Highlight
Systems genetics approaches integrated with traditional rice genetic tools provide a unique opportunity to mine valuable alleles suitable to develop climate-resilient heat-tolerant rice varieties with desirable rice grain quality attributes.
Key words: Chalk, functional genomics, genetics, grain quality, milling and head rice yield, stress tolerance, systems biology, systems genetics.
Abstract
To ensure rice food security, the target outputs of future rice breeding programmes should focus on developing climate-resilient rice varieties with emphasis on increased head rice yield coupled with superior grain quality. This challenge is made greater by a world that is increasingly becoming warmer. Such environmental changes dramatically impact head rice and milling yield as well as increasing chalkiness because of impairment in starch accumulation and other storage biosynthetic pathways in the grain. This review highlights the knowledge gained through gene discovery via quantitative trait locus (QTL) cloning and structural–functional genomic strategies to reduce chalk, increase head rice yield, and develop stable lines with optimum grain quality in challenging environments. The newly discovered genes and the knowledge gained on the influence of specific alleles related to stability of grain quality attributes provide a robust platform for marker-assisted selection in breeding to design heat-tolerant rice varieties with superior grain quality. Using the chalkiness trait in rice as a case study, we demonstrate here that the emerging field of systems genetics can help fast-track the identification of novel alleles and gene targets that can be pyramided for the development of environmentally robust rice varieties that possess improved grain quality.
Introduction
The world’s rice (Oryza sativa L.) production is predicted to be severely affected by the global rise in temperature associated with climate change (Neelin et al., 2006). Furthermore, the global population is expected to grow to 9 billion by 2050 (Godfray et al., 2010). To feed the rapidly growing Asian population (projected to increase from 4.3 billion to 5.2 billion by 2050) that depends on rice as a staple food, paddy yield should not just be enhanced, but grain quality requirements need to be met to ensure consumer acceptance. Consumers primarily assess rice grain quality based on physical (milled yield, translucency, size, shape, and colour) and sensory properties, with a strong emphasis on cooking quality and aroma (Ebron, 2013). Environmental stresses not only affect milling yield, but they also lead to a significantly elevated proportion of chalky grains which in turn alters starch and cooking quality (Cooper et al., 2003; Cheng et al., 2005b ; Fabre et al., 2005; Lanning et al., 2011, 2012; Li et al., 2011; Lanning and Siebenmorgen, 2013). Thus abiotic stresses, in particular high temperatures, have a negative effect on various grain quality traits (Fig. 1) and they also reduce sensory attributes (Chun et al., 2009), which leads to a reduction in overall consumer acceptance. Excellent reviews are available that describe rice grain quality from a traditional biochemistry perspective (Champagne, 2008; Fitzgerald et al., 2008, 2009; Champagne et al., 2010; Cuevas et al., 2010; Huang et al., 2013; Liu et al., 2013; Siebenmorgen et al., 2013; Zhao and Fitzgerald, 2013). However, the implications of abiotic stresses on grain quality and its associated molecular and physiological mechanisms are not yet fully understood. This review will focus on the impact of heat stress perturbations on grain quality with special emphasis on enhancing head rice yield (HRY; intact grain after milling) and reducing chalk (the opaque area in the rice grain).
Fig. 1.
The ontology of seed development covering import phase transitions. High temperature stress-induced perturbations occurring during seed development affect the grain quality, cooking quality, and eating quality of rice.
Compared with that of other crops, the productivity of rice is more accurately measured after milling (Lyman et al., 2013). Although annual world rice production reaches 750 Mt of paddy (http://faostat.fao.org/site/339/default.aspx), the final milled rice yield (MRY; including head rice and broken grains) corresponds to 490 Mt. This implies that as many as 260 Mt year–1 are lost after milling (including hull and bran). Furthermore, the cost of broken rice is significantly undervalued in the world market. This can be avoided with the availability of rice varieties whose grains are not prone to breakage during post-harvest storage and processing. To improve food security and rice quality, rice breeders need to prioritize improving HRY (mass percentage of intact whole rice after milling) over MRY (mass percentage of milled rice including both whole grains and broken grains) at the global production level (Juliano, 1998, 2003). However, improving this parameter in rice varieties is complicated because it is negatively affected by environmental stresses such as exposure to high night temperature, extreme day temperatures, and severe drought (Lyman et al., 2013; Siebenmorgen et al., 2013; Zhao and Fitzgerald, 2013; Usui et al., 2014). The significant environmental effect on HRY is mostly due to high-temperature stress during the sensitive phase of gametogenesis (Cooper et al., 2008; Jagadish et al., 2010; Muthurajan et al., 2011; Lyman et al., 2013). Milling quality attributes are also influenced by unsynchronized flowering and seed filling, chemical properties of the rice grain, as well as abrupt changes in moisture content during harvest and storage (Juliano, 2007).
Chalk is one among many other variable parameters that influence MRY and HRY (Fitzgerald and Resurreccion, 2009; Lanning et al., 2011). It is defined as the opaque portion found in an otherwise translucent white endosperm that is associated with loose packing of storage starch and protein (Lanning et al., 2011; Lyman et al., 2013; Usui et al., 2014). The occurrence of temperature stress during the early to middle stage of seed development triggers non-uniform filling and impairment in storage biosynthesis, leading to chalk formation. The gaps formed due to aborted starch granule formation are thought to be responsible for making chalky grains more brittle and for forming fissures along the grain. As a result, chalky grains crack easily during grain processing, which reduces HRY as a consequence of the elevated amount of broken grains (Lisle et al., 2000). Despite these generalizations, however, a clear association between chalk and breakage susceptibility during milling still needs to be empirically established.
The recent progress made in the area of genetics and genomics, and the body of knowledge surrounding the molecular and physiological mechanisms associated with perturbations to temperature stress during rice seed development in the light of how grain quality in rice is affected have not been systematically reviewed. In this review, we highlight the strategies to develop rice grains with improved grain quality particularly in improving HRY and minimizing chalkiness as key quality parameters to improve rice food security under a warmer climate by integrating the knowledge on genetics and genomics gained thus far using a systems genetics approach.
Unravelling the genetic basis of head rice yield and chalk
Rice HRY is a complex, multigenic trait genetically controlled primarily by the triploid endosperm and the diploid maternal tissues (Pooni et al., 1992; Zhu and Weir, 1994). Several quantitative trait loci (QTLs) have been identified for milling quality in different rice mapping populations using seeds with variable grain shapes (Dong et al., 2004; Jiang et al., 2005; Kepiro et al., 2008; Yuan et al., 2010; Nelson et al., 2011). In addition, 14 QTLs were recorded for grain fissuring (Septiningsih et al., 2003; Pinson et al., 2013). These previous studies revealed that grain size and shape are highly correlated with HRY and quality (Zheng et al., 2007). Although increased grain length is negatively associated with HRY, targeted improvement in grain width and thickness was shown to improve MRY and HRY (Siebenmorgen and Meullenet, 2004). Thus, HRY in slender grains needs to be improved for consumers who prefer long grains.
The genome view of mapped grain quality QTLs suggests that the genetic regions responsible for HRY and chalk overlapped with grain size and shape QTLs on chromosomes 3, 5, and 6 (Supplementary Fig. S1, Table S1 available at JXB online). Among the cloned genes related to grain size and shape, GS3, GW2, and GW5/qSW5 act as negative regulators of grain size, while GS5 and GW8 act as positive regulators of cell proliferation in rice. Genes associated with grain size, shape, and weight belong to selective proteolysis, and key regulators of cellularization and brassinosteroid signalling (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Huang et al., 2013; Sreenivasulu and Wobus, 2013). Until now, the haplotypic variations that contribute to the stability of grain size under abiotic stress exposure in domesticated rice have not been explored. Furthermore, the underlying QTL regions for milling quality have yet to be fine-mapped and cloned to unravel the interconnections between grain dimension genes. Factors such as genotype×environment (G×E) interactions are known to affect rice milling quality (Gravois et al., 1991), and therefore a comprehensive understanding of MRY and HRY requires the identification of QTL alleles and/or G×E interactions. If heterogeneity occurs in grain size and shape because of G×E interactions, mill settings need to be optimized for every variety and production environment. Thus, achieving stability of grain dimension traits continues to remain important for the milling industry.
The genetic components underlying the inter-relationships between various grain quality parameters and milling quality remain unclear. Interestingly, many QTLs for HRY overlapped with QTLs for regions of pre-broken brown rice kernels, seed density, amylose content (AC), kernel whiteness, and chalkiness on chromosome 6 (Fig. 2A). In addition, clusters defined on chromosome 8 are emphasized as the main genetic basis for the effect of rice chalkiness, amylose, protein, and eating quality of cooked rice (Guo et al., 2007; Liu et al., 2011). These different QTLs co-located on the same region influence various grain quality, cooking, and eating quality traits in rice. Fine-mapping these hotspot QTLs can offer a huge potential to identify high-value genes that could be tapped to improve milling, cooking, and sensory and processing quality. This can be done by molecular breeding to address the demands of the rice industry and rice consumers.
Fig. 2.
The grain quality QTL hotspot in rice chromosome 6 and the effect of gene perturbation in the starch biosynthesis pathway. (A) Selected QTLs for grain quality traits on japonica chromosome 6 located in syntenic regions between indica and japonica reference genomes. Chromosome 6 is a grain quality hotspot as multiple grain quality QTLs related to amylose, chalk, head rice yield, gel consistency, gelatinization temperature, and grain dimensions and shape are co-located on both arms of the chromosome. The accession IDs of these grain quality QTLs are listed in Supplementary Table S1 at JXB online. (B) Gene perturbation in the starch metabolism pathway. Perturbations in the starch metabolism pathway involving starch synthase III and starch branching enzyme IIa and IIb lead to alterations in amylose–amylopectin composition and starch structure variation (Butardo, 2011). The proposed molecular and physiological mechanisms of chalk formation are a complex system associated with source–sink disturbances. However, the initial genetic evidence points to the disturbance of the storage pathways in developing seeds.
Genetic basis of chalk
More than 140 QTLs were reported for the chalkiness trait across all 12 chromosomes, mostly among Asian cultivars (Tan et al., 2000; Li et al., 2003; Wan et al., 2005). In a recent study, several QTL clusters related to various chalk phenotypes have been identified using five different mapping populations that are stable across two environments (Peng et al., 2014a ). The percentage of grain with chalkiness (PGWC) has been genetically mapped on chromosomes 5, 8, and 10, which explains 50.8% of the genetic variation (Liu et al., 2012). Stable QTLs with a reproducible chalky phenotype across many environments have been fine-mapped to regions on chromosomes 8 and 9 (Wan et al., 2004, 2005). Furthermore, using a set of chromosome segment substitution lines developed from a cross between cultivar C-51 (chalky endosperm) and the recurrent parent 93-11 (translucent endosperm), two additional loci that control PGWC located on chromosomes 6 and 7 were detected and designated as qPGWC-6 and qPGWC-7, respectively (Zhou et al., 2009). The chalk QTL qPGWC-7 contains 13 genes related to several unknown proteins, including COBRA-like proteins. The qPGWC-6 QTL map position is closer to the Waxy gene, and thus variable AC appears to affect chalk (Zhou et al., 2009). Within the proximity of the Waxy locus, eight chalk QTLs have been co-located (Peng et al., 2014a ).
Chalkiness is reported to be influenced by multiple QTLs, though some of these QTLs may not be reproducible because the genetic backgrounds used in some of the studies to generate the bi-parental mapping populations are variable for grain quality parameters such as AC and grain width. So far, fine-mapping studies identified the following as possible candidate genes responsible for chalk: pyruvate orthophosphate dikinase (Kang et al., 2005), starch synthase IIIa (Fujita et al., 2007a ), UDP-glucose pyrophosphorylase (Woo et al., 2008), cell wall invertase (Wang et al., 2008), and H + -translocating pyrophosphatase (Li et al., 2014). How these genes interact to produce the chalky grain phenotype leading to grains susceptible to cracking and reduced milling potential or lowering HRY has not yet been studied.
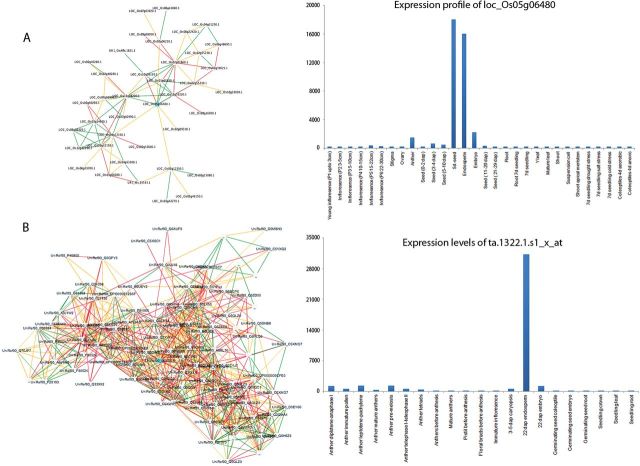
Novel approaches can be employed to fast-track the design of novel rice ideotypes with superior grain quality and exhibiting chalk-free features under stress. The strengths of a systems genetics approach (combining systems biology and traditional genetics approaches) can be used to unravel structural–functional relationships by tapping the genetic diversity represented by rice core collections or the detailed information that can be obtained from high-density mapping populations. As a proof of concept, we explored rice re-sequencing resources for synteny between japonica and indica subspecies using the recently cloned chalk gene H + -translocating pyrophosphatase (Li et al., 2014) as well as several grain quality QTLs identified on chromosome 5. We found that they are located in collinear blocks (Fig. 3). Interestingly, hotspots for higher SNP frequency were observed either between the subtypes or within the indica re-sequencing genetic region (Fig. 3). Overlaying regulatory networks identified using a top-down systems biology approach using the PLANET database (Mutwil et al., 2011) suggested that the expression variation created between cereal species appears to cause functional divergence for the endosperm-specific H + -translocating pyrophosphatase gene responsible for chalk (Fig. 4). The derived gene regulatory network of the H + -translocating pyrophosphatase gene in rice seems to control the selective proteolysis pathway (E3.SCF) during 4–6 days after flowering (DAF). On the other hand, its orthologue in wheat (Ta.1322.1.S1_x_at) regulates thousands of genes involved in starch biosynthesis and protease inhibitors during seed filling (Supplementary Table S2 at JXB online). Such comprehensive information can be mined by combining genome-wide association studies (GWAS) together with gene co-expression networks through systems genetics approaches (Ficklin and Feltus, 2013), to clone both major and minor QTL-encoding genes of grain quality and characterize their functions from diverse populations for stress tolerance.
Fig. 3.

Synteny between japonica chromosome 5 (OsJ5) and related chromosomes in indica (OSI1 and OSI5). This figure shows the syntenic blocks between chromosome 5 of O. sativa japonica with chromosomes 1 and 5 of indica (innermost section of the figure). The bar plot shows SNP density in the genomic region based on SNPs common to five indica re-sequenced genomes. Blue lines represent SNP densities ≥5 per kb, while orange lines represent SNP densities <5 per kb. SNP density was calculated separately for every 100kb. Mapped grain quality QTLs are shown next to the SNP density bar plots. Mapped QTLs whose lengths are between 1Mb and 5Mb are represented as lines, while fine-mapped QTLs (size <1Mb) are represented as triangles. Genomic hotspots that have the highest SNP densities can be found on chromosomes 1 and 5 of indica as well as japonica. These hotspots coincide with several grain quality QTLs, including the recently cloned chalk gene H + -translocating pyrophosphatase.
Fig. 4.
The gene regulatory networks derived from co-expression data of seed development-specific genes. (A) The gene regulatory networks of the endosperm-specific H + -translocating pyrophosphatase gene cloned for chalkiness in rice derived using the PLANET database (Mutwil et al., 2011) seem to control the selective proteolysis pathway (E3.SCF) during 3–5 DAF (Supplementary Table S2 at JXB online). (B) On the other hand, its orthologue in wheat (Ta.1322.1.S1_x_at) regulates thousands of genes involved in starch biosynthesis and protease inhibitors during seed filling. These divergent expression patterns of H + -translocating pyrophosphatase genes between rice and triticale members are likely to trigger functional divergence during seed development.
A molecular understanding of grain chalkiness under elevated day and night temperature and its relationship to seed storage metabolism
High temperature (35/30 °C day/night) reduces grain weight, AC, and flour gel consistency (GC) of rice grains (Lin et al., 2010). If the day temperatures are high beyond the critical levels during the onset of fertilization and post-anthesis period, flag leaf photosynthesis decreases substantially, resulting in disturbed source–sink assimilate transport. As a result, high temperature not only decreases seed set but also affects grain quality (Jagadish et al., 2010; Lyman et al., 2013) due to reduction in endosperm sink strength and incomplete grain-filling events (Fig. 1). Deterioration of grain quality under high temperature is accompanied by the altered expression of starch metabolism-related genes. Functional analysis of the rice GRAIN INCOMPLETE FILLING 1 (GIF1) gene that encodes a cell wall invertase (CWI) required to cleave sucrose into hexoses for carbon partitioning during early grain filling (Wang et al., 2008) has been successfully targeted to improve grain weight during domestication. However, GIF1 expression in wild rice is much wider during seed development and results in a grain weight reduction and enhanced chalkiness (Wang et al., 2008). Ectopic expression of GIF1 with the 35S or rice Waxy promoter resulted in smaller grains. In comparison with sucrose synthase-mediated sucrose cleavage, the GIF1 gene requires a higher ATP content to convert sucrose into hexoses (Sreenivasulu and Wobus, 2013), and thus it will remain as a rate-limiting step under stress conditions. Because of its role as a sink fine-tuning gene, GIF1 is an important factor in driving optimum storage and unimpaired quality even under stress.
Many elite rice cultivars are also susceptible to forming chalky grains under elevated temperature. Transcriptome analysis of contrasting lines differing in chalkiness showed that several key starch biosynthesis genes, such as granule-bound starch synthase I (GBSSI), branching enzyme IIb (BEIIb), starch synthases, and debranching enzymes, and a cytosolic pyruvate orthophosphate dikinase gene were substantially repressed, while a starch-degrading alpha amylase gene was preferentially up-regulated under high temperature (Yamakawa et al., 2007; Liu et al., 2010). In addition, high night temperature affects the activities of many enzymes associated with the conversion of sucrose into starch synthesis during grain filling (Counce et al., 2005; Yamakawa et al., 2007; Yamakawa and Hakata, 2010). Reduced activities of ADP-glucose pyrophosphorylase and starch synthase enzymes have been correlated with reductions in grain weight and starch production at high temperatures (Singletary et al., 1997). Transcriptome analysis, in conjunction with QTL browsing, suggested that many high-temperature-responsive pathways related to starch and storage protein metabolism are co-located along the chalk QTL regions (Yamakawa et al., 2008; Peng et al., 2014a ). High temperature increased the accumulation of all classes of storage proteins (glutelins, prolamins, globulins, and protein disulphide isomerase) at the early filling stage, but it decreased prolamin accumulation during maturation (Lin et al., 2010). Accumulation of prolamins and globulins was more sensitive to high temperature than other seed storage proteins (Lin et al., 2010). Stress-responsive elements such as HSE (heat stress responsive element), ARE (anaerobic induction element), ABRE (abscisic acid response element), and MBS (MYB binding site) were found to be enriched in the promoter regions of prolamin and globulin genes. This suggests that high temperature deregulates the temporal patterns of prolamins and globulins and reduces amylose, leading to the formation of chalk (Li et al., 2011).
It has also been observed that heat stress results in chalky grain production because of changes in starch structure (Yamakawa et al., 2007; Yamakawa and Hakata, 2010; Patindol et al., 2014) which usually results in decreased amylose concentration (Chen et al., 2008; Lanning et al., 2012) and altered gelatinization temperature (GT; Cuevas et al., 2010) in many rice varieties. Alterations in starch structure primarily due to a reduction in AC lead to variation between translucent and chalky grains (Patindol and Wang, 2003). The presence of many air spaces within the loosely packed starch granules in the chalky grain prevents light transmission, which is visible as opaque regions along the translucent grain (Ashida et al., 2009). However, not all opaque grains can be considered as chalky. For example, glutinous rice grains are opaque white due to micropores within the polyhedral starch granules. In contrast, the incidence of chalk in non-glutinous grains is due to air spaces between spherical starch granules (Juliano, 2007). As shown in Fig. 2B, several mutants characterized from the starch biosynthesis pathway showed a pleiotropic effect of chalk (Fujita et al., 2007b ; Yamakawa et al., 2007). For instance, starch synthase IIIa mutants (ss3a-1 and ss3a-2) with elevated amylose and reduced amylopectin resulted in loose starch packing with a chalky phenotype (Fujita et al., 2007b ). Likewise, many other substandard starch grain (ssg) mutants affecting the starch granule structure exhibited chalky phenotypes (Matsushima et al., 2014). The chalky to opaque grains were produced when the activity of starch branching enzyme IIb (SBEIIb) was down-regulated in rice endosperm, and the extent of opacity was found to be directly proportional to the elevation in the proportion of long-chain amylopectin (Butardo et al., 2011). In addition, even minor perturbations in the amylopectin fine structure affected during high night temperature can produce streaks of opacity in rice grains (Patindol et al., 2014).
Hakata et al. (2012) reported that the involvement of a starch-hydrolysing enzyme, α-amylase, triggered grain chalkiness at high temperature. In developing seeds, high temperature induced the α-amylase genes Amy1A, Amy1C, Amy3A, Amy3D, and Amy3E, as well as α-amylase activity (Hakata et al., 2012). Through an RNA interference (RNAi) strategy, temperature-induced α-amylase gene expression is suppressed, leading to fewer chalky grains (Hakata et al., 2012). This implies that the degradation of starch by amylase under elevated temperature is another layer of regulation responsible for altering starch structure and grain chalkiness in rice. Furthermore, systematic metabolite profiling of starch mutants and selected lines from indica and japonica subspecies differing in starch structure and amylose content reveals the importance of phospholipid complexes in starch structure and also emphasizes the mechanisms involved in white cores, opacity, and chalk formation (Kusano et al., 2012; Matsuda et al., 2012).
Based on the inferences drawn from the above studies, we conclude that chalk is the result of poor filling of starch granules in endosperm primarily affecting the amylose to amylopectin pathway and, in addition, imbalances in the finer readjustment with the starch degradation pathway triggered under stress. The formation of chalky grain is also triggered by the disruption of the pH homeostasis in the endomembrane trafficking system during early endosperm development, leading to elevated amounts of small vesicle-like structures and a decrease in the number and size of protein bodies (Li et al., 2014). This results in the formation of air spaces among starch granules and protein bodies, which is responsible for the abnormal shape and spatial rearrangements of these storage compounds, leading to chalky rice grains. Recent additional evidence also suggests that the plant-specific kelch repeat protein interlinks the role of Golgi-associated trafficking involved in the storage protein sorting pathway, leading to the trigger of a floury endosperm and chalky phenotype with a preferential accumulation of glutelin precursor in the mutant grains (Ren et al., 2014). How the altered inter-relationships between storage starch and storage protein biosynthesis affect the formation of the chalky phenotype, leading to an increase in the susceptibility to cracking and reduced HRY, needs further investigation.
Impact of starch metabolism under stress
Aside from improved HRY and reducing chalk, breeders should also focus on other important traits such as cooking, sensory, and processing quality to address the needs of the food industry and consumers. Up to 90% of the rice grain is starch (on a dry basis) and it is therefore a key contributor to many grain quality attributes. The cooking, sensory, and functional properties of rice are highly influenced by the quality and physicochemical properties of starch (Juliano, 2007). It is therefore important to understand how different abiotic stresses can modify starch composition and accumulation in rice endosperm in an era of abrupt and uncertain environmental fluctuations.
High-temperature stress during the grain-filling stage has deleterious effects on starch quality. Major genes involved in the starch biosynthetic pathway such as GBSS, SBEI, and SBEIIb are down-regulated in grains exposed to high-temperature stress. Grains produced under high-temperature conditions also result in aberrant starch, with small granules and reduced amylose and amylopectin content, reflecting a similar phenotype to that observed in floury-endosperm (flo) mutants (Kawasaki et al., 1996; Yamakawa et al., 2007; Satoh et al., 2003). The flo2 mutation in rice affected grain size, with a lower AC, and showed floury features because of the loose filling of starch granules with larger air spaces in the grain compared with their wild-type counterpart (Qiao et al., 2010; She et al., 2010). Mutations in other starch-biosynthesizing enzymes also exhibit similar characters to those of flo2 mutants (Nishi et al., 2001). She et al. (2010) identified the gene responsible for the flo2 mutant as OsCEO1, a novel regulatory cascade of endosperm organogenesis, and it may have an important role in the response to high-temperature stress. Likewise, the FLO6 gene codes for an unknown protein that possesses a C-terminal carbohydrate-binding module that binds to the starch molecule, which alters the physicochemical properties of starch and thus alters complex granule formation (Peng et al., 2014b ).
Both high night temperature and high day temperature affect grain quality by altering starch and storage protein properties (Li et al., 2011). Apart from Wx, numerous QTLs associated with AC and GC have been mapped on different rice chromosomes (Supplementary Fig. S1 at JXB online) (Aluko et al., 2004; Sun et al., 2006; Sabouri et al., 2012). High temperatures affect GBSSI activity in rice (Cheng et al., 2005c), and the response of GBSSI to temperature is due to a single nucleotide polymorphism (SNP) in the 5’ leader intron of the GBSSI gene sequence in rice and barley (Hirano and Sano, 1998; Patron et al., 2002). The GBSSI intron splicing is high in GT-SNP (AGGTATA) genotypes, resulting in high amylose accumulation (Inukai et al., 2000; Mikami et al., 2008) even if the temperature is altered. Under unfavourable environments, genotypes with alleles containing a TT-SNP (AGTTATA) in the leader sequence of the 5′ intron of GBSSI are not properly spliced, and hence GBSSI activity is lowered (Bligh et al., 1998). This ultimately results in a low AC in rice grains (Cai et al., 1998; Isshiki et al., 1998; Sato et al., 2002). Thus, japonica cultivars appear to be more sensitive to high temperature with regard to amylose synthesis than indica cultivars (Inukai et al., 2000; Sun et al., 2011). High-temperature stress also reduced Wx protein expression, leading to lowered amylose with altered starch viscosity and grain quality (Larkin and Park, 2003; Larkin et al., 2003). Some of these QTLs are associated with the stability of AC in various rice varieties grown in high-temperature conditions (Zhang et al., 2014). The chromosome substitution lines carrying the stable major QTLs for amylose (qHAC8a, qHAC8b, and/or qHAC4) have been linked to the high pre-mRNA splicing efficiency of the Wx gene. Thus, increasing the pre-mRNA processing of this gene is identified to be the key factor for maintaining stable amylose in rice seeds at high temperature (Zhang et al., 2014). It is therefore possible to combine the correct alleles to ensure proper mRNA splicing of the GBSSI gene in rice breeding strategies to maintain stable amylose biosynthesis under increasingly warming climatic conditions.
The effect of heat stress during seed development not only reduces AC but also modifies starch structure and thermal properties by affecting the gelatinization temperature (GT) (Lu et al., 2014). GT is associated with the activity of starch synthase IIa (SSIIa), the enzyme responsible for the elongation of amylopectin chains within the crystalline lamella of degree of polymerization (DP) 12–24 (Umemoto et al., 2004; Nakamura et al., 2005; Waters et al., 2006). Functional SNPs in the gene coding for SSIIa have been identified and can be used to group rice samples into high- and low-GT classes (Nakamura et al., 2005; Waters et al., 2006; Cuevas et al., 2010), which influences cooking quality. Apart from SSIIa, QTLs for GT were mapped on other rice chromosomes using different populations (Lanceras et al., 2000; He et al., 2006; Sabouri et al., 2012). Heat stress not only decreases AC but it also increases the overall proportion of longer amylopectin chains in rice starch (Patindol et al., 2014), possibly because of increased SSI and SSIIa activity (Umemoto et al., 1999; Umemoto and Aoki, 2005; Yamakawa et al., 2007). Although SSI preserves the elongation of A and B chains of amylopectin, reduced activity of SBEIIb and SBEI lowers the branching frequency of amylopectin (Jiang et al., 2003). Thus, reduced GBSSI and increased SSI activities under high temperature contribute to the lower ratio of amylose to amylopectin (Cheng et al., 2005a ). These results point to the possibility that a decrease in amylose under heat stress may account for the loss of grain weight as well as impaired cooking quality because of the alteration in starch properties. The inferences drawn above can help in engineering starch quality by maintaining an optimum amylose to amylopectin ratio even under stress.
Present state and future perspectives in designing climate-resilient rice with superior quality grain
Rice grain yield has been reported to decline by 6% and HRY by 9–14% for every 1 °C increase in temperature (Peng et al., 2004; Welch et al., 2010; Lyman et al., 2013). This wasted food could have been made available to help the global food requirement considering that rice feeds roughly half of the world’s population. High night temperatures greatly influence milling quality by modulating endosperm morphology, grain dimensions, and starch-metabolizing enzymes (Counce et al., 2005). This confounding evidence suggests that the global rise in temperature primarily affects HRY and key grain quality attributes such as seed storage biosynthesis and grain chalkiness. To enhance HRY and reduce susceptibility to chalk-mediated grain breakage under stress conditions, there is a need to (i) select better germplasm with reduced chalk under multienvironments as a source of inbreds; (ii) target both parents to have a similar AC and GT in creating hybrid vigour for stress tolerance with higher HRY as a prime target; and (iii) use marker-assisted selection strategies to reduce chalk in the lines suitable for stress-prone environments. Events leading to chalk formation are associated mostly with reduced sink strength and imbalances in carbon and nitrogen partitioning in a growing sink because of photoassimilate limitation under stress. Thus, besides implementing strategies to enhance yield advantages through elevated grain number per panicle and a change in spike architecture with profuse secondary branches in the panicles (Sreenivasulu and Schnurbusch, 2012), source–sink relationships under stress conditions need to be fine-tuned (Supplementary Fig. S2 at JXB online). In addition, strategies need to be developed to synchronize the ability of genotypes to stabilize HRY with reduced chalk under challenging environments.
Based on the inferences drawn from previous published studies, chalk appears to be the result of poor filling of starch granules in the rice endosperm, primarily affecting amylose and amylopectin pathways and disrupting the spatiotemporal packing of starch granules. Triggering of the chalky phenotype may also be due to imbalances in the finer readjustments with the starch degradation pathway during grain filling. However, susceptibility to cracking because of brittle grains must be overcome to mitigate HRY reduction, and its connected links to the chalky phenotype need to be thoroughly dissected. This is because the rice grain is very sensitive to even minor perturbations in starch structure, which does not necessarily result in brittle grains and a reduction in HRY in some instances. The definitive genetic, environmental, and physiological link between chalkiness and breakage susceptibility needs to be established. In addition, the connection between seed storage protein biosynthesis and starch granule formation has to be empirically tested to determine the role of carbon partitioning and allocation in the formation of brittle chalky grains that significantly reduce HRY.
Grain quality traits are controlled by many major QTLs, implying that the genetic mechanisms underlying quality traits are complex. More than 600 QTLs related to grain quality have been reported (see Supplementary Table S1 at JXB onlime) in the Gramene Genome Database (http://www.gramene.org). Additional evidence gathered from meta-QTL mapping studies revealed that stable and major QTL genetic regions identified on chromosomes 3 and 6 have overlapping regions for chalky endosperm, AC, protein content, viscosity properties, and the integrated values of organoleptic evaluation (Supplementary Fig. S1). It appears that this QTL cluster is a novel gene resource for controlling rice grain quality traits. Cloned genes responsible for rice grain quality traits as well as fine-mapped QTLs form a strong base for genomic selection useful in efficient breeding for designing climate-ready lines that are suitable according to regional differences in grain quality preferences. With a much lower sequencing price, genome re-sequencing has been used to accelerate breeding. The genetic and genomic information that can be harnessed from the 3000 whole genomes of cultivated rice relatives (3,000 Rice Genomes Project, 2014) will provide an opportunity to mine different alleles related to grain quality traits involved during adaptation to different climatic conditions at the subspecies level. Because some useful grain quality traits might have been lost during the course of rice domestication, these developments in genotyping technology can also be used to explore the genetic diversity of wild rice (Nock et al., 2011) to find useful genes for starch quality improvement under climate change scenarios. Structural and functional genomics resources available from millet not only offer the prospects of incorporating stress tolerance target traits in other cereals but also offer the value to explore the nutritional benefits (Muthamilarasan and Prasad, 2014). Such comprehensive augmented knowledge obtained that is related to climate-adapted allelic variation can be used to design superior grain quality for stress-prone environments. This can be achieved by exploring valuable alleles that can be targeted to fine-tune grain quality in high-yielding lines that are stable across a variety of stress-prone areas through genomics-assisted selection and marker-assisted breeding. On top of this, employing systems biology strategies such as regulatory networks and flux balance analysis, as well as systems genetics methods, can help to decipher the holistic view of grain quality perturbations to multiple abiotic stress factors (heat stress with drought, humidity, salinity, and elevated carbon dioxide), which co-occur in nature (Fig. 5).
Fig. 5.
Schematic representation of the systems genetics approach to explore the potential of existing intraspecific variation for various grain quality traits on a genetic map using GWAS/QTL. Unravelling a holistic view of grain quality perturbation under stress requires the integration of knowledge from systems biology (regulatory networks and flux balance analysis), systems genetics, and comparative genomics to explore the perspectives of a genomics revolution in breeding to develop climate-resilient lines with superior grain quality.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Summary of grain quality QTLs identified using bi-parental mapping populations depicted on rice chromosomes 3, 4, and 6.
Table S1. A summary of more than 600 grain quality QTLs.
Table S2. Selected super pfam families which are enriched in the vicinity of networks of H+ pyrophosphatase in rice and wheat species.
Acknowledgements
The authors thank Professor T.J. Siebenmorgen (University of Arkansas, USA), and Drs A. Henry and K.S.V. Jagadish (International Rice Research Institute, Philippines) for insightful comments and helpful discussions. This work has been supported under the CGIAR thematic area Global Rice Science Partnership (GRISP), Stress-Tolerant Rice for Africa and South Asia (STRASA) Phase III, and Australian Centre for International Agricultural Research (Project ID CIM/2014/024) funding. The authors have no conflict of interest to declare.
Glossary
Abbreviations:
- Amy
amylase
- AC
amylose content
- DAF
days after flowering
- FLO
floury-endosperm
- GC
gel consistency
- GIF
grain incomplete filling
- GBSS
granule-bound starch synthase
- GS
grain size
- GT
gelatinization temperature
- GW
grain weight
- GWAS
genome-wide association studies
- HRY
head rice yield
- MRY
milled rice yield
- PGWC
percentage of grain with chalkiness
- QTL
quantitative trait locus
- SBE
starch branching enzyme
- SNP
single nucleotide polymorphism
- SS
starch synthase.
References
- 3,000 Rice Genomes Project. 2014. The 3,000 rice genomes project. Gigascience 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluko G, Martinez C, Tohme J, Castano C, Bergman CJ, Oard JH. 2004. QTL mapping of grain quality traits from the interspecific cross Oryza sativa×O. glaberrima . Theoretical and Applied Genetics 109, 630–639. [DOI] [PubMed] [Google Scholar]
- Ashida K, Iida S, Yasui T. 2009. Morphological, physical, and chemical properties of grain and flour from chalky rice mutants. Cereal Chemistry 86, 225–231. [Google Scholar]
- Bligh HFJ, Larkin PD, Roach PS, Jones CA, Fu H, Park WD. 1998. Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Molecular Biology 38, 407–415. [DOI] [PubMed] [Google Scholar]
- Butardo V, Jr, Fitzgerald MA, Bird AR, et al. 2011. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. Journal of Experimental Botany 62, 4927–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo VM. 2011. Exploring rice diversity and biotechnology to develop grains with novel starch properties and altered digestibility. PhD thesis, University of Queensland, Brisbane, Queensland, Australia. [Google Scholar]
- Cai XL, Wang ZY, Xing YY, Zhang J-L, Hong M-M. 1998. Aberrant splicing of intron 1 leads to the heterogeneous 5’ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. The Plant Journal 14, 459–465. [DOI] [PubMed] [Google Scholar]
- Champagne ET. 2008. Rice aroma and flavor: a literature review. Cereal Chemistry 85, 445–454. [Google Scholar]
- Champagne ET, Bett-Garber KL, Fitzgerald MA, et al. 2010. Important sensory properties differentiating premium rice varieties. Rice 3, 270–281. [Google Scholar]
- Chen M-H, Bergman CJ, Pinson SRM, Fjellstrom RG. 2008. Waxy gene haplotypes: associations with apparent amylose content and the effect by the environment in an international rice germplasm collection. Journal of Cereal Science 47, 536–545. [Google Scholar]
- Cheng F, Zhong L, Zhao N, Liu Y, Zhang G. 2005a. Temperature induced changes in the starch components and biosynthetic enzymes of two rice varieties. Plant Growth Regulation 46, 87–95. [Google Scholar]
- Cheng FM, Zhong LJ, Wang F, Zhang GP. 2005b. Differences in cooking and eating properties between chalky and translucent parts in rice grains. Food Chemistry 90, 39–46. [Google Scholar]
- Cheng FM, Zhong LJ, Zhao NC, Liu Y, Zhang GP. 2005c. Temperature induced changes in the starch components and biosynthetic enzymes of two rice varieties. Plant Growth Regulation 46, 87–95. [Google Scholar]
- Chun A, Song J, Kim K-J, Lee H-J. 2009. Quality of head and chalky rice and deterioration of eating quality by chalky rice. Journal of Crop Science and Biotechnology 12, 239–244. [Google Scholar]
- Cooper B, Clarke JD, Budworth P, et al. 2003. A network of rice genes associated with stress response and seed development. Proceedings of the National Academy of Sciences, USA 100, 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper NTW, Siebenmorgen TJ, Counce PA. 2008. Effects of nighttime temperature during kernel development on rice physicochemical properties. Cereal Chemistry 85, 276–282. [Google Scholar]
- Counce PA, Bryant RJ, Bergman CJ, Bautista RC, Wang YJ, Siebenmorgen TJ, Moldenhauer KA, Meullenet J-FC. 2005. Rice milling quality, grain dimensions, and starch branching as affected by high night temperatures. Cereal Chemistry 82, 645–648. [Google Scholar]
- Cuevas RP, Daygon VD, Corpuz HM, Reinke RF, Waters DLE, Fitzgerald MA. 2010. Melting the secrets of gelatinisation temperature in rice. Functional Plant Biology 37, 439–447. [Google Scholar]
- Dong YJ, Tsuzuki E, Lin DZ, Kamiunten H, Terao H, Matsuo M, Cheng SH. 2004. Molecular genetic mapping of quantitative trait loci for milling quality in rice (Oryza sativa L.). Journal of Cereal Science 40, 109–114. [Google Scholar]
- Ebron G. 2013. In search of the perfect grain. Rice Today 12, 15–17. [Google Scholar]
- Fabre D, Siband P, Dingkuhn M. 2005. Characterizing stress effects on rice grain development and filling using grain weight and size distribution. Field Crops Research 92, 11–16. [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. 2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoretical and Applied Genetics 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Ficklin SP, Feltus FA. 2013. A systems-genetics approach and data mining tool to assist in the discovery of genes underlying complex traits in Oryza sativa. PLoS One 8, e68551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MA, McCouch SR, Hall RD. 2009. Not just a grain of rice: the quest for quality. Trends in Plant Science 14, 133–139. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MA, Resurreccion AP. 2009. Maintaining the yield of edible rice in a warming world. Functional Plant Biology 36, 1037–1045. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MA, Sackville-Hamilton NR, Calingacion MN, Verhoeven HA, Butardo V., Jr 2008. Is there a second gene for fragrance in rice? Plant Biotechnology Journal 6, 416–423. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kondo T, Utsumi Y, Nishi A, Tokunaga T, Satoh H, Jane J-L, Park J-H, Nakamura Y. 2007a. The pleiotropic effects of starch synthase IIIa (SSIIIa) mutation on the other SS isozymes in rice endosperm. Plant and Cell Physiology 48, 181. [Google Scholar]
- Fujita N, Kubo A, Suh DS, Wong KS, Jane JL, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y. 2003. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiology 44, 607–618. [DOI] [PubMed] [Google Scholar]
- Fujita N, Toyosawa Y, Utsumi Y, Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, Itoh R, Miyao A, Hirochika H, Satoh H, Nakamura Y. 2009. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. Journal of Experimental Botany 60, 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. 2006. Function and characterization of starch synthase I using mutants in rice. Plant Physiology 140, 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, et al. 2007b. Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiology 144, 2009–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: the challenge of feeding 9 billion people. Science 327, 812–818. [DOI] [PubMed] [Google Scholar]
- Gravois KA, Moldenhauer KA, Rohman PC. 1991. Genetic and genotype×environmental effects for rough rice and head rice yields. Crop Science 31, 907–911. [Google Scholar]
- Guo Y, Mu P, Liu J, Lu Y, Li Z. 2007. QTL mapping and Q×E interactions of grain cooking and nutrient qualities in rice under upland and lowland environments. Journal of Genetics and Genomics 34, 420–428. [DOI] [PubMed] [Google Scholar]
- Hakata M, Kuroda M, Miyashita T, Yamaguchi T, Kojima M, Sakakibara H, Mitsui T, Yamakawa H. 2012. Suppression of alpha-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnology Journal 10, 1110–1117. [DOI] [PubMed] [Google Scholar]
- He Y, Han Y, Jiang L, Xu C, Lu J, Xu M. 2006. Functional analysis of starch-synthesis genes in determining rice eating and cooking qualities. Molecular Breeding 18, 277–290. [Google Scholar]
- Hirano H-Y, Sano Y. 1998. Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant and Cell Physiology 39, 807–812. [Google Scholar]
- Huang RY, Jiang LR, Zheng JS, Wang TS, Wang HC, Huang YM, Hong ZL. 2013. Genetic bases of rice grain shape: so many genes, so little known. Trends in Plant Science 18, 218–226. [DOI] [PubMed] [Google Scholar]
- Inukai T, Sako A, Hirano H-Y, Sano Y. 2000. Analysis of intragenic recombination at wx in rice: correlation between the molecular and genetic maps within the locus. Genome 43, 589–596. [DOI] [PubMed] [Google Scholar]
- Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K. 1998. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5’ splice site of the first intron. The Plant Journal 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Jagadish SV, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. 2010. Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). Journal of Experimental Botany 61, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang GH, Hong XY, Xu CG, Li XH, He YQ. 2005. Identification of quantitative trait loci for grain appearance and milling quality using a doubled-haploid rice population. Journal of Integrative Plant Biology 47, 1391–1403. [Google Scholar]
- Jiang H, Dian W, Wu P. 2003. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 63, 53–59. [DOI] [PubMed] [Google Scholar]
- Juliano BO. 1998. Varietal impact on rice quality. Cereal Foods World 43, 207–222. [Google Scholar]
- Juliano BO. 2003. Rice chemistry and quality. Munoz, Nueva Ecija: Philippine Rice Research Institute. [Google Scholar]
- Juliano BO. 2007. Rice chemistry and quality. Munoz, Nueva Ecija: Philippine Rice Research Institute. [Google Scholar]
- Kang HG, Park S, Matsuoka M, An GH. 2005. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-4-type pyruvate orthophosphate dikinase gene (OsPPDKB). The Plant Journal 42, 901–911. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Mizuno K, Shimada H, Satoh H, Kishimoto N, Okumura S, Ichikawa N, Baba T. 1996. Coordinated regulation of the genes participating in starch biosynthesis by the rice Floury-2 locus. Plant Physiology 110, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepiro JL, McClung AM, Chen M-H, Yeater K, Fjellstrom RG. 2008. Mapping QTLs for milling yield and grain characteristics in a tropical japonica long grain cross. Journal of Cereal Science 48, 477–485. [Google Scholar]
- Kusano M, Fukushima A, Fujita N, Okazaki Y, Kobayashi M, Oitome NF, Ebana K, Saito K. 2012. Deciphering starch quality of rice kernels using metabolite profiling and pedigree network analysis. Molecular Plant 5, 442–451. [DOI] [PubMed] [Google Scholar]
- Lanceras JC, Huang Z-L, Naivikul O, Vanavichit A, Ruanjaichon V, Tragoonrung S. 2000. Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105). DNA Research 7, 93–101. [DOI] [PubMed] [Google Scholar]
- Lanning S, Siebenmorgen T. 2013. Effects of preharvest nighttime air temperatures on whiteness of head rice. Cereal Chemistry Journal 90, 218–222. [Google Scholar]
- Lanning SB, Siebenmorgen TJ, Ambardekar AA, Counce PA, Bryant RJ. 2012. Effects of nighttime air temperature during kernel development of field-grown rice on physicochemical and functional properties. Cereal Chemistry 89, 168–175. [Google Scholar]
- Lanning SB, Siebenmorgen TJ, Counce PA, Ambardekara AA, Mauromoustakos A. 2011. Extreme nighttime air temperatures in 2010 impact rice chalkiness and milling quality. Field Crops Research 124, 132–136. [Google Scholar]
- Larkin PD, McClung AM, Ayres NM, Park WD. 2003. The effect of the Waxy locus (Granule Bound Starch Synthase) on pasting curve characteristics in specialty rices (Oryza sativa L.). Euphytica 131, 243–253. [Google Scholar]
- Larkin PD, Park WD. 2003. Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Molecular Breeding 12, 335–339. [Google Scholar]
- Li H, Chen Z, Hu M, Wang Z, Hua H, Yin C, Zeng H. 2011. Different effects of night versus day high temperature on rice quality and accumulation profiling of rice grain proteins during grain filling. Plant Cell Reports 30, 1641–1659. [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, et al. 2014. Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nature Genetics 46, 398–404. [DOI] [PubMed] [Google Scholar]
- Li Z-F, Wan J, Xia J-F, Zhai H. 2003. Mapping quantitative trait loci underlying appearance quality or rice grains (Oryza sativa L.). Acta Genetica Sinica 30, 251–259. [PubMed] [Google Scholar]
- Lin C-J, Li C-Y, Lin S-K, Yang F-H, Huang J-J, Liu Y-H, Lur H-S. 2010. Influence of high temperature during grain filling on the accumulation of storage proteins and grain quality in rice (Oryza sativa L.). Journal of Agricultural and Food Chemistry 58, 10545–10552. [DOI] [PubMed] [Google Scholar]
- Lisle AJ, Martin M, Fitzgerald MA. 2000. Chalky and translucent rice grains differ in starch composition and structure and cooking properties. Cereal Chemistry 77, 627–632. [Google Scholar]
- Liu L, Waters DLE, Rose TJ, Bao J-s, King GJ. 2013. Phospholipids in rice: significance in grain quality and health benefits: a review. Food Chemistry 139, 1133–1145. [DOI] [PubMed] [Google Scholar]
- Liu X, Guo T, Wan X, et al. 2010. Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice. BMC Genomics 11, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wan X, Ma X, Wan J. 2011. Dissecting the genetic basis for the effect of rice chalkiness, amylose content, protein content, and rapid viscosity analyzer profile characteristics on the eating quality of cooked rice using the chromosome segment substitution line population across eight environments. Genome 54, 64–80. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang Y, Wang SW. 2012. QTL analysis of percentage of grains with chalkiness in Japonica rice (Oryza sativa). Genetics and Molecular Research 11, 717–724. [DOI] [PubMed] [Google Scholar]
- Lu D, Shen X, Cai X, Yan F, Lu W, Shi YC. 2014. Effects of heat stress during grain filling on the structure and thermal properties of waxy maize starch. Food Chemistry 143, 313–318. [DOI] [PubMed] [Google Scholar]
- Lyman NB, Jagadish KSV, Nalley LL, Dixon BL, Siebenmorgen T. 2013. Neglecting rice milling yield and quality underestimates economic losses from high-temperature stress. PLoS One 8, e72157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Okazaki Y, Oikawa A, Kusano M, Nakabayashi R, Kikuchi J, Yonemaru J, Ebana K, Yano M, Saito K. 2012. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. The Plant Journal 70, 624–636. [DOI] [PubMed] [Google Scholar]
- Matsushima R, Maekawa M, Kusano M, Kondo H, Fujita N, Kawagoe Y, Sakamoto W. 2014. Amyloplast-localized SUBSTANDARD STARCH GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiology 164, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano H-Y, Suzuki Y, Sano Y. 2008. Allelic diversification at the wx locus in landraces of Asian rice. Theoretical and Applied Genetics 116, 979–989. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M, Prasad M. 2014. Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theoretical and Applied Genetics 128, 1–14. [DOI] [PubMed] [Google Scholar]
- Muthurajan R, Shobbar ZS, Jagadish SV, Bruskiewich R, Ismail A, Leung H, Bennett J. 2011. Physiological and proteomic responses of rice peduncles to drought stress. Molecular Biotechnology 48, 173–182. [DOI] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. 2011. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. The Plant Cell 23, 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Francisco PB, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N. 2005. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Molecular Biology 58, 213–227. [DOI] [PubMed] [Google Scholar]
- Neelin JD, Munnich M, Su H, Meyerson JE, Holloway CE. 2006. Tropical drying trends in global warming models and observations. Proceedings of the National Academy of Sciences, USA 103, 6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JC, McClung AM, Fjellstrom RG, Moldenhauer KA, Boza E, Jodari F, Oard JH, Linscombe S, Scheffler B, Yeater KM. 2011. Mapping QTL main and interaction influences on milling quality in elite US rice germplasm. Theoretical and Applied Genetics 122, 291–309. [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. 2001. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- Nock CJ, Waters DLE, Edwards M, Bowen SG, Rice NF, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnology Journal 9, 328–333. [DOI] [PubMed] [Google Scholar]
- Patindol JA, Siebenmorgen TJ, Wang Y-J, Lanning SB, Counce PA. 2014. Impact of elevated nighttime air temperatures during kernel development on starch properties of field-grown rice. Cereal Chemistry Journal 91, 350–357. [Google Scholar]
- Patindol J, Wang YJ. 2003. Fine structures and physicochemical properties of starches from chalky and translucent rice kernels. Journal of Agricultural and Food Chemistry 51, 2777–2784. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Smith AM, Fahy B, Hylton C, Naldrett MJ, Rossnagel BG, Denyer K. 2002. The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5’-non-coding region. Plant Physiology 130, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Wang L, Fan C, Jiang G, Luo L, Li Y, He Y. 2014a. Comparative mapping of chalkiness components in rice using five populations across two environments. BMC Genetics 15, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Wang Y, Liu F, et al. 2014b. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. The Plant Journal 77, 917–930. [DOI] [PubMed] [Google Scholar]
- Peng S, Huang JL, Sheehy JE, Laza RC, Visperas RM, Zhong XH, Centeno GS, Khush GS, Cassman KG. 2004. Rice yields decline with higher night temperature from global warming. Proceedings of the National Academy of Sciences, USA 101, 9971–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson SRM, Jia YL, Gibbons JW. 2013. Three quantitative trait loci conferring resistance to kernel fissuring in rice identified by selective genotyping in two tropical japonica populations. Crop Science 53, 2434–2443. [Google Scholar]
- Pooni HS, Kumar I, Khush GS. 1992. A comprehensive model for disomically inherited metrical traits expressed in triploid tissues. Heredity 69, 166–174. [Google Scholar]
- Qiao Y, Lee SI, Piao R, Jiang W, Ham TH, Chin JH, Piao Z, Han L, Kang SY, Koh HJ. 2010. Fine mapping and candidate gene analysis of the floury endosperm gene, FLO(a), in rice. Molecular Cells 29, 167–174. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Liu F, et al. 2014. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. The Plant Cell 26, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri A, Rabiei B, Toorchi M, Aharizad S, Moumeni A. 2012. Mapping quantitative trait loci (QTL) associated with cooking quality in rice (Oryza sativa L.). Australian Journal of Crop Science 6, 808–814. [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y. 2003. Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiology 133, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Suzuki Y, Sakai M, Imbe T. 2002. Molecular characterization of Wx mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.). Breeding Science 52, 131–135. [Google Scholar]
- Septiningsih EM, Trijatmiko KR, Moeljopawiro S, McCouch SR. 2003. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon . Theoretical and Applied Genetics 107, 1433–1441. [DOI] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, et al. 2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. The Plant Cell 22, 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M. 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genetics 40, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Siebenmorgen TJ, Grigg BC, Lanning SB. 2013. Impacts of preharvest factors during kernel development on rice quality and functionality. Annual Review of Food Science and Technolology 4, 101–115. [DOI] [PubMed] [Google Scholar]
- Siebenmorgen T, Meullenet J-FC. 2004. Impact of drying, storage, and milling of rice quality and functionality. In: Champagne ET, ed. Rice: chemistry and technology , 3rd edn. AACC International Press, 301–328. [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL. 1997. Influence of gene dosage on carbohydrate synthesis and enzymatic activities in endosperm of starch-deficient mutants of maize. Plant Physiology 113, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X-J, Huang W, Shi M, Zhu M-Z, Lin H-X. 2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Schnurbusch T. 2012. A genetic playground for enhancing grain number in cereals. Trends in Plant Science 17, 91–101. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Wobus U. 2013. Seed-development programs: a systems biology-based comparison between dicots and monocots. Annual Review of Plant Biology 64, 189–217. [DOI] [PubMed] [Google Scholar]
- Sun M-M, Abdula SE, Lee H-J, Cho Y-C, Han L, Koh H-J, Cho Y-G. 2011. Molecular aspect of good eating quality formation in Japonica rice. PLoS One 6, e18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-y, Hao W, Lin H-x. 2006. Identification of QTLs for cooking and eating quality of rice grain. Rice Science 13, 161–169. [Google Scholar]
- Tan YF, Xing YZ, Li JX, Yu SB, Xu CG, Zhang Q. 2000. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theoretical and Applied Genetics 101, 823–829. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Aoki N. 2005. Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Functional Plant Biology 32, 763–768. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Aoki N, Lin H, Nakamura Y, Inouchi N, Sato Y, Yano M, Hirabayashi H, Maruyama S. 2004. Natural variation in rice starch synthase IIa affects enzyme and starch properties. Functional Plant Biology 31, 671–684. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Terashima K, Nakamura Y, Satoh H. 1999. Differences in amylopectin structure between two rice varieties in relation to the effects of temperature during grain-filling. Starch – Stärke 51, 58–62. [Google Scholar]
- Usui Y, Sakai H, Tokida T, Nakamura H, Nakagawa H, Hasegawa T. 2014. Heat-tolerant rice cultivars retain grain appearance quality under free-air CO2 enrichment. Rice (N Y) 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ. 2005. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theoretical and Applied Genetics 110, 1334–1346. [DOI] [PubMed] [Google Scholar]
- Wan XY, Weng JF, Zhai H, Wan JM. 2004. Fine mapping of pgwc8 gene affecting percentage of grains with chalkiness in rice (Oryza sativa. L). Rice Genetics Newsletter 21, 54–56. [Google Scholar]
- Wang E, Wang J, Zhu X, et al. 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nature Genetics 40, 1370–1374. [DOI] [PubMed] [Google Scholar]
- Waters DLE, Henry RJ, Reinke RF, Fitzgerald MA. 2006. Gelatinization temperature of rice explained by polymorphisms in starch synthase . Plant Biotechnology Journal 4, 115–122. [DOI] [PubMed] [Google Scholar]
- Welch JR, Vincent JR, Auffhammer M, Moya PF, Dobermann A, Dawe DC. 2010. Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proceedings of the National Academy of Sciences, USA 107, 14562–14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M-O, Ham T-H, Ji H-S, et al. 2008. Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). The Plant Journal 54, 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Ebitani T, Terao T. 2008. Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breeding Science 58, 337–343. [Google Scholar]
- Yamakawa H, Hakata M. 2010. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant and Cell Physiology 51, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. 2007. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiology 144, 258–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P-R, Kim H-J, Chen Q-H, Ju H-G, Ji S-D, Ahn S-N. 2010. Mapping QTLs for grain quality using an introgression line population from a cross between Oryza sativa and O. rufipogon . Journal of Crop Science and Biotechnology 13, 205–212. [Google Scholar]
- Zhang G, Cheng Z, Zhang X, Guo X, Su N, Jiang L, Mao L, Wan J. 2011. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 54, 448–459. [DOI] [PubMed] [Google Scholar]
- Zhang H, Duan L, Dai JS, Zhang CQ, Li J, Gu M-H, Liu QQ, Zhu Y. 2014. Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency on Wx pre-mRNA. Theoretical and Applied Genetics 127, 273–282. [DOI] [PubMed] [Google Scholar]
- Zhao X, Fitzgerald MA. 2013. Climate change: implications for the yield of edible rice. PLoS One 8, e66218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng TQ, Xu JL, Li ZK, Zhai HQ, Wan JM. 2007. Genomic regions associated with milling quality and grain shape identified in a set of random introgression lines of rice (Oryza sativa L.). Plant Breeding 126, 158–163. [Google Scholar]
- Zhou L, Chen L, Jiang L, et al. 2009. Fine mapping of the grain chalkiness QTL qPGWC-7 in rice (Oryza sativa L.). Theoretical and Applied Genetics 118, 581–590. [DOI] [PubMed] [Google Scholar]
- Zhu J, Weir BS. 1994. Analysis of cytoplasmic and maternal effects. II. Genetic models for triploid endosperms. Theoretical and Applied Genetics 89, 160–166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.