Highlight
The opinion is put forward here that certain intronic regions of plant genes could be converted to double-stranded RNA precursors for sRNA production through an RDR-dependent pathway.
Key words: Arabidopsis thaliana, double-stranded RNA precursor, intron, phase-distributed, RDR (RNA-dependent RNA polymerase), small RNA.
Abstract
Recent research has linked the non-coding intronic regions of plant genes to the production of small RNAs (sRNAs). Certain introns, called ‘mirtrons’ and ‘sirtrons’, could serve as the single-stranded RNA precursors for the generation of microRNA and small interfering RNA, respectively. However, whether the intronic regions could serve as the template for double-stranded RNA synthesis and then for sRNA biogenesis through an RDR (RNA-dependent RNA polymerase)-dependent pathway remains unclear. In this study, a genome-wide search was made for the RDR-dependent sRNA loci within the intronic regions of the Arabidopsis genes. Hundreds of intronic regions encoding three or more RDR-dependent sRNAs were found to be covered by dsRNA-seq (double-stranded RNA sequencing) reads, indicating that the intron-derived sRNAs were indeed generated from long double-stranded RNA precursors. More interestingly, phase-distributed sRNAs were discovered on some of the dsRNA-seq read-covered intronic regions, and those sRNAs were largely 24 nt in length. Based on these results, the opinion is put forward that the intronic regions might serve as the genomic origins for the RDR-dependent sRNAs. This opinion might add a novel layer to the current biogenesis model of the intron-derived sRNAs.
Introduction
In plants, a large portion of protein-coding genes produce intron-containing transcripts named pre-mRNAs (precursor messenger RNAs) during the initial step of the transcription. The pre-mRNAs are then processed into mature mRNAs through intron excision, which is indispensible for the follow-up translation (Lorkovic et al., 2000). For the genes with multiple exons, the embedded introns could be selectively spliced out during pre-mRNA processing, which is defined as alternative splicing (AS). AS greatly enhances the coding potential of the multi-exon genes, thus increasing the complexity of the plant transcriptome and proteome (Reddy et al., 2013). Although there have been great research efforts for the study of intron-implicated AS, other novel biological roles of the intronic regions remain to be explored.
Interestingly, recent research has linked introns to the biogenesis of small RNAs (sRNAs). In plants, there are two major classes of sRNAs, i.e. microRNAs (miRNAs) and small interfering RNAs (siRNAs). Plant miRNAs are 21–22-nt sRNAs originated from stem-loop structured, single-stranded precursors. The primary precursors called pri-miRNAs (primary microRNAs) are first cropped by Dicer-like 1 (DCL1), and are processed into the secondary precursors called pre-miRNAs (precursor microRNAs). The pre-miRNAs are cropped again by DCL1, resulting in miRNA/miRNA* short duplexes. Finally, the mature miRNAs are selectively incorporated into Argonaute (AGO)-associated silencing complexes (Jones-Rhoades et al., 2006; Voinnet, 2009). Recent discovery of intronic miRNA genes called mirtrons has added a novel layer to the canonical biogenesis cascade of the plant miRNAs. The debranched introns could form hairpin-like precursors for miRNA generation, which bypasses the DCL1-mediated first cleavages on the pri-miRNAs (Zhu et al., 2008; Meng and Shao, 2012; Yang et al., 2012; Rogers and Chen, 2013). Interestingly, mirtrons were also identified in animals (Berezikov et al., 2007; Okamura et al., 2007; Ruby et al., 2007; Ladewig et al., 2012). A recent study in rice showed that certain introns termed sirtrons could form internal hairpin structures encoding siRNAs. The 21-, 22-, and 24-nt sirtron-derived siRNAs are processed by DCL4-, DCL2-, and DCL3, respectively. It was observed that the sirtron-derived siRNAs had a strong tendency to be co-expressed with their host genes. Moreover, the AGO4-associated 24-nt siRNAs could direct site-specific DNA methylation on their host genes (Chen et al., 2011).
In summary, several pieces of experimental evidence have pointed to the emerging role of intronic regions in sRNA production. However, it has been noted that the current discoveries were focusing on the single-stranded introns which were capable of forming hairpin-like precursors for miRNA or siRNA production. Whether the intronic sequences could serve as the templates for double-stranded RNA synthesis through an RDR (RNA-dependent RNA polymerase)-dependent pathway is unclear. This assumption was raised based on the biogenesis models of ta-siRNAs (trans-acting small interfering RNAs) and heterochromatic siRNAs. Specifically, the single-stranded transcripts serve as the templates for double-stranded RNA synthesis through the RDR6- or the RDR2-dependent pathway. Then, the double-stranded precursors are processed by DCL4 and DCL3 for the production of ta-siRNAs and heterochromatic siRNAs, respectively (Jones-Rhoades et al., 2006). To investigate the possibility of intron-involved sRNA production through an RDR-dependent pathway, a genome-wide search was made for the RDR-dependent sRNA loci within the intronic regions in Arabidopsis (Arabidopsis thaliana).
Search for the RDR-dependent sRNAs in Arabidopsis
The sRNA high-throughput sequencing (HTS) data sets prepared from three rdr mutants (rdr1, rdr2, and rdr6) of Arabidopsis were retrieved from the GEO (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2009). These data sets were contributed by two previous studies (Kasschau et al., 2007; Lu et al., 2006). Please refer to the Supplementary Materials and Methods at JXB online for detailed information of the rdr-related HTS data sets. Each group of rdr-related data sets was treated separately to search for the RDR-dependent sRNAs based on the following criterion: the level of the RDR-dependent sRNA should be 10 RPM or higher in ‘Col-0’, and should be undetectable in the ‘rdr’ mutant (Fig. 1). As a result, 55 214 sRNAs were identified to be RDR1-dependent, 65 546 were RDR2-dependent, and 72 860 were RDR6-dependent in Arabidopsis (see Supplementary Data S1 at JXB online).
Fig. 1.
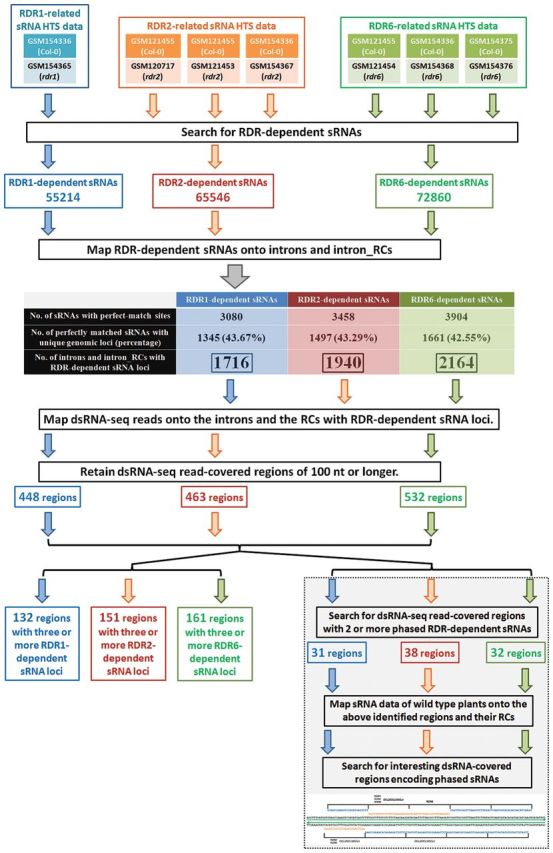
Workflow for the identification of phase-distributed, RDR (RNA-dependent RNA polymerase)-dependent small RNAs within the intronic regions in Arabidopsis.
Identification of RDR-dependent sRNA loci within the intronic regions
To investigate the potential of the introns for the production of the RDR-dependent sRNAs, the above-identified RDR-dependent sRNAs were mapped onto the intron sequences retrieved from the Arabidopsis Information Resource (TAIR, release 10; http://www.arabidopsis.org/) (Huala et al., 2001). Considering the proposed opinion that introns might serve as the templates for RDR-mediated double-stranded RNA synthesis, which was the prerequisite for the generation of RDR-dependent sRNAs, the reverse complementary sequences of the introns named intron_RCs were also included for sRNA mapping. As a result, 3 080 RDR1-dependent sRNAs, 3 458 RDR2-dependent sRNAs, and 3 904 RDR6-dependent sRNAs could find their perfectly matched loci within specific introns or intron-RCs. Next, the genomic origins of these intron-located sRNAs were investigated by mapping them onto the Arabidopsis genome (TAIR 10). The result showed that a large portion of RDR1- (1 345/3 080=43.67%), RDR2- (1 497/3 458=43.29%), and RDR6-dependent (1 661/3 904=42.55%) sRNAs possessed unique genomic loci, indicating that they were indeed encoded within the intronic regions. The remaining portion of the sRNAs had multiple genomic loci. However, the possibility that they were produced from the intronic regions could not be excluded since these sRNAs could find one of their genomic loci within specific introns. Therefore, the introns and RCs containing at least one sRNA locus were retained for the following analysis. It resulted in 941 introns and 775 RCs containing RDR1-dependent sRNA loci, 1 022 introns and 918 RCs containing RDR2-dependent sRNA loci, and 1 154 introns and 1 010 RCs containing RDR6-dependent sRNA loci (see Supplementary Data S2 at JXB online). Based on the above results, it is proposed that the intronic regions might be one of the genomic origins of the RDR-dependent sRNAs. In addition, the sRNA loci show an evenly distributed pattern between the sense and the RC sequences of the introns, indicating that the sRNAs are likely to be generated from the long double-stranded precursors.
A portion of the intronic regions containing RDR-dependent sRNA loci were covered by dsRNA-seq (double-stranded RNA sequencing) reads
To obtain further evidence supporting the idea that certain intronic regions could be converted to double-stranded precursors for the production of RDR-dependent sRNAs, dsRNA-seq data sets contributed by Zheng et al. (2010) were included in the following analysis. First, the dsRNA-seq reads were mapped onto the introns and the intron_RCs containing RDR-dependent sRNA loci. A search was then made for the dsRNA-seq read-covered regions of 100 nt or longer. Every nucleotide of these regions should be covered by at least one dsRNA-seq read. As a result, 234 and 214 dsRNA-seq read-covered regions were identified on the introns and the RCs containing RDR1-dependent sRNA loci, respectively. A total of 239 and 224 dsRNA-seq read-covered regions were identified on the introns and the RCs containing RDR2-dependent sRNA loci, respectively. A total of 284 and 248 dsRNA-seq read-covered regions were identified on the introns and the RCs containing RDR6-dependent sRNA loci, respectively (Fig. 1; see Supplementary Data S3 at JXB online).
To investigate the potential of the dsRNA-seq read-covered regions for sRNA production, a search was made for the dsRNA-seq read-covered regions containing three or more RDR-dependent sRNA loci. As a result, 132 [132/(234+214)=29.5% of the above identified regions] dsRNA-seq read-covered regions contain three or more RDR1-dependent sRNA loci. A total of 151 [151/(239+224)=32.6%] dsRNA-seq read-covered regions contain three or more RDR2-dependent sRNA loci. A total of 161 [161/(284+248)=30.3%] dsRNA-seq read-covered regions contain three or more RDR6-dependent sRNA loci (Fig. 1). Notably, many of these regions tend to be the hot spots for sRNA generation, because more than ten sRNA loci were identified from each of these regions (see Supplementary Data S1 at JXB online). Taken together, the above results indicate that certain intronic sequences could be converted to double-stranded precursors, and a significant portion of these precursors possess a great potential of generating RDR-dependent sRNAs.
Phase-distributed sRNAs identified within the dsRNA-seq read-covered regions
Based on the biogenesis model of the ta-siRNAs, the double-stranded precursors synthesized by RDR6 are processed into short siRNA duplexes by DCL4 (Jones-Rhoades et al., 2006). The ta-siRNAs of 21 nt were phase-distributed on both strands of the precursors. In this regard, we questioned whether the intron-originated double-stranded precursors could be processed by specific DCL(s) for the production of phase-distributed sRNAs.
To address this possibility, a search was first made for the dsRNA-seq read-covered regions with two or more phase-distributed, RDR-dependent sRNA loci. As a result, 31 regions were identified to contain two or more phase-distributed, RDR1-dependent sRNA loci. Thirty-eight regions contain two or more phase-distributed, RDR2-dependent sRNA loci. Thirty-two regions contain two or more phase-distributed, RDR6-dependent sRNA loci (see Supplementary Data S5 at JXB online). However, it was observed that most of the identified phases were constituted by only two RDR-dependent sRNA loci. If these dsRNA-seq read-covered regions indeed produced phase-distributed sRNAs, it was thought that the absent phased sRNAs might be unstable in planta which could result in low abundances in ‘Col-0’ (the wild-type plants of Arabidopsis) data sets. According to the criterion for the identification of the RDR-dependent sRNAs, the sRNAs with abundances lower than 10 RPM in ‘Col-0’ would be excluded.
Therefore, the sRNA sequences from ‘Col-0’ (GSM121455, GSM154336, and GSM154375) were mapped onto the sequence regions with two or more phase-distributed, RDR-dependent sRNA loci identified above. Both the sense and the RC strands of these regions were included for mapping. When the search was made for the phased sRNA duplexes, the 2-nt 3′ overhangs of the short sRNA duplexes resulting from the DCL-mediated processing were taken into account. As a result, six dsRNA-seq read-covered regions were discovered. Specifically, three different phases were identified on the region within the 14th intron of AT1G40104.1 (encodes an unknown protein according to TAIR10 annotation). Two different phases were identified on the region within the 5th intron of AT4G03380.1 (homologous to nuclear assembly factor 1). Two different phases were identified on the region within the 5th intron of AT5G36180.1 (serine carboxypeptidase-like 1). One phase was identified on the region within the 7th intron of AT4G04710.1 (calcium-dependent protein kinase). One phase was identified on the region within the 3rd intron of AT5G27606.1 (unknown protein). One phase was identified on the region within the 1st intron of AT2G18530.1 (protein kinase superfamily protein) (see Supplementary Fig. S1 at JXB online). Notably, most of the phased sRNAs were 24 nt in length. For each phase, there were three RDR-dependent sRNAs (see Supplementary Fig. S1 at JXB online and see expression levels of these sRNAs in Supplementary Data S6 at JXB online).
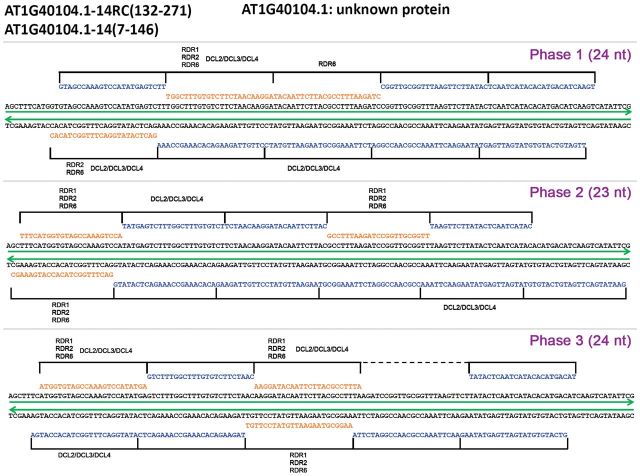
Taken the 14th intron of AT1G40104.1 as an example, three different phases (two of 24 nt and one of 23 nt) were discovered on the dsRNA-seq read-covered region within this intron (Fig. 2). Each phase is constituted by five continuous sRNA duplexes with 2-nt overhangs at the 3′ ends. Three RDR-dependent sRNAs were identified within each phase and the rest of the phase-distributed sRNAs with low abundances were identified from ‘Col-0’. These low-abundant sRNAs might be subjected to rapid degradation once they were processed from the double-stranded precursors by specific DCL(s). In addition, it was observed that a portion of the phase-distributed sRNAs had unique genomic loci. For example, all of the phase-distributed sRNAs identified from the 5th intron of AT4G03380.1 and the 3rd intron of AT5G27606.1 possess unique genomic loci (see Supplementary Fig. S1 at JXB online), supporting the reliability of these intron-derived sRNAs.
Fig. 2.
Example of phase-distributed sRNAs (small RNAs) identified within a dsRNA-seq (double-stranded RNA sequencing) read-covered intronic region of Arabidopsis. Three phases were discovered on this region. The ID of the host intron is shown on the top left, and the location of this region is provided in parentheses. ‘RC’ indicates the reverse complementary strand of the sense intronic strand. The gene annotation retrieved from the Arabidopsis Information Resource (TAIR, release 10; http://www.arabidopsis.org/) is provided on the top. The RDR (RNA-dependent RNA polymerase)-dependent sRNAs are highlighted in red and the other phase-distributed sRNAs identified from the sRNA sequencing data sets of wild type plants of Arabidopsis are highlighted in blue. The phased sRNAs are clearly denoted by solid lines and the positions which lack sequencing-evidenced sRNAs are denoted by dashed lines. The dsRNA-seq read-covered regions are marked by green lines on both sense and RC strands. The sRNAs dependent on RDR or DCL (Dicer-like) or both are marked.
To find evidence to support the idea that the phased sRNAs were processed by specific DCL(s), the sRNA HTS data sets prepared from the dcl mutants were retrieved from GEO. Specifically, two groups of data sets were included: (i) GSM366868 (wild-type plant: Col-0), GSM366869 (mutant: dcl1) and GSM366870 (triple mutant: dcl234); (ii) GSM121455 (Col-0), GSM121456 (dcl1) and GSM121457 (dcl234). Because DCL1 is mainly involved in the processing of miRNA precursors (Jones-Rhoades et al., 2006), it was thought that the production of the phased sRNAs from the long double-stranded precursors might depend on DCL2, DCL3 or DCL4, but not DCL1. Comparing the abundances of the phased sRNAs among ‘Col-0’, ‘dcl1’, and ‘dcl2dcl3dcl4’, a total of ten phased sRNAs (six of the ten sRNAs are RDR-dependent) showed clear dependence on DCL2, DCL3 or DCL4 (see Supplementary Fig. S1 and Supplementary Data S6 at JXB online). It was not possible to tell which DCL(s) was (were) critical for the generation of these phased sRNAs since the HTS data sets available for this analysis were prepared from the triple mutants dcl2dcl3dcl4. However, according to the reported sequence features of the sRNAs processed by different DCLs (Jones-Rhoades et al., 2006; Chen et al., 2011), and the fact that most of the phased sRNAs identified in this study were 24 nt, it was deduced that DCL3 might be the important enzyme for the generation of these phased sRNAs.
Conclusions and perspectives
Based on the above results, the opinion is put forward that certain intronic regions of plant genes could be converted to double-stranded precursors for RDR-dependent sRNA production. However, some questionable aspects need to be addressed before an elaborate biogenesis model of these intron-derived sRNAs can be presented. First, at which stage are the intronic regions converted to the double-stranded RNA precursors, before or after the excision of the introns from the pre-mRNAs? Second, as introduced in the ‘Introduction’, alternative splicing is widely adopted by plants to increase the complexity of the transcriptome. In some cases, the introns, along with their flanking exons, are excised from the pre-mRNAs during alternative splicing. In the case study of this paper, the focus was on the ability of the intronic regions for the production of sRNAs through an RDR-dependent pathway. Thus, whether the identified introns could generate RDR-dependent sRNAs in a way which is independent on the flanking exons is an interesting research topic. Third, based on the analysis of the RDR-dependence of the sRNAs generated from the dsRNA-seq read-covered intronic regions, many sRNAs showed simultaneous dependence on RDR1, RDR2, and RDR6 (Fig. 2; see Supplementary Fig. S1 at JXB online). In other words, the biogenesis of these sRNAs will be inhibited when any of the three RDRs is inactivated. This observation points to the possibility that the three RDRs might co-operate together for double-stranded RNA synthesis from the intronic regions; this needs further investigation. Fourth, limited data sets were available for us to investigate the DCL-dependence of the phase-distributed sRNAs within the double-stranded intronic regions. It was observed that the levels of some of the phased sRNAs were dramatically reduced in the mutant dcl2dcl3dcl4 (see Supplementary Data S6 at JXB online). Thus, it could only be deduced that DCL3 might be the critical enzyme since most of the phased sRNAs were 24 nt in length, but this needs experimental clarification. Finally, in this study, a search was only made for the intron-derived, RDR-dependent sRNAs in Arabidopsis owing to the limited availability of dsRNA-seq data in public databases. However, it will be interesting to see whether this kind of sRNAs exist in the other plant species. In summary, this opinion paper might add a novel layer to the current biogenesis model of the intron-derived sRNAs. It is hoped that our primary observations could inspire further research efforts for the studies onthe biological functions of the introns and on the biogenesis and the classification of the plant sRNAs.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Data S1. RDR-dependent sRNAs identified in Arabidopsis.
Supplementary Data S2. RDR-dependent sRNAs mapped onto introns and intron_RCs.
Supplementary Data S3. dsRNA-covered regions on introns and intron_RCs with RDR-dependent sRNAs.
Supplementary Data S4. dsRNA-covered regions with three or more RDR-dependent sRNAs.
Supplementary Data S5. dsRNA-covered regions with two or more phased RDR-dependent sRNAs.
Supplementary Data S6. Expression-based evidence supporting RDR- and DCL-dependent sRNAs.
Supplementary Fig. S1. Phase-distributed sRNAs identified within the dsRNA-seq read-covered intronic regions of Arabidopsis.
Supplementary Materials and Methods.
Acknowledgements
We would like to thank all the publicly available datasets and the scientists behind them. This work was supported by the National Natural Sciences Foundation of China [31100937], the Starting Grant funded by Hangzhou Normal University to Yijun Meng [2011QDL60], the Science and Technology Development Program of Hunan Province [2013FJ2012], and the Young Scientist Research Foundation of Education Bureau of Hunan Province [14B088].
Glossary
Abbreviations:
- AGO
Argonaute
- AS
alternative splicing
- DCL1
Dicer-like 1
- dsRNA-seq
double-stranded RNA sequencing
- GEO
Gene Expression Omnibus
- HTS
high-throughput sequencing
- miRNA
microRNA
- pre-miRNA
precursor microRNA
- pre-mRNA
precursor messenger RNA
- pri-miRNA
primary microRNA
- RC
reverse complementary
- RDR
RNA-dependent RNA polymerase
- RPM
reads per million
- siRNA
small interfering RNA
- sRNA
small RNA
- TAIR
the Arabidopsis Information Resource
- ta-siRNA
trans-acting small interfering RNA.
References
- Barrett T, Troup DB, Wilhite SE, et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Research 37, D885–D890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. 2007. Mammalian mirtron genes. Molecular Cell 28, 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Meng Y, Yuan C, Bai L, Huang D, Lv S, Wu P, Chen LL, Chen M. 2011. Plant siRNAs from introns mediate DNA methylation of host genes. RNA 17, 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, et al. 2001. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Research 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biology 5, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. 2012. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Research 22, 1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W. 2000. Pre-mRNA splicing in higher plants. Trends in Plant Science 5, 160–167. [DOI] [PubMed] [Google Scholar]
- Lu C, Kulkarni K, Souret FF, et al. 2006. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Research 16, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Shao C. 2012. Large-scale identification of mirtrons in Arabidopsis and rice. PLoS One 7, e31163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila . Cell 130, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A. 2013. Complexity of the alternative splicing landscape in plants. The Plant Cell 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. The Plant Cell 25, 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Yang GD, Yan K, Wu BJ, Wang YH, Gao YX, Zheng CC. 2012. Genome-wide analysis of intronic microRNAs in rice and Arabidopsis. Journal of Genetics 91, 313–324. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD. 2010. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genetics 6, e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. 2008. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research 18, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.