (A=borehole of Iosif Mine; B=from Big Hall of Rudolf Mine; PA=Phosphatase Activity; ADA=Actual Dehydrogenase Activity; PDA=Potential Dehydrogenase Activity; CA=Catalase Activity)
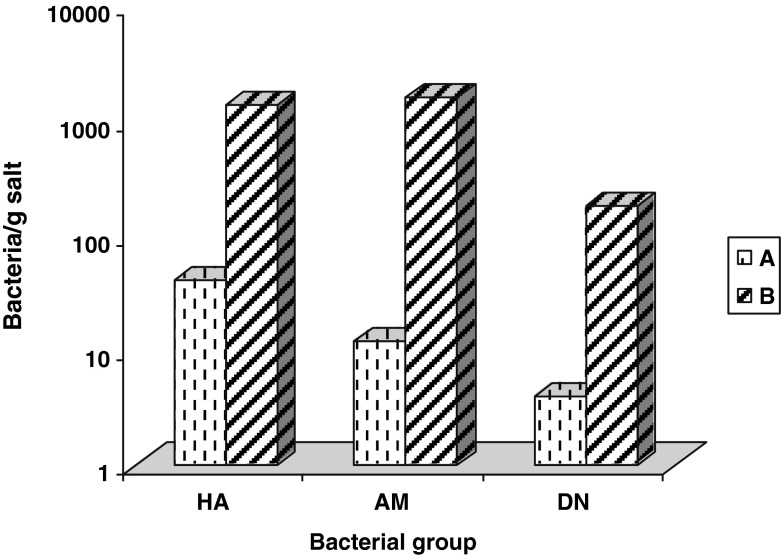
Also, bacterial eco-physiological groups were more abundant in samples B from Big Hall of Rudolf Mine. In this sampling site all identified bacteria groups (HA = heterotrophs aerobes: 1400 bacteria/g salt, AM = ammonifiers: 1600 bacteria/g salt, DN = denitrifiers: 180 bacteria/g salt) have reached relative higher numbers than bacteria groups from site A (Fig. 2). The presence of the studied groups in site C was very low (HA = 23 bacteria/g salt, AM = 9 bacteria/g salt, DN = 2 bacteria/g salt).
Fig. 2.
Bacteriological analyses of salt samples from A and B sites (Turda Salt Mine). (A=borehole of Iosif Mine; B=from Big Hall of Rudolf Mine; HA=heterotrophs aerobes; AM=ammonifiers; DN=denitrifiers)
The high values of enzymatic and microbial activities in the samples collected from Big Hall of Rudolf Mine may be caused by the contamination from the recreational area situated nearby.
The general bacterial potential of the salt was assessed using the formula (Muntean 1995–1996):
where BISQ = the bacterial indicator of salt quality;
- n
The number of the eco-physiological groups considered within the calculation
- N
The bacteria number belonging to each eco-physiological group.
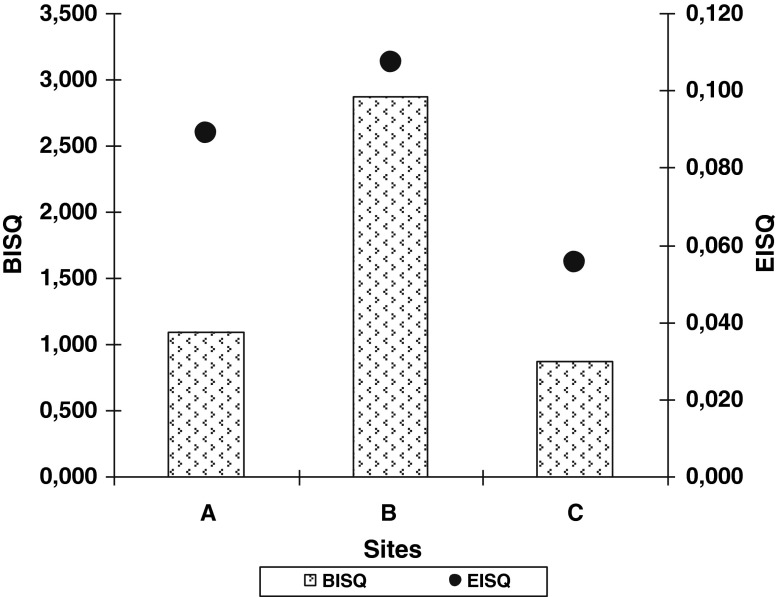
The BISQ values were relatively low. At the samples from Iosif Mine (A) we obtained a BISQ value of 1.094. At Big Hall of Rudolf Mine (B) we obtained a BISQ = 2.869 and the lowest value of the BISQ was in water samples from Maria Theresia Mine (C) (0.872).
On the base of the absolute values of each enzymatic activity, the enzymatic indicator of sediment quality (EISQ) was calculated, according with Muntean et al. (1996). The values of the EISQ follow an increasing curve along sampling sites, having the lowest value (EISQ = 0.056) in C site and the highest value (EISQ = 0.107) in B site (Fig. 3). The values of EISQ are between 0 and 1.
Fig. 3.
Bacterial (BISQ) and enzymatic (EISQ) indicators of salt quality. (A=borehole of Iosif Mine; B=from Big Hall of Rudolf Mine; C=from lake of Maria Theresia Mine)
Biological quality of a sample can be established, base on both indicators. In these samples, both, enzymatic and bacterial indicators of salt quality are having low values, indicating a low enzymatic and bacterial potential (Fig. 3).
A total of 15 samples from the 3 hypersaline sites from Turda Salt Mine were investigated by PCR analyses using the 16S rRNA genes. In these 15 samples only 7 species were identified.
According to the 16S rRNA analysis, sequences from the studied samples were obtained, with the phylogenetical similarity ranged from 93 to 100 %.
Table 2 summarizes the closest related species capable of tolerating the extreme conditions from the salt mine. The sequence data obtained were submitted to GenBank database under accession number: NR-041929 for Paenibacillus wynnii; NR-074290 for Bacillus megaterium QM B1551 strain; NR-044571 for Massilia niabensis strain 5420S-26; NR-025058 for Burkholderia fungorum strain LMG 16225; NR-042967 for Virgibacillus halodenitrificans strain DSM 10037; NR-075056 for Aquifex aeolicus strain VF5; NR-029172 for Aquifex pyrophilus strain Kol5a (Table 2).
Table 2.
Phylogenetic affiliation of sequences obtained from the Turda Salt Mine and the origin of most closely related isolates
| Isolate | Closest relatives (Accession number) | Simi-larity | Environmental preferences | References |
|---|---|---|---|---|
| A(1) | Paenibacillus wynnii NR-041929 | 93 % | Extreme conditions | La Duc et al. 2007 |
| A(5) | Bacillus megaterium QM B1551 strain NR-074290 | 95 % | Halotolerant | Khan 2011 |
| B(9) | Massilia niabensis strain 5420S-26 NR-044571 | 97 % | Air samples | Weon et al. 2010 |
| B(10) | Burkholderia fungorum strain LMG 16225 NR-025058 | 96 % | Environment samples, soil, groundwater, Arctic soil | Coenye et al. 2001 |
| C(11) | - Virgibacillus halodenitrificans strain DSM 10037 NR-042967 | 93 % | - A facultatively anaerobic halotolerant (salt-tolerant) denitrifier. | - Sang-Jae et al. 2012 |
| -Aquifex aeolicus strain VF5 NR-075056 | 100 % | - grows in mineral salt | - Deckert et al. 1998 | |
| -Aquifex pyrophilus strain Kol5a NR-029172 | 100 % | - grows in mineral salt | - Reysenbach et al. 2009 |
A borehole of Iosif Mine, B big hall of rudolf mine, C lake of Maria Theresia Mine
It can be noticed that these bacteria belong to very diverse eco-physiological groups. The occurrence of psychro-tolerant bacteria Burkholderia fungorum, thermophiles as Aquifex pyrophilus, obligate anaerobes as Paenibacillus wynnii, halophile as Bacillus megaterium beside the halotolerant bacteria as Virgibacillus halodenitrificans, Aquifex aeolicus, Aquifex pyrophilus in low alkaline pH environments, supports the assertion that “everything is everywhere, the environment selects” (de Wit and Bouvier 2006).
Conclusions
The determined bacteria groups in salt samples from Turda Salt Mine were: aerobic heterotrophs, ammonifiers and denitrifiers. Considering their abundance, the heterotrophs (10–103 bacteria/g salt) were followed by the ammonifying bacteria (1–103 bacteria/g salt) and denitrifying bacteria (1–102 bacteria/g salt).
The four studied quantitative enzymatic activities (phosphatase, catalase, actual and potential dehydrogenase) were present in all the salt samples, having different intensity, depending on the sampling place and the enzyme substrate.
Both, the enzymatic and bacterial indicators of salt quality registered low values: EISQ = maximum 0.1 and BISQ = maximum 2.8, values which are indicating low enzymatic and bacterial potential of the samples.
PCR amplification of 16S rRNA gene from bacterial isolates indicated a phylogenetic similarity ranging from 93 % to 100 %.
Acknowledgments
This work was financially supported by a grant from Romanian Space Agency (ROSA), STAR Technology, Romania, PCDI 1/2012.
References
- Alef K, Nannipieri P. Methods in applied soil microbiology and biochemistry. London: Academic; 1995. [Google Scholar]
- Alexander M (1965) Most probable-number method for microbial populations. In: Black CA, Evans DD, White JL, Ensminger LE, Clark FE (eds), Methods of Soil Analysis, Ed. Am Soc Agron Madison
- Atlas RM. Handbook of microbiological media. 3. New York: CRC Press; 2004. [Google Scholar]
- Atlas RM. Handbook of microbiological media. 4. New York: CRC Press; 2010. [Google Scholar]
- Birbir M, Ogan A, Calli B, Mertoglu B. Enzyme characteristics of extremely halophilic archaeal community in Tuzkoy Salt Mine, Tukey. World J Microbiol Biotechnol. 2004;20:613–21. doi: 10.1023/B:WIBI.0000043185.06176.b8. [DOI] [Google Scholar]
- Carpa R, Butiuc-Keul A. Microbial activity in the subterranean environment of Dârninii Cave, Bihor Mountains. ELBA Bioflux. 2009;1(1):13–22. [Google Scholar]
- Carpa R, Drăgan-Bularda M, Muntean V. General microbiology-laboratory practice. Cluj-Napoca: Ed. Presa Universitara Clujeana; 2014. [Google Scholar]
- Coenye T, Laevens S, Willems A, Ohlen M, Hannant W, Govan JRW, Gillis M, Falsen E, Vandamme P. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int J Syst Evol Microbiol. 2001;51:1099–1107. doi: 10.1099/00207713-51-3-1099. [DOI] [PubMed] [Google Scholar]
- Crăciunaş C, Butiuc-Keul A, Flonta M, Brad A, Sigarteu M (2010) Application of molecular techniques to the study of Pseudomonas aeruginosa clinical isolate in Cluj-Napoca, Romania. Annals of Oradea University, Biology Fascicle, 243-247
- de Wit R, Bouvier T. Everything is everywhere, but, the environment selects; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–8. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- Kamekura M. Diversity of extremely halophilic bacteria. Extremophiles. 1998;2:289–95. doi: 10.1007/s007920050071. [DOI] [PubMed] [Google Scholar]
- Khan JA. Biodegradation of azo dye by moderately halotolerant Bacillus megaterium and study of enzyme azoreductase involved in degradation. Adv Bio Tech. 2011;10(07):21–27. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73(8):2600–2611. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Lee Y-J, Jeong H, Lee SJ, Lee H-S, Pan J-G, Kim B-C, Lee D-W. Draft genome sequence of Virgibacillus halodenitrificans 1806. J Bacteriol. 2012;194(22):6332–6333. doi: 10.1128/JB.01280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang M, Feng J, Zhang W, Wang Y, Gu Y, Song C, Wang S. Treatment of high-salinity chemical wastewater by indigenous bacteria – bioaugmented contact oxidation. Bioresour Technol. 2013;144:380–386. doi: 10.1016/j.biortech.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Lupan I, Ianc MB, Kelemen BS, Carpa R, Rosca–Casian O, Chiriac MT, Popescu O. New and old microbial communities colonizing a seventeenth-century wooden church. Folia Microbiol. 2014;59:45–51. doi: 10.1007/s12223-013-0265-3. [DOI] [PubMed] [Google Scholar]
- Margesin R, Schinner S. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles. 2001;5:73–83. doi: 10.1007/s007920100184. [DOI] [PubMed] [Google Scholar]
- Muntean V (1995–1996) Bacterial indicator of mud quality. Contrib Bot 73–76
- Muntean V, Crişan R, Paşca D, Kiss S, Drăgan-Bularda M. Enzymological classification of salt lakes in Romania. Int J Salt Lake Res. 1996;5(1):35–44. doi: 10.1007/BF01996034. [DOI] [Google Scholar]
- Reysenbach AL, Hamamura N, Podar M, Griffiths E, Ferreira S, et al. Complete and draft genome sequences of six members of the Aquificales. J Bacteriol. 2009;191:1992–3. doi: 10.1128/JB.01645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Porro C, Martín S, Mellado E, Ventosa A. Diversity of moderately halophilic bacteria producting extracellular hydrolytic enzymes. J Appl Microbiol. 2003;94:295–300. doi: 10.1046/j.1365-2672.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Ștefănescu M, Dicea O, Tari O (2000) Influence of extension and compression on salt diapirism in its type area, East Carpathians Bend Area, Romania, Geological Society London, Special Publications, 174
- Vreeland RH, Piselli AF, Jr, McDonnough S, Meyers SS. Distribution and diversity of halophilic bacteria in a subsurface salt formation. Extremophiles. 1998;2:321–233. doi: 10.1007/s007920050075. [DOI] [PubMed] [Google Scholar]
- Wang CY, Hsieh YR, Ng C-C, Chan H, Lin HT, Tzeng WS, Shyu YT. Purification and characterization of a novel halostable cellulase from Salinivibrio sp. strain NTU-05. Enzym Microb Technol. 2009;44:373–379. doi: 10.1016/j.enzmictec.2009.02.006. [DOI] [Google Scholar]
- Weon HY, Yoo SH, Kim SJ, Kim YS, Anandham R, Kwon SW. Massilia jejuensis sp. nov. and Naxibacter suwonensis sp. nov., isolated from air samples. Int J Syst Evol Microbiol. 2010;60:1938–1943. doi: 10.1099/ijs.0.015479-0. [DOI] [PubMed] [Google Scholar]
- Williams DW. Guidelines of lake management, vol 6. Management of inland saline waters. Japan: International Lake Environment Committee Foundation; 1998. [Google Scholar]